Abstract
Gram-negative bacteria, such as Escherichia coli, utilize efflux resistance systems in order to expel toxins from their cells. Heavy-metal resistance is mediated by resistance nodulation cell division (RND)-based efflux pumps composed of a tripartite complex that includes an RND-transporter, an outer-membrane factor (OMF), and a membrane fusion protein (MFP) that spans the periplasmic space. MFPs are necessary for complex assembly and have been hypothesized to play an active role in substrate efflux. Crystal structures of MFPs are available, however incomplete, as large portions of the apparently disordered N and C termini are unresolved. Such is the case for CusB, the MFP of the E. coli Cu(I)/Ag(I) efflux pump, CusCFBA. In this work, we have investigated the structure and function of the N-terminal region of CusB, which includes the metal-binding site and is missing from previously determined crystal structures. Results from mass spectrometry and X-ray absorption spectroscopy show that the isolated N-terminal 61 residues (CusB-NT) bind metal in a 1:1 stoichiometry with a coordination site composed of M21, M36, and M38, consistent with full-length CusB. NMR spectra show that CusB-NT is mostly disordered in the apo state; however, some slight structure is adopted upon metal binding. Much of the intact protein’s function is maintained in this fragment as CusB-NT binds metal in vivo and in vitro, and metal is transferred between the metallochaperone CusF and CusB-NT in vitro. Functional analysis in vivo shows that full-length CusB is necessary in an intact polypeptide for full metal resistance, though CusB-NT alone can contribute partial metal resistance. These findings reinforce the theory that the role of CusB is not only to bind metal, but also to play an active role in efflux.
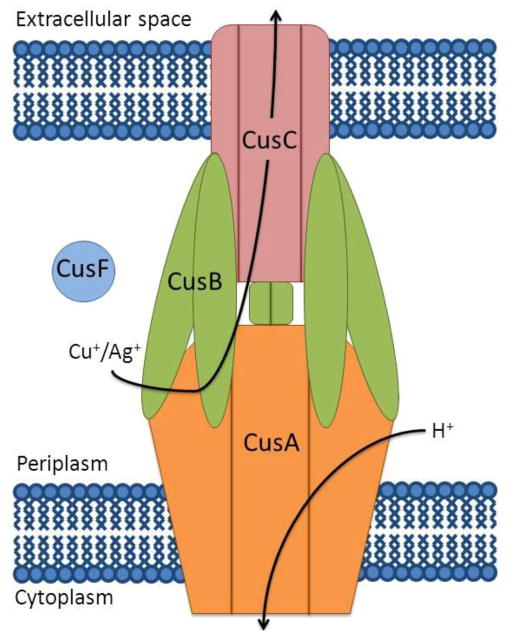
Transition metal homeostasis is of great importance in biology. While transition metals are essential for many biological functions in the cell and life would not be possible without them, they can cause cellular damage and eventually cell death if their concentrations are not properly regulated (1, 2). In the Gram-negative bacterium, Escherichia coli, copper and silver ions are extruded from the cell by the metal efflux pump, CusCFBA, whose corresponding operon is up-regulated in response to elevated metal levels in the periplasm (3). CusCBA is believed to form a tripartite complex (Figure 1) that spans the periplasmic space between the inner and outer membranes, similar to the assembly of tripartite multidrug efflux systems (4, 5). However, unlike the multidrug efflux pumps, which demonstrate broad substrate specificity, CusCFBA is highly specific for Cu(I)/Ag(I) (6–8).
Figure 1.
Schematic of the periplasmic Cu(I)/Ag(I) efflux pump, CusCFBA: trimeric RND-transporter CusA (orange), trimeric OMF CusC (pink), MFP CusB (green) and metallochaperone CusF (blue).
Studies of the individual components of the Cus system have revealed important structural and functional details (reviewed in (9)). CusF, the novel fourth component of the Cus system, is a small soluble periplasmic metallochaperone that is essential for maximal Cus metal resistance (3). Metal transfer occurs between CusF and the membrane fusion protein (MFP), CusB (10). CusB interacts with both CusA and CusC, most likely stabilizing the intermembrane tripartite complex, similar to AcrA in the well-characterized multidrug efflux pump AcrAB-TolC (5, 11–13). CusB and CusF transiently interact in a metal-dependent manner and distribute metal approximately equally between the two proteins when mixed in vitro in equimolar concentrations (10), representative of their similar metal-binding affinities (7, 14). The inner membrane transporter protein, CusA, belongs to the resistance nodulation cell division (RND) superfamily of proteins and utilizes the proton motive force to drive substrate efflux (15). Over the last few years, crystal structures of CusB (13), CusA (16), and the outer-membrane factor (OMF) protein, CusC (17), have been solved individually, along with a co-crystal structure of CusBA (18), suggesting a ratio of 3:6:3 for the intact CusCBA complex. Evidence for the same stoichiometry within the MtrCDE multidrug efflux complex has recently been demonstrated and supports this hypothesis (19). While these structures provide a framework for our understanding of the stoichiometry and orientation of the components of the pump upon assembly, they do not elucidate the dynamic mechanism of metal efflux nor do they reveal the specific protein-protein interactions necessary for metal transfer within the pump.
CusB belongs to a protein family termed the membrane fusion protein (MFP) family (20–22). While many MFPs contain a single N-terminal transmembrane α-helix (N-TMS) (23) or lipid modification embedding them in the inner membrane (24), CusB demonstrates no association with either membrane, as it lacks the N-TMS motif and is soluble during purification (14). CusB plays a central structural role in the Cus complex by linking the inner and outer membrane proteins and is essential for metal resistance (3). The crystal structure of CusB shows four domains (25): the membrane proximal (MP), β-barrel, lipoyl, and α-helical domains. However, the N-terminal region of CusB is missing from both crystal structures (13, 18). This region of the protein, CusB-NT, consisting of the first 61 amino acids of the mature protein, is of great importance because it includes the three conserved Met residues (M21, M36, and M38) determined to comprise the metal-binding site of CusB (14). Previous studies showed that mutation of any one of these methionines to isoleucine in the full-length protein resulted in loss of metal binding in vitro and a loss of metal resistance in vivo (14). Chemical cross-linking/mass spectrometry experiments have determined that CusB and CusF interact at their metal-binding regions (26), implicating CusB-NT as the important region for protein-protein interaction and metal transfer.
In this manuscript, we describe experiments performed to probe the structure and function of this essential N-terminal region of CusB. In vitro experiments, including nuclear magnetic resonance (NMR) spectroscopy, native mass spectrometry, and X-ray absorption spectroscopy (XAS), were used to investigate metal binding, metal transfer, and structural changes upon metal binding. Native mass spectrometry has become a powerful technique to study intact proteins and other large biological molecules (27). With the gentle ionization conditions of nanoelectrospray (nanoESI) (28), the disruption of native protein structures is minimized during transfer of proteins from solution into the gas phase (29) and therefore this technique is effective for the study of protein-ligand binding (30–36). In vivo function was examined through bacterial growth on copper-containing plates.
Materials and Methods
Plasmid construction for expression of CusB-NT
The N-terminal sequence of CusB from Escherichia coli, residues Glu1-Thr61 (using the numbering without the leader sequence, hereafter referred to as CusB-NT) was PCR amplified using taq DNA polymerase (Fermentas) from a plasmid containing the gene of the full-length CusB sequence using the nucleotide primer pair: 5′AAAAAGGATCCGGGGAAAACCTGTATTTTCAGGGCGAACCGCCTGCAGAAAAAACGTCG3′ and 5′AAAAAGAATTCTCACGTCTGAGTCGGGTCAATGC3′ (37). The forward primer was designed to include a region encoding for the Tobacco Etch Virus (TEV) protease cut site (ENLYFQG), which, once cleaved, leaves an N-terminal Gly residue. The resulting fragment was purified using the QIAquick PCR purification kit (Qiagen) prior to digestion with BamHI and EcoRI (Fermentas). This insert was ligated into the pGEX-2TK vector (GE Healthcare) which introduces a glutathione S-transferase (GST) tag at the N-terminus of the protein. The cloned gene was sequenced and verified for accuracy (The University of Michigan DNA Sequencing Core). The resulting plasmid pGEX_CusB-NT was transformed into E. coli BL21-(λDE3) cells (Invitrogen). Mutation of M36 to isoleucine in pGEX_CusB-NT was performed using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene).
Cell growth and protein purification
For preparation of CusB-NT, E. coli BL21-(λDE3) cells containing pGEX_CusB-NT were grown in LB media containing 100 μg/mL ampicillin at 37 °C until they reached an O.D.600 of 0.8–1.0, then induced with 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG). Growth was continued at 37 °C for 4–5 hours before the cells were harvested by centrifugation and frozen. For preparation of uniformly 15N–labeled CusB-NT, E. coli BL21-(λDE3) cells containing pGEX_CusB-NT were grown in M9 minimal media (38) containing 1.0 g/L 15NH4Cl (Cambridge Isotopes Laboratories) as the sole nitrogen source. Cells were grown in 20 ml of Luria Bertani (39) media overnight, then centrifuged and transferred to 1 L of M9 media and grown at 37 °C until they reached an O.D.600 of 0.8–1.0, then induced with 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG). Growth was continued at 37 °C for 4–5 hours. The purification protocol was the same for unlabeled and 15N-labeled CusB-NT and was as follows. Pelleted cells were resuspended in 10 mL/g of PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3). Lysozyme (final concentration 12.5 μg/mL), phenylmethyl-sulfonyl fluoride (final concentration 0.5 mM), leupeptin and pepstatin (final concentration 2 μg/mL), and DNAse (~150 units) were added and cells were lysed using a French press. The lysate was centrifuged at 31000g for 1 hour to remove cell debris and the supernatant was loaded onto a GST resin (GE Healthcare) affinity column, equilibrated with PBS. After washing with PBS, the CusB-NT fusion protein was eluted with 10 mM reduced glutathione in 50 mM Tris, 5 mM DTT, pH 8.0. Fractions containing the fusion protein were combined and TEV protease was added (1 mg protease:10 mg substrate) along with EDTA (final concentration, 0.5 mM). TEV proteolysis of the fusion protein was performed at room temperature for 2 hours without agitation. The resulting mixture was concentrated using a 3-kDa molecular weight cutoff concentrator (Amicon). CusB-NT was separated from the GST-tag and TEV protease using an S100 gel filtration column (GE Healthcare) equilibrated with the appropriate buffer, depending on the experiment. Aliquots of the fractions were run on SDS-polyacrylamide gels and stained with Coomassie. Fractions judged to be >95% pure were pooled and concentrated. Protein concentrations were determined using the Bradford assay (40) (Bio-Rad). The intact molecular weight of the protein was confirmed using mass spectrometry. CusB-NT M36I was purified as described for the wild-type CusB-NT. CusF was grown and purified as previously described (8). Ubiquitin for mass spectrometry was obtained from Sigma-Aldrich (St. Louis, MO).
Circular dichroism spectroscopy
Circular dichroism spectra of 28 μM CusB-NT in 50 mM Tris buffer, pH 8.0, were acquired on an Olis DSM 20 CD Spectrophotometer with a path length of 1 mm. For the Ag(I)-CusB-NT spectrum, a two-fold molar excess of AgNO3 was added. For both spectra, the instrument was set to scan between 200–260 nm in 60 increments with an integration time of 8 sec.
NMR analysis
15N-labeled CusB-NT was expressed in minimal media containing 15NH4Cl as the sole nitrogen source and purified as described above. Samples were concentrated to ~1 mM in 20 mM MES pH 5.7. 1H-15N Heteronuclear Single Quantum Coherence (HSQC) spectra were acquired at 298 K on a 600 MHz Varian Inova instrument using pulse sequences from Varian Biopack of the following samples: (i) apo-CusB-NT and (ii) Ag(I)-CusB-NT (1:1 molar amount of AgNO3 added to the apo sample). All samples contained 10% D2O and 0.02% NaN3. Spectra were processed with NMRPIPE (41) and analyzed with NMRView (42).
Mass spectrometry
All protein samples were buffer exchanged into 100 mM ammonium acetate (pH 7) using size exclusion chromatography spin columns (BioRad) before analysis. Non-specific binding of excess ligand, which is a common problem in the analysis of protein-ligand binding by electrospray mass spectrometry, was minimized by removing excess silver through multiple stages of dialysis prior to MS analysis.
Mass spectra were acquired on a Synapt G2 quadrupole/time-of-flight mass spectrometer (Waters Corporation, Manchester, UK). 3–5 μL protein solution was injected each time into a glass capillary, and a platinum wire was inserted into the capillary to apply high voltage (1–1.5 kilovolts) for nanoelectrospray. The source temperature was kept at room temperature without any desolvation gas. All the conditions in the mass spectrometer were tuned to minimize activation and disruption of the non-covalent interactions in the protein samples. Backing pressure was set to 4.4 mbar. The pressure in the time-of-flight mass analyzer was at 5×10−7 mbar. Mass calibration was performed with cesium iodide clusters in m/z range 50–5000. Data were analyzed with MassLynx v4.1 without further smoothing. The spectra were interpreted manually. Abundances of apo/Ag(I) species were obtained by integrating the identified protein peaks. For simplicity, none of the buffer adducts (non-specific addition of sodium, potassium, HEPES, etc.) were taken into consideration.
X-ray absorption spectroscopy sample preparation
EXAFS samples were prepared in an anaerobic chamber. CuCl2 was added to CusB-NT in 50 mM Tris, pH 8.0 (1:1 metal:protein concentration) in the presence of 50 mM ascorbate buffered at pH 8.0. 75 μL of Cu(I)-CusB-NT was mixed with 25 μL of ethylene glycol, transferred to an EXAFS vial, and flash-frozen in liquid nitrogen. The final protein concentration was ~500 μM.
XAS data collection and analysis
Cu K-edge (8.9 keV) extended X-ray absorption fine structure (EXAFS) and X-ray absorption near edge structure (XANES) data were collected at the Stanford Synchrotron Radiation Lightsource operating at 3 GeV and 300 mA under continuous top-up mode on beam line (BL) 7-3 using a Si[220] monochromator and a Rh-coated mirror upstream of the monochromator with a 13 keV energy cutoff to reject harmonics. Data were collected in fluorescence mode using a high-count-rate Canberra 30-element Ge array detector with maximum count rates below 120 kHz. A Z-1 Ni oxide filter and Soller slit assembly were placed in front of the detector to reduce the elastic scatter peak. Energy calibration was achieved by reference to the first inflection point of a copper foil (8980.3 eV) placed between the second and third ionization chamber. The samples (80 μL) were measured as aqueous glasses (> 20% ethylene glycol) at 10 – 15 K. Six scans of a sample containing only sample buffer were collected, averaged and subtracted from the averaged data for the protein samples to remove Z-1 Kβ fluorescence and produce a flat pre-edge baseline. Data reduction and background subtraction were performed using the program modules of EXAFSPAK (43). Data from each detector channel were inspected for glitches, drop-outs, or other nonlinear behavior before inclusion in the final average. Spectral simulation was carried out using the program EXCURVE 9.2 (43–46) as previously described (14, 47, 48). The parameters refined in the fit included shell occupancy N, Cu-scatterer distance R, and Debye-Waller factor (2σ2) for each shell, and the threshold energy for photoelectron ionization (E0), which was constrained to be the same for all shell of scatterers.
Constructs for complementation assay
The pETDuet-1 (Novagen) vector was used for co-expression of two constructs. A schematic illustration of the eight constructs is provided in Figure S1. To make the constructs expressing full-length CusB, the cusB gene (including the region encoding the signaling peptide, which targets the protein to the periplasm) was amplified from a plasmid containing the gene of the full-length CusB sequence and cloned into the multiple cloning site 1 (MCS1) of pETDuet-1 using the restriction enzymes BamHI and HindIII (Fermentas). To generate a truncated CusB lacking the N-terminus, the full-length cusB gene in MCS1 was used as a template for site-directed mutagenesis (QuikChange Kit, Stratagene) to remove the region of the gene encoding for the protein sequence CusB-NT (residues Glu1-Thr61) while keeping the signaling peptide, creating the construct referred to as cusBΔNT. Constructs expressing CusB-NT (Glu1-Thr61) (including the signaling peptide) were created by PCR amplifying this region of the gene and cloning into the MCS2 of the plasmid containing either full-length cusB, cusBΔNT, or no gene inserted in MCS1, using the restriction enzymes NdeI and XhoI (Fermentas). Constructs in which methionine 36 is replaced by isoleucine were generated by site-directed mutagenesis. The DNA sequences of all constructs were verified for accuracy (University of Arizona Genetics Core). These constructs were each transformed into E. coli strain EC950-(λDE3). The strain EC950, which contains a ΔcusBΔcueO background, was chosen for the integration of λDE3 prophage into the chromosome using a λDE3 lysogenization kit (Novagen). Appropriate induction of each plasmid was tested by growing cells in 5 mL LB media containing 100 μg/ml ampicillin at 37 °C until they reached an O.D.600 of 0.8–1.0, then each was induced with 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG). Growth was continued at 37 °C for 4 hours before performing a whole-cell protein preparation of 1 ml of each culture. Cell concentrations were normalized, then induction of CusB proteins of the appropriate size for each construct was verified by Western Blot analysis using anti-CusB antibodies (Figure S2).
Functional analysis of growth on copper plates
LB agar plates were prepared containing 100 μg/mL ampicillin, 1 mM IPTG and a range of CuSO4 concentrations (0–2 mM). 5 ml cultures of EC950-(λDE3) E. coli cells containing each construct were grown in LB medium at 37 °C to an O.D.600 of ~0.5, normalized, streaked on plates and placed in a 37 °C incubator for 24 hours. Growth was observed and allowed to continue for up to 24 hours more at room temperature. Cell growth was scored on a scale of 0–4, with 0 representing no growth and 4 representing the most growth. Two individuals scored the plates independently and the analysis was performed three times for the same set of plates.
Results
CusB-NT is mostly disordered but adopts some structure upon metal binding
To examine the function of the N-terminal region of CusB, this portion of the protein (CusB-NT) was expressed and purified for in vitro studies. In order to probe the secondary structure of CusB-NT, circular dichroism spectra were obtained of apo-CusB-NT and Ag(I)-CusB-NT (Figure S3). The resulting spectra match well to that predicted for a random coil (49). Upon the addition of metal, no significant change was observed by CD spectroscopy.
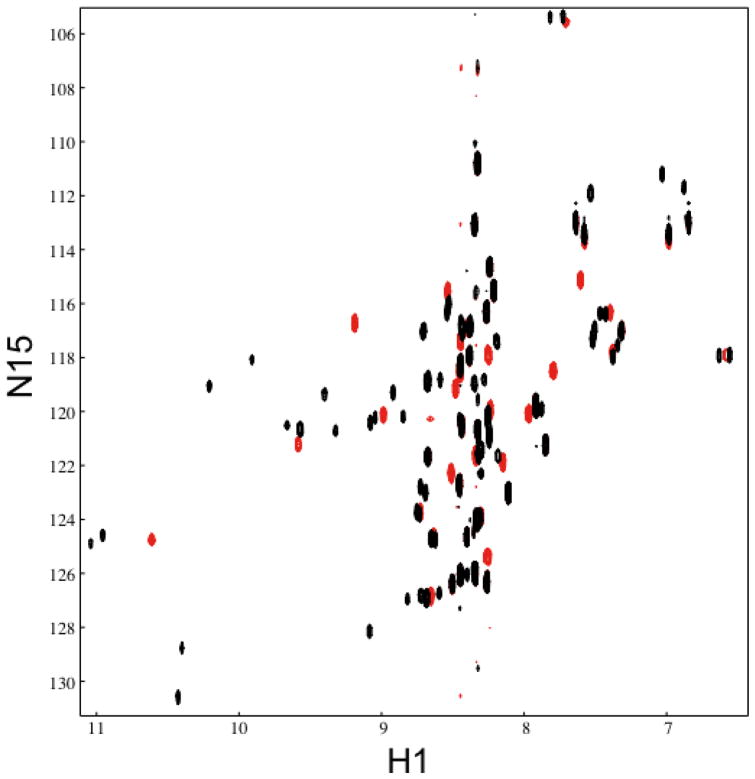
To further investigate the structure of CusB-NT, NMR 1H-15N correlation (HSQC) spectra were collected of apo-CusB-NT and Ag(I)-CusB-NT (Figure 2). The lack of dispersion in the peaks of the apo-CusB-NT HSQC spectrum is indicative of a mostly disordered protein, consistent with the CD results. Upon the addition of metal, however, a significant number of peaks shift and, in general, the peaks become slightly more disperse. More peaks are observed in the Ag(I)-CusB-NT spectrum compared to the apo-CusB-NT spectrum, which may be due to multiple conformations in the Ag(I)-bound state. In the Ag(I)-CusB-NT spectrum, approximately 72 peaks can be counted (excluding probable peaks arising from side chain NH2 groups), though only 54 non-proline peaks are expected.
Figure 2.
Overlay of NMR 1H-15N correlation (HSQC) spectra of apo-CusB-NT (red) and Ag(I)-CusB-NT (black).
CusB-NT binds metal ion with a 1:1 stoichiometry
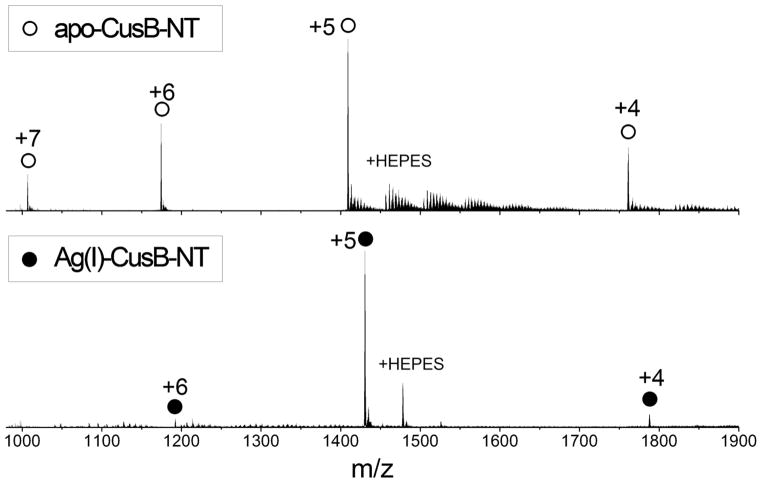
Mass spectrometry was used to investigate the metal-binding properties of CusB-NT. The spectra of apo-CusB-NT and Ag(I)-CusB-NT are displayed in Figure 3. All the peaks have isotopic resolution and can be easily assigned based on the experimental masses calculated from the charge state and measured m/z ratios. In the apo-CusB-NT spectrum, the deconvoluted isotopic molecular weight is measured to be 7036.6 Da. The deconvoluted isotopic molecular weight from the Ag(I)-CusB-NT spectrum is determined to be 7143.6 Da. This mass is 107 Da larger than the measured molecular weight of apo-CusB-NT, which matches the mass of one Ag(I) atom. The peak at m/z 1478, next to the strongest peak (m/z 1430) in the Ag(I)-CusB-NT spectrum, is of the appropriate mass for CusB-NT binding one Ag(I) and one HEPES, suggesting that the experimental conditions are gentle enough to preserve non-covalent interactions in the gas phase. No peaks indicative of secondary Ag(I) binding were observed in the spectrum.
Figure 3.
NanoESI mass spectra of 40 μM apo-CusB-NT (top) and 40 μM Ag(I)-CusB-NT (bottom). Peaks corresponding to apo-CusB-NT and Ag(I)-CusB-NT are labeled with open and filled circles, respectively, along with their corresponding charge states.
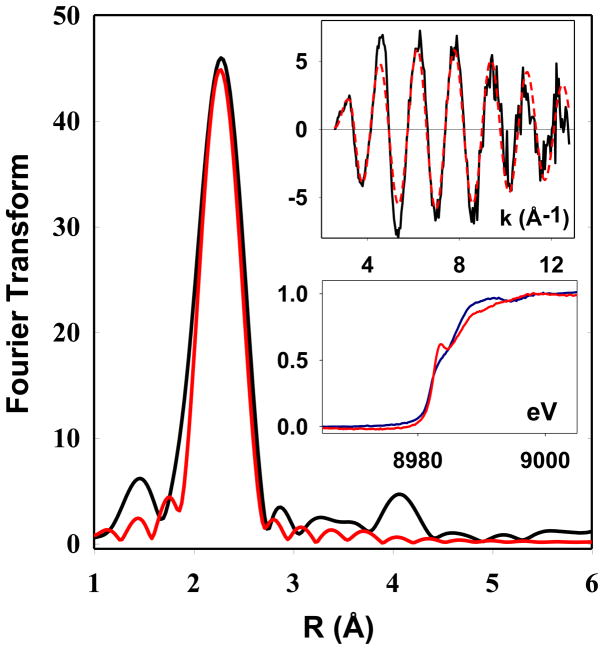
The Cu(I) center of CusB-NT was studied by X-ray absorption spectroscopy. Figure 4 shows the Fourier transform and EXAFS (top insert) for Cu(I)-CusB-NT. In simulations, the best fit to the experimental data was obtained using three sulfur atoms with Cu-S bond lengths of 2.29 Å. The lower insert in Figure 4 compares the absorption edges of Cu(I)-CusB-NT (blue trace) with the full-length Cu(I)-CusB protein (red trace).
Figure 4.
X-ray absorption spectroscopy of CusB-NT. Main panel shows the Fourier transform with the EXAFS plotted in the top insert. Black traces are experimental data and red traces are simulated data. The bottom insert shows a comparison of CusB-NT (blue) with the full-length protein (red). The Cu-S(Met) bond length determined from simulation is 2.29 Å for CusB-NT.
Metal transfer occurs between CusF and CusB-NT
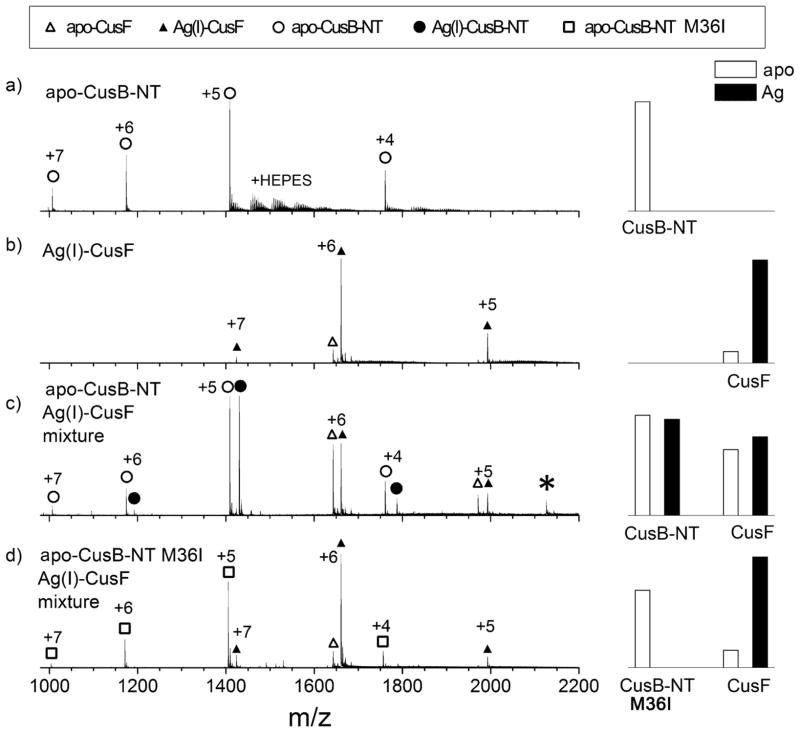
Mass spectrometry was utilized to monitor metal transfer between CusB-NT and CusF. In the starting samples, apo-CusB-NT is free of Ag(I) before mixing (monoisotopic molecular weight is measured to be 7036.7 Da) (Figure 5a), and the majority of CusF is bound to Ag(I) in the Ag(I)-CusF sample before mixing (monoisotopic molecular weight is measured to be 9954.3 Da) (Figure 5b). When Ag(I)-CusF and apo-CusB-NT are mixed at 1:1 molar ratio in solution, then analyzed by mass spectrometry, additional peaks are present in the spectrum (Figure 5c). The monoisotopic molecular weights of these peaks, calculated from the charge and m/z measured in the spectrum, are 9847.5 Da and 7143.6 Da, which are appropriate for apo-CusF and Ag(I)-CusB-NT, respectively. As a control for non-specific loss of Ag(I) from Ag(I)-CusF, a mutated CusB-NT with one of the methionine residues altered to isoleucine (M36I) was mixed with Ag(I)-CusF. This spectrum does not show peaks corresponding to apo-CusF or Ag(I)-bound CusB-NT-M36I (Figure 5d). As further controls, ubiquitin and cytochrome c were separately mixed with Ag(I)-CusF and no loss of silver from Ag(I)-CusF was observed (Figure S4).
Figure 5.
NanoESI mass spectra of (a) apo-CusB-NT at 40 μM, (b) Ag(I)-CusF at 40 μM, (c) 1:1 mixture of apo-CusB-NT and Ag(I)-CusF, both at 40 μM, and (d) 1:1 mixture of Apo-CusB-NT-M36I and Ag(I)-CusF. Apo-CusB-NT and Ag(I)-CusB-NT are labeled with open and filled circles respectively. Apo-CusF and Ag(I)-CusF are represented with open and filled triangles respectively. Apo-CusB-NT-M36I is labeled with open squares. Relative abundances of the apo vs. Ag(I)-bound proteins after deconvolution are shown as bar graphs in the column on the right for corresponding mass spectra on the left. The asterisk at m/z 2125 in (c) corresponds to the Ag(I)/CusF/CusB complex.
A small fraction of a Ag(I)/CusF/CusB-NT complex is formed in the mixture of Ag(I)-CusF with wild-type apo-CusB-NT, which is apparent by a minor peak at m/z 2125 with a charge state of +8 (Figure 5c). However, in the mixture of CusB-NT-M36I and Ag(I)-CusF, no peak indicative of a complex is observed (Figure 5d).
Deconvolution of the apo and Ag(I)-bound protein peaks allows an estimation of the abundance of each species, as illustrated in the bar graphs next to the spectra (Figure 5). The relative abundances of the apo and Ag(I)-bound forms of CusF and CusB-NT suggest that the metal distributes approximately equally between the two proteins (Figure 5c). Spectra of mixtures of 2:1 apo-CusB-NT and Ag(I)-CusF or 1:2 apo-CusB-NT and Ag(I)-CusF show distribution of metal between the two proteins, and both apo and Ag(I)-bound peaks for both proteins are apparent in the spectra (Figure S5). In each case, the available metal ion also distributes approximately equally between the two proteins.
CusB functional analysis in vivo
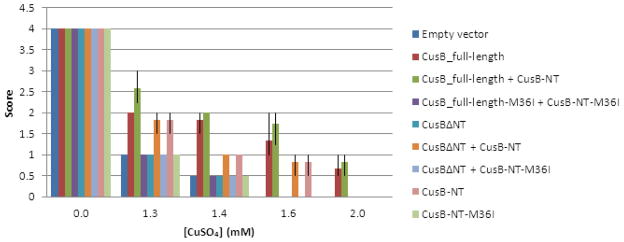
To investigate metal binding and functional metal resistance by CusB in vivo, E. coli cells expressing various CusB constructs were evaluated for survival on copper-containing media (Figure 6). These experiments were performed in cells in which the chromosomal genes for cusB and the multicopper oxidase cueO have been deleted. Cells were transformed with plasmids containing genes that encode CusB-NT, full-length CusB, CusB with the N-terminal region deleted (CusBΔNT), and dual combinations of these proteins. Analysis of E. coli growth showed that cells containing CusBΔNT grew the same as cells without CusB (empty vector) and did not grow at higher copper concentrations (above 1.6 mM CuSO4). Cells co-expressing both full-length CusB and CusB-NT showed slightly better growth than those containing full-length CusB alone. Cells containing both of these constructs (full-length CusB and full-length CusB plus CusB-NT) grew at the highest copper concentration evaluated (2 mM CuSO4). Interestingly, cells dually expressing both CusB in which the N-terminal region was deleted (CusBΔNT) and CusB-NT grew in a similar manner as those cells that only express CusB-NT, and in both cases these cells grew to a copper concentration of 1.8 mM. This copper concentration is higher than that of cells containing empty vector or CusBΔNT, but lower than that of cells containing full-length CusB alone or full-length CusB plus CusB-NT. All constructs in which one of the metal-binding methionines of CusB was mutated to isoleucine (M36I) exhibited the same growth as the empty vector control. Specifically, no increased copper tolerance is achieved in cells containing CusB-NT constructs with M36I mutations.
Figure 6.
Functional analysis of E. coli cells expressing different CusB constructs grown on copper-containing LB-agar plates. Copper sulfate concentrations in the media are given on the x-axis and relative growth is scored on the y-axis.
Discussion
N-terminal metal-binding site of CusB
Previous work proposed that the metal-binding site of CusB consists of three methionine residues (M21, M36, and M38) (14), all of which are contained within the N-terminal region of CusB, which is the region that interacts with CusF (26). However, further efforts to understand the structure and function of this region of CusB have been hampered by its absence from the crystal structures of CusB. Therefore, we sought to characterize this region in isolation. Our results show that the metal-binding function of CusB is maintained in CusB-NT. We have shown by mass spectrometry that CusB-NT binds metal with a 1:1 stoichiometry. Furthermore, the CusB-NT metal coordination site identified by EXAFS consists of three Cu(I)-S bonds. Because there are only three Met residues and no Cys residues in CusB-NT, the three S atoms of M21, M36, and M38 are coordinating the metal. Since the MS data show that CusB-NT-M36I was unable to acquire silver from Ag(I)-CusF, this finding further supports the identification of M36I as involved in metal binding. The Cu-S bond lengths of 2.29 Å match well with those reported previously for full-length CusB (14). However, small differences exist in the absorption edges suggesting that the N-terminal fragment may have subtle differences in coordination geometry of the Cu(I) bound at the Met triad. We note that the crystal structures of CusB and of the CusA/CusB complex reveal four different conformations of the full-length CusB molecule (13, 18), although the N-terminal Cu(I) binding site is missing from these structures. Therefore, it is possible that CusB-NT may be in a different conformational state to that measured previously (14). However, on the whole, metal binding by CusB-NT is consistent with metal binding by the full-length protein and consists of the same three Met residues.
Metal-binding induces slight structural alterations in CusB-NT
Apo-CusB-NT is mostly disordered, as demonstrated by circular dichroism and NMR. This finding is not surprising, as this region of CusB has been predicted to be disordered and is missing from two previously determined crystal structures of full-length CusB (60 residues in the CusB structure and 51 residues in the CusBA co-crystal structure), along with the C terminus (22 residues in the CusB structure and 7 residues in the CusBA co-crystal structure) (13, 18). Similarly, early crystal structures of multidrug efflux MFPs, such as MexA and MacA, are also missing their N and C termini (50–52); the apparent disorder of these portions of MFPs suggests that these regions are flexible and dynamic in nature. However, our NMR experiments demonstrate that CusB-NT adopts some structure upon metal binding, as evidenced by the moderately increased dispersion in the Ag(I)-CusB-NT spectrum. The NMR spectral changes may reflect ordering of specific metal-binding residues, though more extensive changes are unlikely.
ZneB, another MFP of the heavy-metal efflux subclass of RND-driven efflux systems from C. metallidurans CH34, has been structurally characterized in the apo and Zn2+-bound forms. The ZneB metal coordination site is located at the flexible interface between the β-barrel and membrane proximal (MP) domains, which is in a different region than the metal binding site in CusB. However, the MP domain (composed of the N-/C-terminal regions of this protein) is only visible in the metal-bound form and is unresolved in the apo form of the crystal structure (33). Furthermore, the structures of apo and holo ZneB, observed in the same crystal, adopt different conformations. The apo form is arranged in a more linear, extended shape while the holo form bends to adopt a crescent shape upon metal binding. Metal-induced conformational changes have also been observed in full-length CusB by analytical gel filtration, which suggest conformational flexibility (14). Structural changes upon metal-binding as observed here in the HSQC spectra of CusB-NT could potentially contribute to this conformational change.
Metal transfer between CusB-NT and CusF
The native mass spectrometry experiments show that CusB-NT can acquire metal from CusF. Based on the estimation of the abundance of each species in the protein mixtures (with some possible error due to different ionization efficiencies or inexact protein concentrations), the Ag(I) ions partition approximately equally between the two proteins. This result is similar to what has been observed in the full-length proteins when they are in equimolar concentrations (10), which is expected for proteins with similar affinities. This finding supports the proposal that the N-terminal region of CusB binds metal ions independently of the rest of CusB, since the affinity and metal transfer ability are identical to that of the full-length protein.
In the native MS spectra of the protein mixture, a small peak at m/z 2125 (charge state +8) was detected, which is appropriate for a complex of Ag(I)/CusF/CusB-NT. The observation of a minor fraction of Ag(I)/CusF/CusB-NT complex is consistent with previous studies of the full-length proteins which only weakly interact (10, 26). Metal ions are crucial for the interactions between CusF and CusB, since previous studies have shown that the apo proteins do not interact, and they also do not interact when both are fully bound to metal ions (10). Alteration of methionine 36 to isoleucine in the full-length protein removes its ability to bind metal ions (10). In our studies, a mutant of CusB-NT (CusB-NT-M36I), which lacks one of the metal binding residues, neither acquires Ag(I) from CusF nor forms a complex with CusF, since peaks at the m/z appropriate for Ag(I)-CusB-NT-M36I or a ternary complex are not observed in the mixture of CusB-NT-M36I and Ag(I)-CusF. Thus in addition to its metal binding properties, the N-terminal portion of CusB also appears to behave identically to the full-length protein in interactions with CusF.
Comparison of N-terminal regions of MFPs
While the four domains of CusB observed in the crystal structure (25) are conserved among all characterized MFPs, MFPs show variability in their N- and C-terminal regions adjacent to the MP domain. The N termini of all MFPs are unresolved (presumably disordered) and therefore missing from their corresponding crystal structures. In AcrA, MexA, and ZneB, the missing portion consists of 9–13 residues; however, in CusB, the N-terminal portion adjacent to the MP domain consists of 60 residues (CusB-NT), enough to constitute an additional domain and it is conserved in putative CusB homologs. We hypothesize that the N-terminal region of CusB is an additional domain that is unique to monovalent metal efflux systems. This domain contains a metal binding function, and likely plays a role in selectivity for specific metals or regulation of CusCBA activity. Promiscuous multidrug transporters which show little substrate specificity may not need this additional substrate binding site.
The role of CusB
How CusB contributes to metal resistance is still unknown. Previous work suggests that metal binding by CusB is essential for function and that metal binding induces a conformational change (14), potentially altering CusA and/or CusC and activating the efflux pump. These findings have suggested that CusB plays an active role in metal transport. Two models have been proposed which incorporate these findings (53). In one model, the funnel model, it is proposed that metal is transferred to the periplasmic cleft of the inner membrane protein CusA from the CusB metal-binding site and then is subsequently exported from the cells. Because the N-terminal region of CusB, including the metal-binding site, is missing from the co-crystal structures of CusBA, it is difficult to hypothesize if metal transfer from CusB to CusA is feasible (18). The N-terminal tails of both CusB molecules are located outside the periplasmic cleft between subdomains PC1 and PC2 of CusA; however, the tail of molecule 1 appears to be closer and potentially more accessible to the metal-binding site of CusA. Interestingly, previous in vitro chemical cross-linking between CusA and CusB resulted in the identification of one cross-link between K67 of CusB and K150 of CusA with the 11.4 Å cross-linker DSS (13). Examination of CusBA together with the cross-linking distance constraint suggests the cross-link is between molecule 1 of CusB and CusA. Thus, the N-terminal region of at least one of the CusB molecules could be localized near the metal-binding site of CusA and possibly funnel metal to the antiporter for extrusion. Previous structure determination of the MP domain of MexA together with in vivo cross-linking of AcrA and AcrB have been successful in mapping the surface contacts between the MFP and RND-transporter in these multidrug systems. These results show that the MP domain of the MFP directly interacts with the antiporter subdomains PN2 and PC1 (4). Furthermore, modeling of the MexA/MexB interaction localizes the MP domain of MexA near the MexB substrate binding pocket and the flexible MP domain has been hypothesized to release substrate to MexB.
The other model, the switch model, proposes that metal binding to CusB is associated with a regulatory function that induces an active conformation in the Cus efflux pump to allow metals to be exported. However, in this model, the metal bound by CusB is not directly transported out of the cell, but must enter CusA through a different path. The 2:1 stoichiometry of CusBA may support this model. A regulatory role has been hypothesized for the zinc binding MFP ZneB, whose unique metal binding site between the MP and β-barrel domains is not predicted to be within the periplasmic cleft of ZneA, suggesting that metal transfer is unlikely (33).
In the N-terminal CusB construct (CusB-NT) described here, we have been able to uncouple the metal binding and complex assembly functions of CusB. We have found that while CusB-NT alone has some contribution to metal resistance, it is not fully functional. This finding rules out the possibility that the function of CusB is solely to act as a metal chelator, since metal binding is retained in the CusB-NT construct. When CusB-NT is expressed independently along with the remainder of the CusB protein, the resistance is the same as when CusB-NT is expressed alone. This finding supports the idea that metal binding triggers a functional conformational change in full-length CusB, which cannot be replicated when the components are not part of an intact polypeptide.
The residual metal resistance that is provided by CusB-NT alone is likely a result of metal chelation by this fragment. This interpretation is corroborated by the observation that metal resistance increases when CusB-NT is expressed in addition to full-length CusB, beyond the resistance level seen for full-length CusB alone. Furthermore, in any of the constructs in which one of the metal-binding methionines is altered to isoleucine (M36I), the copper tolerance is the same as the empty vector control. This finding indicates that metal-binding is functionally important in providing resistance by this region. Another interpretation of the increased metal resistance observed when CusB-NT and full-length CusB are expressed together may be because the efflux system remains “on” longer if metal is not removed from full-length CusB in the Cus complex. If CusB functions as a regulator, the system is postulated to turn off upon metal dissociation from CusB due to release to the periplasm or transfer to CusF. However, since metal transfer can occur between CusB-NT alone and CusF, as demonstrated here by mass spectrometry and EXAFS, this may eliminate much of this route of metal removal from full-length CusB when CusB-NT is present.
Supplementary Material
Acknowledgments
We thank Dr. Christian Roessler for assistance with NMR analysis and Dr. Eun-Hae Kim for thoughtful scientific discussions.
Abbreviations
- NMR
nuclear magnetic resonance
- RND
resistance nodulation cell division
- OMF
outer membrane factor
- MFP
membrane fusion protein
- N-TMS
N-terminal transmembrane segment
- XAS
X-ray absorption spectroscopy
- EXAFS
extended X-ray absorption fine structure
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LB
Luria Bertani
- HSQC
heteronuclear single quantum coherence
- CD
circular dichroism
- ESI
electrospray ionization
- MP
membrane proximal
- HME
heavy-metal efflux
- MS
mass spectrometry
Footnotes
We gratefully acknowledge support from the National Institutes of Health GM079192 to M.M.M., GM054803 to N. J. B., and the National Science Foundation 0923551 to V.H.W.
Supporting Information Available: Figure S1: Schematic of pETDuet-1 constructs; Figure S2: CusB expression from pETDuet-1 constructs probed by Western blotting; Figure 3S: Circular dichroism spectra of apo-CusB-NT and Ag(I)-CusB-NT. Figure S4: NanoESI mass spectra of Ag(I)-CusF and ubiquitin. Figure S5: NanoESI mass spectra of Apo-CusB-NT and Ag(I)-CusF mixtures at different ratios. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 2.Nies DH. Transition metal homeostasis in bacteria as a flow equilibrium of uptake and efflux processes. Amino Acids. 2009;37:22–22. [Google Scholar]
- 3.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 6.Conroy O, Kim EH, McEvoy MM, Rensing C. Differing ability to transport nonmetal substrates by two RND-type metal exporters. FEMS Microbiol Lett. 2010;308:115–122. doi: 10.1111/j.1574-6968.2010.02006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kittleson JT, Loftin IR, Hausrath AC, Engelhardt KP, Rensing C, McEvoy MM. Periplasmic metal-resistance protein CusF exhibits high affinity and specificity for both CuI and AgI. Biochemistry. 2006;45:11096–11102. doi: 10.1021/bi0612622. [DOI] [PubMed] [Google Scholar]
- 8.Loftin IR, Franke S, Roberts SA, Weichsel A, Heroux A, Montfort WR, Rensing C, McEvoy MM. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry. 2005;44:10533–10540. doi: 10.1021/bi050827b. [DOI] [PubMed] [Google Scholar]
- 9.Mealman TD, Blackburn NJ, McEvoy MM. Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of E. coli. Current Topics in Membranes. 2012 doi: 10.1016/B978-0-12-394390-3.00007-0. in press. [DOI] [PubMed] [Google Scholar]
- 10.Bagai I, Rensing C, Blackburn NJ, McEvoy MM. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry. 2008;47:11408–11414. doi: 10.1021/bi801638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zgurskaya HI, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zgurskaya HI, Nikaido H. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J Bacteriol. 2000;182:4264–4267. doi: 10.1128/jb.182.15.4264-4267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su CC, Yang F, Long F, Reyon D, Routh MD, Kuo DW, Mokhtari AK, Van Ornam JD, Rabe KL, Hoy JA, Lee YJ, Rajashankar KR, Yu EW. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J Mol Biol. 2009;393:342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem. 2007;282:35695–35702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 15.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 16.Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467:484–U140. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulathila R, Kulathila R, Indic M, van den Berg B. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PloS one. 2011;6:e15610. doi: 10.1371/journal.pone.0015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011;470:558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janganan TK, Bavro VN, Zhang L, Matak-Vinkovic D, Barrera NP, Venien-Bryan C, Robinson CV, Borges-Walmsley MI, Walmsley AR. Evidence for the assembly of a bacterial tripartite multidrug pump with a stoichiometry of 3:6:3. J Biol Chem. 2011;286:26900–26912. doi: 10.1074/jbc.M111.246595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinh T, Paulsen IT, Saier MH., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of Gram-negative bacteria. J Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saier MH, Jr, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 22.Zgurskaya HI, Yamada Y, Tikhonova EB, Ge Q, Krishnamoorthy G. Structural and functional diversity of bacterial membrane fusion proteins. Biochim Biophys Acta. 2009;1794:794–807. doi: 10.1016/j.bbapap.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. 1999;287:695–715. doi: 10.1006/jmbi.1999.2630. [DOI] [PubMed] [Google Scholar]
- 24.Ge Q, Yamada Y, Zgurskaya H. The C-terminal domain of AcrA is essential for the assembly and function of the multidrug efflux pump AcrAB-TolC. J Bacteriol. 2009;191:4365–4371. doi: 10.1128/JB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reffay M, Gambin Y, Benabdelhak H, Phan G, Taulier N, Ducruix A, Hodges RS, Urbach W. Tracking membrane protein association in model membranes. PloS One. 2009;4:e5035. doi: 10.1371/journal.pone.0005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mealman TD, Bagai I, Singh P, Goodlett DR, Rensing C, Zhou H, Wysocki VH, McEvoy MM. Interactions between CusF and CusB identified by NMR spectroscopy and chemical cross-linking coupled to mass spectrometry. Biochemistry. 2011;50:2559–2566. doi: 10.1021/bi102012j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heck AJ. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 28.Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 29.Sobott F, Robinson CV. Principles of Mass Spectrometry Applied to Biomolecules. John Wiley & Sons, Inc; 2006. Understanding protein interactions and their representation in the gas phase of the mass spectrometer; pp. 147–175. [Google Scholar]
- 30.Hu P, Ye QZ, Loo JA. Calcium stoichiometry determination for calcium binding proteins by electrospray ionization mass spectrometry. Anal Chem. 1994;66:4190–4194. doi: 10.1021/ac00095a013. [DOI] [PubMed] [Google Scholar]
- 31.Pan J, Xu K, Yang X, Choy WY, Konermann L. Solution-phase chelators for suppressing nonspecific protein-metal interactions in electrospray mass spectrometry. Anal Chem. 2009;81:5008–5015. doi: 10.1021/ac900423x. [DOI] [PubMed] [Google Scholar]
- 32.Shirran SL, Barran PE. The use of ESI-MS to probe the binding of divalent cations to calmodulin. J Am Soc Mass Spec. 2009;20:1159–1171. doi: 10.1016/j.jasms.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 33.De Angelis F, Lee JK, O’Connell JD, 3rd, Miercke LJ, Verschueren KH, Srinivasan V, Bauvois C, Govaerts C, Robbins RA, Ruysschaert JM, Stroud RM, Vandenbussche G. Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems. Proc Natl Acad Sci U S A. 2010;107:11038–11043. doi: 10.1073/pnas.1003908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erba EB, Zenobi R. Mass spectrometric studies of dissociation constants of noncovalent complexes. Ann Rep Sect “C” (Phys Chem) 2011;107:199–228. [Google Scholar]
- 35.Kitova EN, Soya N, Klassen JS. Identifying specific small-molecule interactions using electrospray ionization mass spectrometry. Anal Chem. 2011;83:5160–5167. doi: 10.1021/ac200244u. [DOI] [PubMed] [Google Scholar]
- 36.Deng L, Sun N, Kitova EN, Klassen JS. Direct quantification of protein-metal ion affinities by electrospray ionization mass spectrometry. Anal Chem. 2010;82:2170–2174. doi: 10.1021/ac902633d. [DOI] [PubMed] [Google Scholar]
- 37.Culotta VC, Lin SJ, Schmidt P, Klomp LW, Casareno RL, Gitlin J. Intracellular pathways of copper trafficking in yeast and humans. Adv Exp Med Biol. 1999;448:247–254. doi: 10.1007/978-1-4615-4859-1_22. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Vol. 1. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- 39.Stroebel D, Sendra V, Cannella D, Helbig K, Nies DH, Coves J. Oligomeric behavior of the RND transporters CusA and AcrB in micellar solution of detergent. Biochim Biophys Acta. 2007;1768:1567–1573. doi: 10.1016/j.bbamem.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Compton SJ, Jones CG. Mechanism of dye response and interference in the Bradford protein assay. Anal Biochem. 1985;151:369–374. doi: 10.1016/0003-2697(85)90190-3. [DOI] [PubMed] [Google Scholar]
- 41.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 43.George GN. Exafspak. Stanford Synchrotron Radiation Laboratory; 1990. [Google Scholar]
- 44.Gurman SJ, Binsted N, Ross I. A rapid, exact curved-wave theory for EXAFS calculations. J Phys C. 1984;17:143–151. [Google Scholar]
- 45.Gurman SJ, Binsted N, Ross I. A rapid, exact curved-wave theory for EXAFS calculations. II. The multiple-scattering contributions. J Phys C. 1986;19:1845–1861. [Google Scholar]
- 46.Binsted N, Gurman SJ, Campbell JW. Excurve 9.2. Daresbury Laboratory; 1998. [Google Scholar]
- 47.Siluvai GS, Mayfield M, Nilges MJ, Debeer George S, Blackburn NJ. Anatomy of a red copper center: spectroscopic identification and reactivity of the copper centers of Bacillus subtilis Sco and its Cys-to-Ala variants. J Am Chem Soc. 2010;132:5215–5226. doi: 10.1021/ja910759v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siluvai GS, Nakano M, Mayfield M, Blackburn NJ. The essential role of the Cu(II) state of Sco in the maturation of the Cu(A) center of cytochrome oxidase: evidence from H135Met and H135SeM variants of the Bacillus subtilis Sco. J Biol Inorg Chem. 2011;16:285–297. doi: 10.1007/s00775-010-0725-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson WC., Jr Secondary structure of proteins through circular dichroism spectroscopy. Ann Rev Biophys Biophysical Chem. 1988;17:145–166. doi: 10.1146/annurev.bb.17.060188.001045. [DOI] [PubMed] [Google Scholar]
- 50.Akama H, Matsuura T, Kashiwagi S, Yoneyama H, Narita S, Tsukihara T, Nakagawa A, Nakae T. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J Biol Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 51.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci U S A. 2004;101:9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yum S, Xu Y, Piao S, Sim SH, Kim HM, Jo WS, Kim KJ, Kweon HS, Jeong MH, Jeon H, Lee K, Ha NC. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J Mol Biol. 2009;387:1286–1297. doi: 10.1016/j.jmb.2009.02.048. [DOI] [PubMed] [Google Scholar]
- 53.Kim EH, Nies DH, McEvoy MM, Rensing C. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol. 2011;193:2381–2387. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.