Abstract
Objectives
This study tested the hypothesis that two common polymorphisms in the chromosome 4q25 region that have been associated with atrial fibrillation (AF) contribute to the variable penetrance of familial AF.
Background
Although mutations in ion channels, gap junction proteins, and signaling molecules have been described for Mendelian forms of AF, penetrance is highly variable. Recent studies have consistently identified 2 common single nucleotide polymorphisms (SNPs) in the chromosome 4q25 region as independent AF susceptibility alleles.
Methods
We studied 11 families in which AF was present in ≥2 individuals who also shared a candidate gene mutation. These mutations were identified in all subjects with familial lone AF (n=33) as well as apparently unaffected family members (age >50 yrs with no AF; n=17).
Results
Mutations were identified in SCN5A (n=6); NPPA (n=2); KCNQ1 (n=1); KCNA5 (n=1) and NKX2.5 (n=1). In genetic association analyses, un-stratified and stratified according to age of onset of AF and unaffected age > 50 yrs, there was a highly statistically significant association between the presence of both common (rs2200733, rs10033464) as well as rare variants and AF (un-stratified P=1×10−8; stratified [age of onset <50 yrs and unaffected age >50 yrs], P=7.6×10−5) (un-stratified P<0.0001; stratified [age of onset <50 yrs and unaffected age >50 yrs], P<0.0001). Genetic association analyses showed that the presence of common 4q25 risk alleles predicted whether carriers of rare mutations developed AF (P = 2.2×10−4).
Conclusions
Common AF-associated 4q25 polymorphisms modify the clinical expression of latent cardiac ion channel and signaling molecule gene mutations associated with familial AF. These findings support the idea that the genetic architecture of AF is complex and includes both rare and common genetic variants.
Atrial fibrillation (AF) is an important and increasing public health problem. The prevalence of AF doubles for each advancing decade of life and there is widespread agreement that the prevalence is increasing over time (1,2). The risk factors for AF are multi-factorial and include male sex, advancing age, coronary artery disease, congestive heart failure and valvular heart disease. However, a substantial portion of the variability in risk for AF remains unexplained, leading investigators to search for genetic factors.
Investigators at the Framingham Heart Study have observed that the odds ratio (OR) of developing AF was 1.8 times higher for individuals with at least one parent diagnosed with AF compared to those without such a parental history (3). The OR increased further (3.2) if one parent was affected before 75 years of age. In a population-based cohort of over 5,000 AF patients from Iceland, first-degree relatives of AF patients were 1.77-fold more likely to have AF than the general population, with a relative risk of 4.67 in first-degree relatives of patients less than 60 years of age (4). Familial aggregation of AF is particularly prominent in individuals with idiopathic or so-called lone AF, i.e., early-onset AF without structural heart disease, for which as many as 30% of probands have a first-degree relative with the arrhythmia (5–7).
Although a Mendelian pattern of inheritance has been reported, large AF kindreds such as those used to identify disease genes in other inherited arrhythmia syndromes, e.g., congenital long QT syndrome, are unusual. A common presentation of the Mendelian form of the arrhythmia is a proband with familial lone AF (6). Mutations in genes encoding cardiac ion channels, gap junction proteins, atrial natriuretic peptide (ANP) and nucleoporins (NUP155) have been reported in isolated cases and small kindreds (8). Although traditional linkage analysis or candidate gene approaches have been successful in identifying monogenic forms of familial lone AF, the mode of transmission for most forms of AF remains unclear supporting the idea that AF inheritance is complex.
In 2007, a genome-wide association study (GWAS) in Icelanders identified a locus on chromosome 4q25 associated with AF in subjects of all ages (9). Within this locus, two non-coding single nucleotide polymorphisms (SNPs) were independently associated with AF and these findings were replicated in two populations of European descent and one of Asian descent. The SNP most strongly associated with AF, rs2200733, conferred a 1.71-fold increased risk of AF while the other SNP, rs10033464, conferred a 1.42-fold increased risk. Recently, this association was replicated in a study of 4 large populations with ambulatory AF (10). This association has also been reported for post-cardiac surgery AF a setting thought to be related to inflammation (11) and has recently been reported to predict the likelihood of successful AF ablation (12). Although mutations in ion channels, gap junction proteins and signaling molecules have been identified in isolated kindreds with two or more individuals affected with familial lone AF, penetrance in these families is highly variable. One potential explanation for this phenomenon is the coexistence of modifier gene alleles, possibly common SNPs altering AF susceptibility. Here we tested the hypothesis that 4q25 genotypes contribute to the variable penetrance of the AF phenotype in familial AF.
METHODS
Vanderbilt AF Registry
Between November 2002 and July 2009, subjects with AF were prospectively enrolled in the Vanderbilt AF Registry, which comprises clinical and genetic databases (13). At enrollment a detailed medical and drug history was obtained in all patients; patients are also asked to complete a symptom questionnaire. Patients were recruited from the Vanderbilt Cardiology and Arrhythmia Clinics, the emergency department, and in-patient services. An echocardiogram is obtained on all patients at time of enrollment into the Registry. The upper limits of normal for cardiac chamber dimensions are based on age and body surface area. Individuals who were greater than 18 years of age with a diagnosis of AF, confirmed by ECG, who presented because of symptoms or who were diagnosed during a routine physical examination are included in the AF Registry. Subjects were excluded if AF was diagnosed in the setting of recent cardiac surgery or were unable to give informed consent or report for follow-up. The study protocol was approved by the Vanderbilt University Institutional Review Board and participants were enrolled following informed written consent.
AF probands and their relatives were clinically classified by a consistently applied set of definitions. AF is defined as replacement of sinus P waves by rapid oscillations or fibrillatory waves that varied in size, shape, and timing and were associated with an irregular ventricular response when atrioventricular conduction was intact. Documentation of AF on an electrocardiogram (ECG), rhythm strip, event recorder, or Holter monitor recording was necessary. Paroxysmal AF was defined as AF lasting more than 30 seconds that terminated spontaneously. Persistent AF was defined as AF lasting more than seven days and requiring either pharmacologic therapy or electrical cardioversion for termination. AF that was refractory to cardioversion or that was allowed to continue was classified as permanent.
Study Cohort
The relationship between clinical phenotype (familial lone AF) and genotype was determined for probands in whom ion channel and other gene mutations were identified, together with their relatives. A total of 70 individuals in 11 kindreds had complete phenotype and genotype data available for analysis (Table 1) and this group forms the study cohort.
Table 1.
Clinical characteristics of familial lone AF probands enrolled in the Vanderbilt AF Registry identified to have mutations or rare variants in ion channel or signaling proteins.
| AF kindred and pedigree number |
Age at onset (yr) |
Age at diagnosis (yr) |
Rhythm | Symptoms | Echo | LA size (mm) |
|---|---|---|---|---|---|---|
| AF 027 | ||||||
| II-1 | 41 | 54 | PerstAF | Yes | LVH | 46 |
| III-1 | 31 | 36 | PAF | Yes | Normal | 42 |
| AF 063 | ||||||
| II-1 | 33 | 39 | PAF | Yes | Normal | 44 |
| II-2 | 41 | 48 | PAF | Yes | Normal | 42 |
| AF 119 | ||||||
| I-1 | 51 | 54 | PerstAF | No | Normal | 46 |
| II-1 | 28 | 38 | PAF | Yes | Normal | 42 |
| AF 240 | ||||||
| II-1 | 70 | 72 | PerstAF | No | LVH | 50 |
| III-1 | 42 | 42 | PAF | Yes | Normal | 43 |
| III-2 | 39 | 42 | PAF | Yes | Normal | 42 |
| AF 527 | ||||||
| I-1 | 52 | 54 | PerstAF | Yes | LVH | 46 |
| II-1 | 56 | 56 | PAF | Yes | Normal | 44 |
| III-1 | 17 | 25 | PAF | yes | Normal | 42 |
| AF 313 | ||||||
| II-1 | 48 | 56 | PermAF | Yes | MR; LVH | 52 |
| II-1 | 44 | 46 | PerstAF | Yes | Normal | 42 |
| II-2 | 42 | 43 | PAF | Yes | Normal | 41 |
| II-4 | 44 | 45 | PAF | Yes | Normal | 42 |
| AF 579 | ||||||
| I-1 | 56 | 64 | PerstAF | No | LVH | 46 |
| II-2 | 48 | 49 | PAF | Yes | Normal | 43 |
| II-3 | 49 | 50 | PAF | No | Normal | 41 |
| AF 673 | ||||||
| II-1 | 48 | 50 | PAF | Yes | Normal | 41 |
| AF 242 | ||||||
| II-1 | 38 | 40 | PAF | Yes | Normal | 44 |
| II-9 | 42 | 44 | PAF | Yes | Normal | 43 |
| III-2 | 20 | 26 | PAF | Yes | Normal | 39 |
| AF 406 | ||||||
| I-1 | 78 | 78 | PermAF | No | LVH | 48 |
| II-4 | 61 | 62 | PerstAF | No | LVH | 49 |
| II-5 | 52 | 58 | PAF | Yes | Normal | 46 |
| III-1 | 17 | 22 | PAF | Yes | Normal | 39 |
| III-3 | 39 | 42 | PAF | Yes | Normal | 42 |
| III-5 | 42 | 46 | PAF | Yes | Normal | 44 |
| III-7 | 44 | 45 | PAF | Yes | Normal | 46 |
| AF 1111 | ||||||
| I-1 | 58 | 64 | PermAF | No | LVH | 50 |
| II-4 | 56 | 60 | PAF | Yes | Normal | 44 |
| II-5 | 46 | 48 | PAF | Yes | Normal | 45 |
PAF = paroxysmal atrial fibrillation; Perst = persistent; Perm = permanent; LA = left atrial; LVH = left ventricular hypertrophy; MR = mitral regurgitation.
Clinical phenotype
The clinical phenotype evaluated in this study is lone familial AF defined as AF occurring in individuals less than 66 years of age without hypertension, overt structural heart disease or thyroid dysfunction (as determined by clinical examination, ECG, echocardiography and normal thyroid function tests) and AF in one or more first-degree relatives (5,6). Left ventricular hypertrophy (LVH) was defined as LV mass index (LVMI) ≥150 g/m2 (14). All familial lone AF probands had a LVMI <150 g/m2.
Familial AF was defined as the presence of AF in one or more first-degree relatives of the index case. Although the proband in each family had to have lone AF, other family members were classified as affected if they had AF, despite structural heart disease. Family history information was initially obtained from the medical record and was supplemented by a questionnaire detailing past medical history, family history, and clinical symptoms. For lone AF probands with a positive family history, family members were contacted and a more detailed personal medical history was obtained. Family members were then sent a blood kit for DNA extraction and written permission was obtained to review their medical records. We routinely evaluate for the presence of asymptomatic AF by providing all unaffected family members who consent to participate in the study with a 7-day full disclosure continuous monitor. In addition, individuals with symptoms suggestive for AF are also given a 30-day event recorder that can be activated whenever they develop symptoms.
Screening for mutations in ion channel and other genes
Whole blood was collected for genomic DNA extraction and analysis from lone AF probands and as many first-degree relatives as would consent to the study. Mutational analyses of SCN5A,(15) KCNQ1,(16) NPPA,(16) KCNA5(17) and NKX2.5 genes were performed in lone AF probands and all consented family members. The coding and flanking regions were amplified by polymerase chain reaction (PCR) using primers designed to obtain fragments of appropriate size as previously described (15),(16,17). Briefly, PCR-amplified DNA fragments were analyzed using the Reveal Discovery System (based on temperature gradient capillary electrophoresis) to identify aberrant conformers, which were then directly sequenced. Table 2 lists the number of lone and typical (non-lone) AF cases enrolled in the Vanderbilt AF Registry and controls that were screened for genetic variants in the ion and non-ion channel genes.
Table 2.
Probands with atrial fibrillation (AF) enrolled in the Vanderbilt AF Registry (cases) and controls that were resequenced for variants in ion channel and non-ion channel genes.
| Gene | Lone AF Cases | Controls |
|---|---|---|
| SCN5A | 118 257 (typical AF) |
460 |
| KCNQ1 | 231 | 500 |
| NPPA | 98 133 (typical AF) |
500 |
| KCNA5 | 231 200 (typical AF) |
300 |
| NKX2.5 | 160 | 500 |
4q25 genotypes
Genotyping of the 2 SNPs (rs2200733, rs10033464) was performed using real-time PCR, iPlex single base primer extension and MALDI-TOF mass spectrometry in a 384-well-format (Sequenom, San Diego, CA) as previously described (18). Seventy individuals (11 familial lone AF probands) and 69 family members underwent genotyping at the 2 common 4q25 SNPs.
Statistical Analysis
The goal of these analyses was to determine if there is a statistically significant association between the presence of both common and rare AF variants and development of AF (i.e., AF disease status). We hypothesized that the common variants on 4q25 influence the development of AF by modulating the rare ion-channel and non-ion channel mutations. To test this hypothesis, first we performed a series of chi-square tests (2×2 tables) of association (using Yates’ correction test for small sample size) with one degree of freedom. We initially performed an un-stratified analysis, followed by five stratified analyses based on the age at onset of AF and the age of the controls. As most lone AF probands develop AF before the age of 50 years, our first age cut-off for AF age of onset was <50 years but we also stratified based on an AF age of onset <40 years. Thus we stratified by: AF age-at-onset < 50 years, AF age-at-onset < 40 years, unaffected individuals > 50 years only, and the two possible combinations of these three criteria. We collapsed the common variant genotypes into an allelic test of presence and absence of the variant allele. The nominal P-values as well as P-values generated with permutation testing were determined.
Permutation testing was performed to correct for multiple testing, while controlling for the familial relationships in the kindreds. The process involved removing the AF affection status within each family and randomly re-assigning in the same proportion of affected and unaffected individuals. This was carried out in each family, 1000 times, to create 1000 null datasets. In each random dataset, we performed association analysis and recorded the chi-square statistic. After 1000 analyses, we had an empirical distribution from which to assign a permutation P-value, which is adjusted for the relatedness in the dataset. In addition, we performed the analyses by way of the presence/absence of rare variants (ignoring the common variant).
As a follow-up, we performed a second series of chi-square analyses (Yates’ correction where appropriate). Here, we restricted the dataset to include only those individuals that possessed a rare variant in the ion channel or non-ion channel genes sequenced; these analyses included 48 individuals. In this set, we performed the identical series of chi-square analyses as described above although here, we were looking at the association of the common variant with AF status; all individuals possessed a rare variant in this set. We performed the un-stratified analysis first, followed by five stratified analyses. We again stratified by: AF age-at-onset < 50 years, AF age-at-onset < 40 years, unaffected individuals > 50 years only, and the two possible combinations of those three criteria. All chi-square tests of association were performed in JMP v8.0.
Results
Study cohort
Our candidate gene approach to identifying novel mutations or rare variants associated with AF identified 11 familial lone AF probands who had one or more first degree relatives with the arrhythmia. The total study cohort consists of 70 individuals in 11 kindreds who had complete phenotype and genotype data available for analysis (Table 2).
Novel SCN5A variants
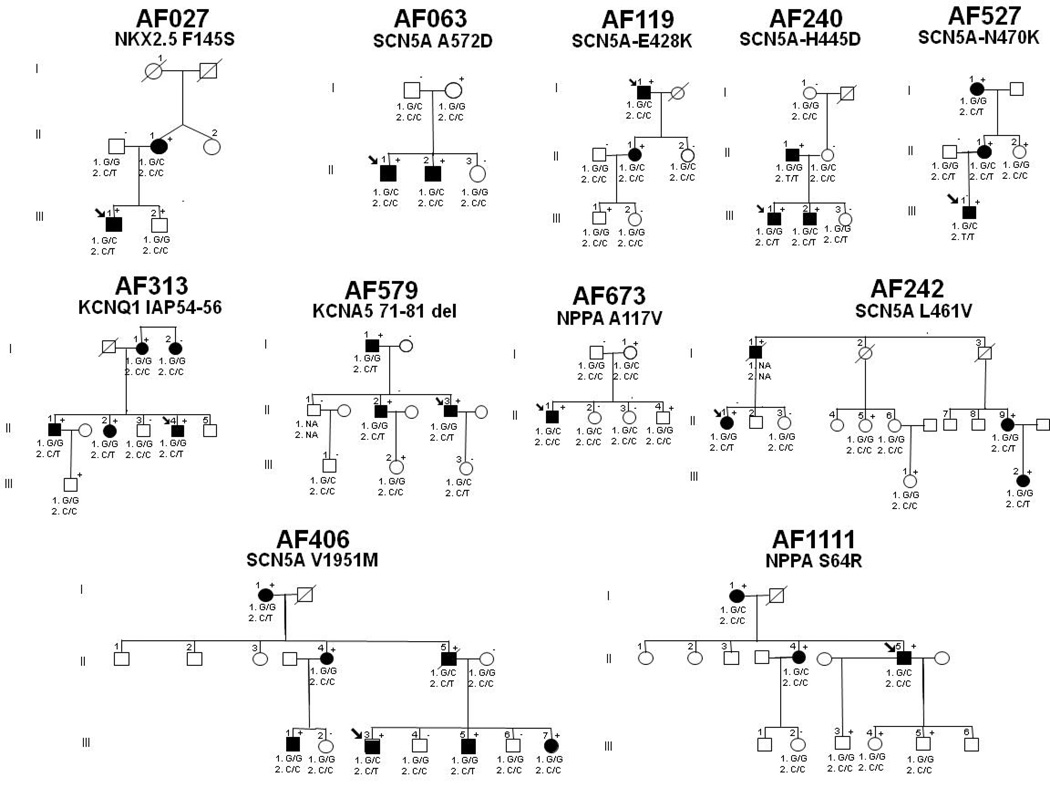
During the initial 3-year enrollment period, 396 patients with AF were approached and 375 (95%) subjects agreed to participate in the AF Registry (15). Within this cohort, 118 patients (31%) had familial lone AF. As previously reported, resequencing identified 8 novel variants in 10 probands (2.7%) that were not found in the population-based controls (0%; P=0.001) (15). Genotyping was performed in 6 of these kindreds with 2 or more affected individuals with AF (Figure 1). The novel variants identified affect highly conserved residues in the SCN5A protein. All probands identified were heterozygous for the mutation.
Figure 1. Pedigrees of twelve familial atrial fibrillation (AF) kindreds with rare ion channel and signaling molecule variants.
Squares indicate male family members, circles female family members, and symbols with a slash mark deceased family members. Arrows indicate the probands. Solid symbols indicate the presence of AF and open symbols indicate unaffected family members. The presence (+) or absence (−) of a rare variant is indicated for persons with DNA samples available for testing. 1. rs10033464 and 2. rs2200733 refer to the genotype for the single nucleotide polymorphisms on chromosome 4q25 that been associated with AF. NA = genotype not available.
KCNQ1 variant
In this study cohort of 231 individuals with familial lone AF, screening for KCNQ1 mutations in genomic DNA identified a unique sequence variant in one case of familial AF (Vanderbilt AF 313).(16) In the proband, a 9-bp duplication resulting in insertion of the amino acids isoleucine (I), alanine (A) and proline (P) at locations 54 to 56 (IAP54–56) in the N-terminus of the KCNQ1 protein was identified. The variant was also confirmed in three affected family members. The sequence change was not found in Caucasian, Han Chinese, or Asian population controls but was identified in 2.1% (2/94) of African American individuals. As the controls were obtained from the anonymous Coriell repository, no clinical information about these individuals is available.
NPPA variants
Genomic DNA sequencing of NPPA identified two novel missense mutations in two Caucasian AF kindreds; S64R in AF 313 (16) and A117V in 1111 (Figure 1). The S64R mutation was also confirmed in two affected family members but was absent in unaffected family members. Furthermore, both mutations were not identified in Caucasian, Han Chinese, Asian, or African-American population controls. The A117V mutation was identified in a family with lone AF (Figure 1). The kindred includes 6 family members, 3 of which were heterozygous for the mutation with the probands presenting with early onset paroxysmal lone AF.
KCNA5 variant
We resequenced KCNA5 in 231 individuals with familial lone AF and identified a novel 33-bp coding region deletion in 2 Caucasian probands with early onset lone AF. The variant results in deletion of 11 amino acids at positions 71–81 of the N-terminus (71–81del) (17). The sequence change was not found in 200 patients with typical AF and 300 ethnically-matched population controls. The proband in this kindred (Vanderbilt AF 579) presented with symptomatic paroxysmal familial lone AF and his AF is currently managed with sotalol. A family history also showed that most of the affected family members developed familial lone AF at a relatively young age and most presented with symptomatic paroxysmal AF.
NKX2.5 variant
NKX2.5 was screened in 160 familial lone AF patients. We identified 1 novel non-synonymous variant (F145S) that was not identified in over 300 control patients with no history of AF. The proband in this kindred (AF 027) presented with early onset symptomatic familial lone paroxysmal AF. The proband’s father also presented with familial lone AF in his 40’s and his brother, who carries the variant, has not been diagnosed with AF at age 52.
4q25 genotypes
Table 3 shows a significant interaction between common and rare genetic variants based on an AF age of onset <50 years. However, when we stratified based on AF age of onset <40 years and age of unaffected individuals in the families, there remained a significant association between the presence of common and rare genetic variants (Table 4). Furthermore, when we examined genetic association of rare variants alone (ignoring common variants) un-stratified and stratified by age of onset and age of unaffected individuals in the families, there remained a highly statistically significant association between the presence of both common and rare variants and AF disease status significant interaction (Table 5). Odds ratios (ORs) were calculated for the test of co-occurrence of rare and common variants, in the un-stratified and stratified analyses. The ORs range from 4–452 for the different strata; these values are far in excess of the 1.4–1.7 reported for studies in the general population, thus supporting the idea that these common variants modulate penetrance of the AF phenotype in these kindreds. ORs could not be calculated for the presence of common variants in those individuals with rare variants only because the contingency table cells contained zero counts.
Table 3.
An interaction between rare (ion channel and non-ion channel) and common genetic variants to determine susceptibility to the development of familial AF.
| AF by age 50 yrs | No AF by age 50 yrs | |
|---|---|---|
| Chr4q25 SNPs absent | 0 | 13 |
| Chr4q25 SNPs present | 21 | 8* |
P = 1.21 × 10−5 comparing presence of both common and rare variants in 11 kindreds with familial AF.
Chr,chromosome; SNPs, single nucleotide polymorphisms.
Table 4.
Genetic association between common and rare variants un-stratified and stratified by age of onset and age of unaffected individuals in the families.
| Sample size |
Presence of both common and rare variants |
OR (95% CI) | Permutation P-value |
|
|---|---|---|---|---|
| Un-stratified | 70 | P = 9.21 × 10−10 | 53 (10–273) | <0.001 |
| Age of onset <50 yrs | 64 | P = 9.41 × 10−8 | 39 (8–205) | <0.001 |
| Age of onset <40 yrs | 57 | P = 4.13 × 10−6 | 51 (6–452) | <0.001 |
| Unaffected age >50 yrs | 48 | P = 8.63 × 10−7 | 41 (7–239) | <0.001 |
| Age of onset <50 yrs, Unaffected age >50 yrs |
42 | P = 1.21 × 10−5 | 31 (5–179) | <0.001 |
| Age of onset <40 yrs, Unaffected age >50 yrs |
35 | P = 1.62 × 10−4 | 40 (4–385) | <0.001 |
OR, odds ratio; CI, confidence interval.
Table 5.
Genetic association of rare variants alone (ignoring common variants) un-stratified and stratified by age of onset and age of unaffected individuals in the families.
| Sample size | Presence/absence of rare variants only (P-value) |
OR (95% CI) | |
|---|---|---|---|
| Un-stratified | 70 | 2.2 × 10−4 | 19 (2.4–155) |
| Age of onset<50yrs | 64 | 0.003 | 14 (1.7–117) |
| Age of onset<40 yrs | 57 | 0.04 | 8.8 (1.1–74) |
| Unaffected age>50 yrs | 48 | 2.1 × 10−4 | 26 (3.0–227) |
| Age of onset<50 yrs Unaffected age>50 yrs |
42 | 0.002 | 20 (2.2–173) |
| Age of onset<40 yrs Unaffected age>50 yrs |
35 | 0.013 | 12 (1.3–109) |
We performed a logistic regression analysis with two main effect terms (presence/absence of rare variants and presence/absence of common variants) in addition to a multiplicative interaction term (presence/absence of rare variant × presence/absence of common variant). Based on the likelihood ratio test, there was a highly significant interaction (P= 0.002).
Discussion
The present findings provide strong evidence that two common AF-associated polymorphisms act as genetic modifiers of the clinical expression of latent cardiac ion channel and signaling gene mutations associated with familial AF. Furthermore, our findings strongly support the idea that AF inheritance is complex and non-Mendelian.
Although the potential mechanism of action of variants at the chromosome 4q25 locus tagged by the two non-coding SNPs remains unknown, it is quite likely mediated through effects on the paired-like homeodomain transcription factor 2 (PITX2) gene. PITX2 plays a critical role in left-right asymmetry and loss of the pitx2c isoform in mice can lead to right atrial isomerization and failure to suppress a default pathway for sinus node development of the pulmonary myocardium (19–21). Importantly, this area is now well-recognized as a common source for ectopic atrial activity necessary for the initiation and propagation of AF (22). Microarray analysis of pitx2 null-mutant and control mice hearts have revealed up-regulation of Kcnq1, a potassium channel gene that has been associated with gain-of-function mutations in familial AF (19,20). More recently, Kirchhof et al (23) performed expression arrays in pitx2+/− mice demonstrating alteration in a number of cellular and molecular pathways in addition to potassium channels, which might provide an explanation for why mutations in ion channels are associated with familial AF. This recent data strongly supports our hypothesis that the 4q25 AF susceptibility alleles likely mediate their effects by modulation of cardiac ion channel expression and signaling proteins in the heart.
Investigating the genetic basis for AF presents a number of important challenges (8). The paroxysmal nature and variable symptoms in AF, a high prevalence in the general population, and a late age of onset in many individuals all make assignment of the correct clinical phenotype challenging. This complexity has compelled a search for new more effective methods for investigating the genetics of AF. One approach is to use the families of affected individuals as an enriched target population for the definition and evaluation of intermediate or endophenotypes, such as prolonged signal-averaged P-wave duration (24), which are causally related to the poorly penetrant classical clinical syndromes (25). Because of these limitations, researchers have begun to apply other approaches to identify genes that may be associated with AF. A logical consequence of the availability of comprehensive genomic maps is the advent of high-density genome-wide searches for modest gene effects using large scale testing of SNPs. This approach has identified common variants on the chromosome 4q25 locus and more recently common SNPs on chromosome 16q22 (26) and 1q21 (27) which confer an increased risk (OR: 1.2–1.4) of AF across multiple different populations.
One of the most vexing problems with non-Mendelian genetic approaches is the differentiation of signal from noise. Association studies may be confounded by etiologic heterogeneity, population stratification, or false positives, while the bias toward small effects may miss major genetic contributions relevant only in a subset of study subjects. It may also prove difficult to define the fundamental mechanisms of any true associations, as the responsible gene may be remotely linked to the locus identified. The development of more efficient models for the rapid validation of genome-wide association study results, and improved understanding of the underlying biology will facilitate the interpretation of such studies. In our study however, variants identified in our screening are likely to be causative because we demonstrated cosegregation of the variants with AF in multiple kindreds (where available), absence in a large control population and the fact that all novel variants are located in highly conserved regions of the ion channel and signaling protein. This supports a link between disturbed ion channel and protein function and AF.
Studies of kindreds with AF suggest a genetic basis for the condition, and mutations in several cardiac potassium channel genes have been linked to familial AF (28,29). Although specific mutations in the KCNQ1 gene have been observed in families with AF, the role of such mutations in AF remains unclear; often such isolated or “private” mutations are in residues of unknown function, effects on channel conductance are variable, and in most cases it may be difficult to discriminate rare polymorphisms of no functional significance from true mutations. However, the low prevalence of KCNQ1 mutations in large AF cohorts, suggests that mutations in this gene are not a major cause of AF (30,31). In AF313, multiple family members presented with early onset lone AF, and our segregation analysis supports the novel KCNQ1 mutation as being causative for AF. Furthermore, the absence of the KCNQ1 variant (IAP54–56) in non African American control groups excludes the possibility that this mutation is a common polymorphism in this population; this finding is consistent with a disease-associated mutation. The KCNQ1-IAP54–56 variant however was identified in 2.1% of healthy African American individuals, suggesting it may be a common risk allele in this population.
Several limitations of the present study warrant consideration. First, most of the AF kindreds are small and thereby formal segregation analysis was limited. Data for some unaffected family members are not available. Second, no novel mutations were identified in the ethnically-defined controls. However, comprehensive resequencing of the individual gene in the population-based controls was not performed and this may condition interpretation of the results of this study. Although it is possible that additional novel ion channel and other gene mutations may have been identified, and our focused strategy of control genotyping may underestimate the true prevalence of rare polymorphisms, the association of the mutations with AF is supported primarily by segregation analysis and the fact that the mutations occurred in a highly conserved region of the protein. Third, population stratification may confound our results especially across families. However, as our cohort comes from a fairly homogeneous population of individuals of European descent, we feel that this is unlikely to bias our results. Furthermore, data on the common 4q25 allele frequencies (as shown in Table 7) would suggest otherwise.
Table 7.
Allele frequencies of chromosome 4q25 single nucleotide polymorphisms (SNPs).
| Variant | Genotype | # alleles | Allele frequency |
|---|---|---|---|
| rs10033464 | C | 22 | 0.16 |
| G | 114 | 0.83 | |
| T | 2 | 0.01 | |
| rs2200733 | C | 114 | 0.83 |
| T | 24 | 0.17 |
Clinical Implications
The idea that a combination of common and rare variants contribute to the very common phenotype of AF may have significant impact on the assessment and management of patients not only with familial AF but also the more common forms of the arrhythmia. Our study has demonstrated that the penetrance of genes encoding rare ion channel and signaling proteins is modulated by common variants on chromosome 4q25 supporting the idea that AF inheritance is complex, i.e., a single genetic variant does not predict the phenotype. Currently there is no clinical role for genetic testing in most patients with AF (32). However, a case can be made to screen candidate AF genes in those individuals who present with early onset lone AF especially if there is a strong family history of the arrhythmia. Identification of a rare genetic variant in lone AF probands may be followed by consideration of screening family members to identify other carriers and also genotyping at the 4q25 AF locus. Identification of family members who are at risk for developing AF is important as therapies known to reduce the incidence of AF such as angiotensin converting-enzyme inhibitors, could be considered.
Conclusions
We have shown that two common AF-associated polymorphisms can act as genetic modifiers of the clinical severity of familial AF. One interpretation of our findings is that in kindreds with familial AF, the arrhythmia develops in individuals who have the rare AF-associated variants as well as susceptibility alleles determined by the 4q25 locus. Our findings support the general concept that common variants may not only act as significant modifiers of the effects of rare variants, but that the genetic architecture of common human phenotypes includes both rare and common variants (33).
Table 6.
Genetic association of common variants in the subset of data with all individuals harboring rare variants: un-stratified and stratified by age of onset and age of unaffected individuals in the families.
| Sample size | Common variants in individuals with rare variants only (P-value) |
|
|---|---|---|
| Un-stratified | 48 | 2.79 × 10−7 |
| Age of onset <50 yrs | 42 | 4.41 × 10−6 |
| Age of onset <40 yrs | 36 | 1.46 × 10−4 |
| Unaffected age >50 yrs | 34 | 3.4 × 10−4 |
| Age of onset <50 yrs, Unaffected age >50 yrs |
28 | 0.001 |
| Age of onset <40 yrs, Unaffected age >50 yrs |
22 | 0.01 |
Acknowledgments
Funding Sources: This work was supported by HL65962, HL075266, and an American Heart Association Established Investigator Award (0940116N).
Abbreviations and Acronyms
- AF
atrial fibrillation
- ANP
atria natriuretic peptide
- GWAS
genome wide association study
- OR
odds ratio
- SNP
single nucleotide polymorphism
Footnotes
Disclosures
None.
References
- 1.Lloyd-Jones DM, Wang TJ, Leip EP, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110:1042–1046. doi: 10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 2.Miyasaka Y, Barnes ME, Gersh BJ, et al. Secular trends in incidence of atrial fibrillation in Olmsted County, Minnesota, 1980 to 2000, and implications on the projections for future prevalence. Circulation. 2006;114:119–125. doi: 10.1161/CIRCULATIONAHA.105.595140. [DOI] [PubMed] [Google Scholar]
- 3.Fox CS, Parise H, D'Agostino RB, Sr, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 4.Arnar DO, Thorvaldsson S, Manolio TA, et al. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J. 2006;27:708–712. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 5.Darbar D, Herron KJ, Ballew JD, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 6.Ellinor PT, Yoerger DM, Ruskin JN, Macrae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–184. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 7.Marcus GM, Smith LM, Vittinghoff E, et al. A first-degree family history in lone atrial fibrillation patients. Heart Rhythm. 2008;5:826–830. doi: 10.1016/j.hrthm.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darbar D. Genetics of atrial fibrillation: Rare mutations, common polymorphisms, and clinical relevance. Heart Rhythm. 2008;5:483–486. doi: 10.1016/j.hrthm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;488:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 10.Kaab S, Darbar D, van Noord C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813–819. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Body SC, Collard CD, Shernan SK, et al. Variation in the 4q25 Chromosomal Locus Predicts Atrial Fibrillation After Coronary Artery Bypass Graft Surgery. Circ Cardiovasc Genet. 2009;2:499–506. doi: 10.1161/CIRCGENETICS.109.849075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2010;55:747–753. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 13.Darbar D, Motsinger AA, Ritchie MD, Gainer JV, Roden DM. Polymorphism modulates symptomatic response to antiarrhythmic drug therapy in patients with lone atrial fibrillation. Heart Rhythm. 2007;4:743–749. doi: 10.1016/j.hrthm.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy D, Savage DD, Garrison RJ, Anderson KM, Kannel WB, Castelli WP. Echocardiographic criteria for left ventricular hypertrophy: the Framingham Heart Study. Am J Cardiol. 1987;59:956–960. doi: 10.1016/0002-9149(87)91133-7. [DOI] [PubMed] [Google Scholar]
- 15.Darbar D, Kannankeril PJ, Donahue BS, et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–1935. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abraham RL, Yang T, Blair M, Roden DM, Darbar D. Augmented potassium current is a shared phenotype for two genetic defects associated with familial atrial fibrillation. J Mol Cell Cardiol. 2010;48:181–190. doi: 10.1016/j.yjmcc.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang T, Yang P, Roden DM, Darbar D. Novel KCNA5 mutation implicates tyrosine kinase signaling in human atrial fibrillation. Heart Rhythm. 2010;7:1246–1252. doi: 10.1016/j.hrthm.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeufer A, Jalilzadeh S, Perz S, et al. Common variants in myocardial ion channel genes modify the QT interval in the general population: results from the KORA study. Circ Res. 2005;96:693–701. doi: 10.1161/01.RES.0000161077.53751.e6. [DOI] [PubMed] [Google Scholar]
- 19.Tessari A, Pietrobon M, Notte A, et al. Myocardial Pitx2 differentially regulates the left atrial identity and ventricular asymmetric remodeling programs. Circ Res. 2008;102:813–822. doi: 10.1161/CIRCRESAHA.107.163188. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mommersteeg MT, Brown NA, Prall OW, et al. Pitx2c and Nkx2–5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 22.Haissaguerre M, Jais P, Shah DC, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–666. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- 23.Kirchhof P, Kahr PC, Kaese S, et al. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 24.Darbar D, Hardy A, Haines JL, Roden DM. Prolonged signal-averaged P-wave duration as an intermediate phenotype for familial atrial fibrillation. J Am Coll Cardiol. 2008;51:1083–1089. doi: 10.1016/j.jacc.2007.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamin EJ, Chen PS, Bild DE, et al. Prevention of atrial fibrillation: report from a national heart, lung, and blood institute workshop. Circulation. 2009;119:606–618. doi: 10.1161/CIRCULATIONAHA.108.825380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;8:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YH, Xu SJ, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 29.Olson TM, Alekseev AE, Liu XK, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–2191. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 30.Ellinor PT, Moore RK, Patton KK, Ruskin JN, Pollak MR, Macrae CA. Mutations in the long QT gene, KCNQ1, are an uncommon cause of atrial fibrillation. Heart. 2004;90:1487–1488. doi: 10.1136/hrt.2003.027227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellinor PT, Petrov-Kondratov VI, Zakharova E, Nam EG, Macrae CA. Potassium Channel Gene Mutations Rarely Cause Atrial Fibrillation. BMC Med Genet. 2006;7:70. doi: 10.1186/1471-2350-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 33.Johansen CT, Wang J, Lanktree MB, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]