Abstract
Nine neurodegenerative disorders are caused by the abnormal expansion of polyglutamine (polyQ) regions within distinct proteins. Genetic and biochemical evidence has documented that the molecular chaperone, heat shock protein 70 (Hsp70), modulates polyQ toxicity and aggregation, yet it remains unclear how Hsp70 might be used as a potential target in polyQ-related diseases. We have utilized a pair of membrane-permeable compounds that tune the activity of Hsp70 by either stimulating or by inhibiting its ATPase functions. Using these two pharmacological agents in both yeast and PC12 cell models of polyQ aggregation and toxicity, we were surprised to find that stimulating Hsp70 solubilized polyQ conformers and simultaneously exacerbated polyQ-mediated toxicity. By contrast, inhibiting Hsp70’s ATPase activity protected against polyQ toxicity and promoted aggregation. These findings clarify Hsp70’s role as a possible drug target in polyQ disorders and suggest that Hsp70 uses ATP hydrolysis to help partition polyQ proteins into structures with varying levels of proteotoxicity. Our results thus support an emerging concept in which certain kinds of polyQ aggregates may be protective, while more soluble polyQ species are toxic.
Keywords: polyQ, protein misfolding, molecular chaperones, heat shock protein 70 (Hsp70), proteostasis, chemical genetics, chemical probes
INTRODUCTION
Protein misfolding is associated with many incurable diseases, including a large number of neurodegenerative diseases associated with the abnormal expansion of polyglutamine (polyQ) regions (1). For instance, Huntington’s disease (HD) is caused by the abnormal expansion of a polyQ region within the protein huntingtin (Htt) (2). This polyQ-expansion leads to the misfolding and aggregation of Htt, which impairs an array of cellular functions, such as transcriptional regulation (3, 4), protein quality control (5–7) and mitochondrial function (8). Thus, one promising approach to develop new therapies for this disease is to better understand how polyQ-expanded proteins are identified by the cell as misfolded, and how they are subsequently processed, sequestered, detoxified and degraded.
Protein quality control, or proteostasis, describes all cellular processes that execute the proper synthesis, folding, refolding, localization, complex formation, and degradation of proteins (9). Hence, proteostasis can be regarded as one of the first lines of defense against protein misfolding, including the misfolding and aggregation of polyQ-expansion proteins. Accordingly, manipulating cellular proteostasis with small molecules has been proposed to be a promising avenue toward correcting protein misfolding and alleviating its detrimental consequences in many different diseases (9), including HD and other polyQ-expansion diseases (10). The major challenge in this area of research remains our relatively poor understanding of which proteins within the proteostasis network will make effective therapeutic targets and, moreover, how the activity of these factors can be rationally controlled to safely produce the desired outcomes.
Among the central elements of proteostasis are the molecular chaperones, particularly the heat shock proteins (Hsps) (11). Heat shock protein 70 (Hsp70) is a major triage chaperone that facilitates many crucial aspects of proteostasis, including the proper folding of polypeptide substrates (12). Additionally, Hsp70s facilitate clearance of misfolded substrates through both the autophagic pathway and the ubiquitin-proteasome system (UPS) (13).
The numerous chaperone activities of Hsp70 are thought to be linked to its enzymatic activity and its ability to directly bind to misfolded or unfolded proteins (14). In brief, Hsp70 is composed of a nucleotide-binding domain (NBD) that is attached to a substrate-binding domain (SBD). Hydrolysis of ATP in the NBD triggers an inter-domain allosteric change that enhances substrate affinity in the SBD. This conversion is stimulated by co-chaperones of the Hsp40 (or J protein) class, which stimulate Hsp70’s ATPase activity and promote substrate binding (15). Thus, members of the Hsp70 family function together with co-chaperones, using cycles of ATP hydrolysis to regulate substrate binding. Despite the wealth of information about Hsp70 structure and biochemistry, the link between ATP turnover and the ultimate cellular fate of Hsp70-bound substrates is still not clear (16).
Numerous genetic and biochemical studies have implicated Hsp70s in regulating misfolding and toxicity in HD and other polyQ expansion diseases (17). For instance, increasing the expression of Hsp70s reduces polyQ-associated toxicity in mouse, fly and tissue culture models (18–21). While these are intriguing observations, it is not yet clear how they might inform potential pharmacological treatments. One possibility is to identify drug-like molecules that enhance Hsp70 expression; for example, by stimulating a stress response and elevating chaperone expression (18, 19). This approach might be expected to phenocopy the favorable effects of Hsp70 over-expression, but the long-term consequences of elevating chaperone levels are not yet known. A complementary, and possibly synergistic, approach may be to “tune” the activity of Hsp70, by directly targeting its ATPase activity or its interactions with co-chaperones (22). It still remains unclear, however, how the activities of Hsp70 might be best harnessed to combat toxicity associated with polyQ expansion proteins. Should ATPase activity be reduced or increased? Should binding to substrates be increased or decreased? Uncovering the mechanistic details of the relationship between Hsp70 and polyQ-expansion proteins will be critical for enabling translational opportunities.
Recent chemical biology efforts have offered intriguing new research tools for addressing a subset of these questions by allowing the tuning of Hsp70’s ATPase activity. For example, high-throughput chemical screens have identified membrane-permeable small molecules that can either enhance or reduce the ATPase activity of Hsp70 without altering its expression level (16, 23, 24). Other compounds have been identified that change the way Hsp70 interacts with its co-chaperones and substrates (25–29). These compounds have been useful in exploring the relationships between Hsp70 and misfolded proteins (22).
In the present study, we utilize a pair of chemical modulators of Hsp70, SW02 and CE12, to specifically investigate the relationships between the ATPase activity of Hsp70 and the proteotoxicity of a misfolded polyQ-expansion protein. SW02 is a stimulator of the ATPase activity of Hsp70 (30, 31) and it belongs to a family of dihydropyrimidines that promote the interaction of J protein co-chaperones with Hsp70s (25). In contrast, a structurally related compound CE12 (32) was identified as an inhibitor of Hsp70 (see below). We rationalized that using SW02 and CE12 side-by-side might provide details about Hsp70’s potential as a drug target in polyQ diseases. Further, it might reveal how Hsp70 makes triage decisions related to the accumulation of misfolded polyQ substrates. Towards those goals, we employed polyQ-expansion proteins consisting of an amino-terminal fragment of Htt with a range of polyglutamine lengths in a well-established yeast model of polyQ misfolding and polyQ toxicity (33–36).
We found that pharmacologically stimulating ATPase activity with SW02 increased polyQ toxicity and decreased aggregation, while CE12 inhibited ATPase activity, exacerbated aggregation and reduced toxicity. CE12 also protected against polyQ-associated toxicity in a mammalian PC12 model (37). Together, these studies suggest that the ATPase activity of Hsp70 is a potential drug target and an important modulator of polyQ toxicity in the context of the intact proteostasis network in living cells. This work also exemplifies how focused pairs of small molecules can be exploited to decipher complex mechanisms of Hsp70 biology.
RESULTS AND DISCUSSION
SW02 and CE12 modulate Hsp70’s ATPase activity
In this study, we focus on the dihydropyrimidine SW02 and the related dihydropyridine CE12 (Figure 1a) (25, 38, 39). We first evaluated their impact on J protein-stimulated Hsp70 activity, using human Hsp72 (HSP1A1). In vitro, SW02 (100 μM) stimulated ATPase activity to about 140% of untreated levels, while CE12 (100 μM) reduced ATPase activity by ~50% (Figure 1b). These results are consistent with the known activities of these and similar compounds against both prokaryotic and other eukaryotic Hsp70s (23, 25). Also consistent with previous findings (39), SW19 had no effect and, thus, it was used as a control in the ensuing experiments (Figure 1b).
Figure 1. Chemical modifiers of Hsp70 ATPase activity.
a) The chemical structures of SW02, SW19 (inactive control), and CE12 are show. b) Human Hsp72 (HSPA1a) ATPase assays reveal that SW02 stimulates nucleotide hydrolysis, while the related compounds SW19 and CE12 are inactive or inhibitory, respectively. Results are the average of triplicates ± SEM.
SW02 and CE12 are not toxic and they do not elicit stress responses in yeast
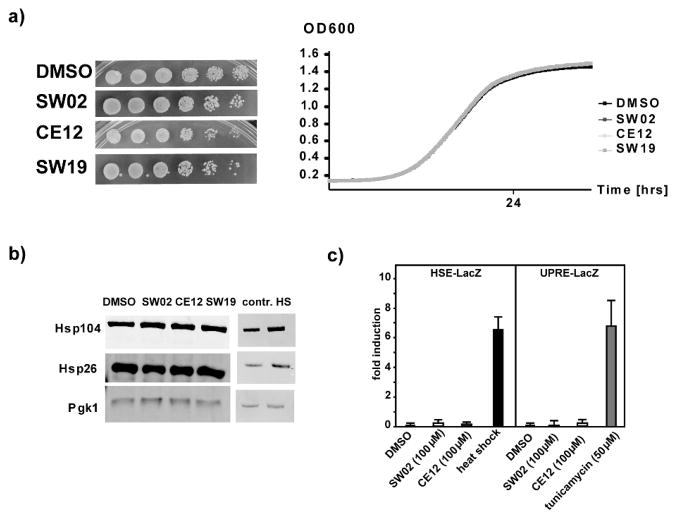
Hsp70 is central to proteostasis and genetic manipulation of Hsp70 protein levels often impairs cellular functions, complicating experiments to explore its mechanisms. We therefore sought to determine whether acute treatments with SW02 and CE12 had toxic effects on yeast cells. We found that even high concentrations of SW02 and CE12 (up to 100 μM) did not alter yeast growth on plates or in liquid cultures (Figure 2a). Thus, these chemical probes were not toxic to yeast cells at the concentration and time regimes used in these studies.
Figure 2. SW02, SW19, and CE12 are not toxic to yeast cells and do not elicit cellular stress responses.
a) Left panel: yeast cells were spotted in six fivefold dilutions (from left to right) on SD plates containing DMSO as a control or 100μM of the indicated small molecules. Right panel: growth of liquid yeast cultures containing DMSO as a control or 100μM of the indicated small molecules was monitored by BioscreenC b) Western blots of protein lysates from yeast cells that were grown in the presence of DMSO as a control or 100μM of the indicated small molecules for 8 hours. The blots were probed with anti-Hsp104 and anti-Hsp26 antibodies as indicators of a heat shock response. Anti-Pgk1 antibody served as a loading control. The right panel shows Western blots of protein lysates from heat shocked yeast cultures. c) β-galactosidase reporter assays (HSE for heat shock and UPRE for unfolded protein response) of yeast cultures treated with the indicated concentrations of small molecule or heat shocked.
We next tested whether the treatment of yeast cells with SW02 or CE12 elicited cellular stress responses. This question was important because inhibitors of the C-terminal domain of the chaperone, Hsp90, cause a strong stress response (40), thereby complicating the exploration of mechanistic chaperone activities in living cells. First, we tested whether SW02 or CE12 induced a heat shock response (HSR), a highly conserved response to protein folding stress in the cytosol of eukaryotic cells (41). In yeast, the proteins Hsp104 and Hsp26 are strongly increased in response to stress, providing a convenient measure of the HSR (42). We found that treatment of yeast cells with either SW02 or CE12 did not elevate the levels of these chaperones, suggesting that they do not trigger a HSR (Figure 2b). Next, these results were confirmed using a commonly used reporter plasmid that allows expression of β-galactosidase (encoded by the lacZ gene) under the transcriptional control of heat shock elements (HSE-lacZ) (43). As shown in Figure 2c, neither SW02 (100 μM) nor CE12 (100 μM) triggered an increase in β-galactosidase activity. Similar findings (Figure 2c) were observed in cells harboring the UPRE-lacZ plasmid that reports on the unfolded protein response (UPR) in the endoplasmic reticulum (ER) (44). In agreement with previous studies (23, 25), these results suggest that SW02 or CE12 did not have general toxic or stress-inducing effects, supporting their suitability for exploring the relationships between Hsp70 activity and polyQ-related processes while avoiding the complications of toxicity and global alterations in chaperone levels.
Pharmacological manipulation of Hsp70 regulates polyQ toxicity
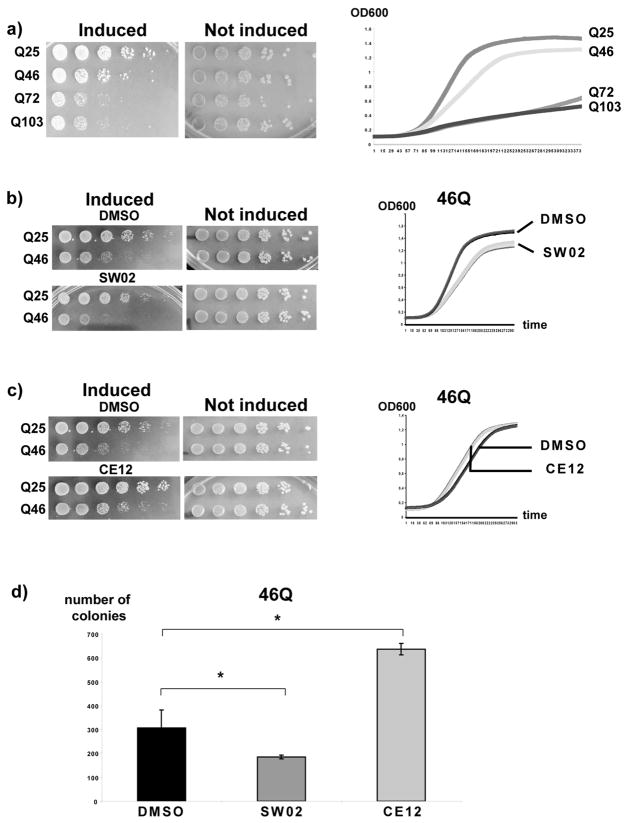
We next used SW02 and CE12 to investigate how Hsp70 might modulate polyQ toxicity. For these experiments, a yeast model was used that recapitulates major hallmarks of polyQ pathology, such as a graded polyQ length-dependent aggregation and toxicity (33, 34, 36) (Figure 3a).
Figure 3. SW02 exacerbates and CE12 antagonizes polyQ toxicity.
a) Left panel: yeast cells expressing 25Q, 46Q, 72Q, or 103Q polyQ Htt were spotted in six fivefold dilutions (left to right) on plates containing galactose as sole carbon source for the induction of the expression of the polyQ Htt proteins (induced) or on plates with glucose as sole carbon source for the repression of the polyQ Htt proteins (not induced). Right panel: growth of liquid yeast cultures expressing 25Q, 46Q, 72Q, or 103Q polyQ Htt was monitored by BioscreenC. b) Left panel: yeast cells expressing 25Q or 46Q polyQ Htt were spotted in six fivefold dilutions (left to right) on plates containing galactose and DMSO or 100μM of SW02. Right panel: growth of liquid yeast cultures expressing 25Q or 46Q polyQ Htt in the presence of DMSO or 100μM of SW02 was monitored by BioscreenC. The growth curves for four different cultures for each condition are shown. c) Left panel: yeast cells expressing 25Q or 46Q polyQ Htt were spotted in six fivefold dilutions (left to right) on plates containing galactose and DMSO or 50μM of CE12. Right panel: growth of liquid yeast cultures expressing 25Q or 46Q polyQ Htt in the presence of DMSO or 50μM of CE12 was monitored by BioscreenC. The growth curves for four different cultures for each condition are shown. d) Yeast cells were plated on SD plates after 8 hour incubation in media containing galactose (for the induction of 46Q) and DMSO, 50μM Ce12, or 100μM SW02. The number of colony forming units was determined from three independent cultures. The error bars represent SEM. *p<0.05 (two-tailed T-test).
We found that SW02, the stimulator of ATPase activity, enhanced the toxicity of 46QHtt on both plates (Figure 3b; left panel) and in liquid cultures (Figure 3b; right panel). This effect was specific for the longer length polyglutamine, 46QHtt, because identical treatments had no effect on cells expressing the control protein 25QHtt. By contrast, the inhibitor CE12 protected against 46QHtt toxicity to a reproducible albeit modest extent (Figure 3c). Like SW02, CE12 did not alter the growth of the cells expressing 25QHtt, suggesting that it did not generally promote cell growth. Similar conclusions were reached by quantifying colony number (Figure 3d): CE12 protected against toxicity and SW02 increased toxicity. Together, these experiments suggest that pharmacologically promoting Hsp70’s ATPase activity increased polyQ toxicity, while inhibiting this activity suppressed polyQ toxicity.
SW02 and CE12 modulate polyQ aggregation
To determine if these striking changes in polyQ toxicity correlated with any changes in polyQ aggregation, the effects of SW02 and CE12 on formation of polyQ foci were explored by fluorescence microscopy. By quantifying the number and type of fluorescent puncta, we found no clear correlation with how SW02 and CE12 modulate polyQ toxicity (Supplementary Figure 1). Thus, we concluded that visible aggregate size (as determined by light microscopy) is not a good indicator of proteotoxicity, consistent with some other recent models.
Because of the limits of fluorescence microscopy, we instead measured the solubility of polyQ aggregates using both SDD-AGE (semi-denaturating detergent agarose gel electrophoresis) and filter retardation assays. These methods are particularly effective in differentiating between soluble and insoluble protein species, which we reasoned might be a better predictor of toxicity. In the SDD-AGE platform, insoluble aggregates form in a polyQ length-dependent manner, with 25Q being mostly soluble, 72Q and 103Q being largely insoluble and the intermediate lengths, 46Q being mixtures of soluble and insoluble (Figure 4a, left panel). Based on SDD-AGE experiments, cells expressing 46Q had more soluble polyQ protein when treated with SW02 (Figure 4a, middle panel). Moreover, even the strongly aggregation-prone 103Q was rendered more soluble by this compound (Figure 4a, right panel). Likewise, in the filter retardation assays, SW02 decreased the amount of insoluble 46Q Htt protein by ~50%. When CE12 was tested in these same experimental platforms, CE12 showed no strong effect in the SDD-AGE experiments, but it greatly enhanced the amount of insoluble 46Q by about 2- to 3-fold in the filter trap assays (Figure 4b).
Figure 4. SW02 and CE12 alter the aggregation of polyQ-expanded Htt.
a) SDD-AGE prepared with protein lysates from yeast cells expressing the indicated polyQ Htt constructs induced by growth in medium containing galactose as sole carbon source for 8 hours. Left panel: SDD-AGE prepared with protein lysates from yeast cells expressing 25Q, 46Q, 72Q, or 103Q polyQ. SDD-AGE prepared with protein lysates from yeast cells expressing 46QHtt (middle panel) or 103QHtt (right panel) in the presence of 100μM SW02 or 50μM CE12 or DMSO. b) Filter retardation assay of undiluted and 1:5 diluted protein lysates of yeast cells expressing 46Q Htt treated as in a). The lower panels in a) and b) show the quantification of the signals in the SDD-AGE experiments or the filter retardation assay, respectively.
Together, these data suggest that SW02 enhanced the solubility of polyQ-expanded Htt, whereas CE12 promoted insolubility. Thus, we speculate that SW02 redistributes polyQ proteins into more toxic, soluble material, while CE12 shuttles polyQ proteins into less toxic and less soluble aggregates. These findings point to a model in which Hsp70 and its ATPase activity play an important role in partitioning polyQ proteins into various types of inclusions with discrete physical properties and distinct effects on cellular viability (45, 46).
SW02 and CE12 modulate polyQ toxicity and aggregation in PC12 cells
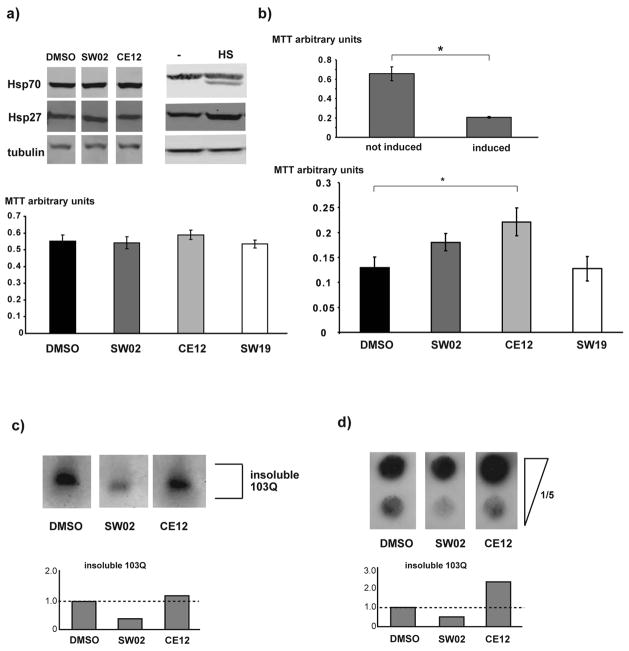
To further explore this model, the effects of SW02 and CE12 on polyQ aggregation and toxicity were tested in rat PC12 cells. This model allows the inducible expression of 103QHtt fused to GFP and it recapitulates major aspects of polyQ aggregation and toxicity (37). We tested whether SW02 and CE12 (10 μM) elicited a HSR in PC12 cells by measuring Hsp70 and Hsp27 levels. SW02 and CE12 did not elicit a HSR (Figure 5a, upper panel). Using an MTT assay for cell viability, we determined that SW02 and CE12 do not produce any toxicity in PC12 cells at concentrations of 20 μM and below (Figure 5a lower panel) suggesting that (as was observed in yeast) these compounds are suitable for studying Hsp70-polyQ interactions in mammalian cells.
Figure 5. SW02 and CE12 modulate polyQ toxicity and aggregation in PC12 cells.
a) Upper panel: Western blots with protein lysates from PC12 cells that were treated with DMSO, 20μM CE12, or 20μM SW02. The blots were probed with anti-Hsp27 or anti Hsp70 antibodies to monitor the heat shock response. Anti-tubulin antibody served as a loading control. The right panel shows Western blots with protein lysates from heat shocked PC12 cells. Lower panel: MTT assays of PC12 cells treated with 20μM of the indicated drugs and DMSO as a control. b) Upper panel: MTT assay of PC12 cells that were not induced or induced for the expression of 103Q for 14 hrs. Lower panel: MTT assays of PC12 cells that were induced for the expression of 103Q Htt for 12 hours and treated with DMSO, 10 μM CE12, 10 μM SW02, or 10 μM SW19. The average of three independent experiments is shown. The error bars represent SEM. * p<0.05 (two-tailed T-test). c) and d) SDD-AGE and filter retardation assays prepared with protein lysates from PC12 expressing 103QHtt (8 hours induction) in the presence of DMSO, 10 μM SW02, and 10 μM CE12.
Treatment of PC12 cells (Figure 5b, upper panel) revealed that neither SW02 nor the control compound SW19 had a statistically significant effect on polyQ toxicity, while treatment with CE12 significantly improved survival (Figure 5b, lower panel). Notably, CE12 only reduced polyQ toxicity when added simultaneously with the induction of 103QHtt (data not shown), consistent with previous kinetic studies suggesting a relatively early role for Hsp70 in polyQ processing in yeast (47).
In addition to effects on viability, the PC12 model allows insight into effects of CE12 and SW02 on polyQ aggregation. As we observed with the yeast studies, there was poor correlation between toxicity and aggregation by fluorescence microscopy (Supplemental Figure 1f). However, SDD-AGE experiments and filter retardation assays demonstrated that SW02 decreased the fraction of insoluble 103QHtt protein in both assays, while CE12 showed an increase in insoluble 103QHtt in the filter trap experiments (Figure 5c and d). Thus, in the PC12 model, CE12 reduced polyQ toxicity and increased aggregation, while SW02 did not significantly alter toxicity and decreased aggregation. Notably, other recent studies also suggest a protective role for aggregates (48, 49), consistent with the CE12 findings. However, we hypothesize that the polyglutamine expansion in the PC12 model may be already too dramatic for its phenotypes to be further enhanced by this compound. In that context, it was exciting to find that CE12 was still able to partially suppress toxicity in this mammalian cell model.
Conclusions
Polyglutamine expansion disorders are a collection of untreatable neurodegenerative diseases characterized by the accumulation of misfolded proteins that contain a stretch of consecutive glutamines (2). Although genetic experiments have linked the molecular chaperone, Hsp70, to the cellular processing of polyQ-containing proteins, the mechanistic details of this interaction, particularly in the complex setting of a living cell, remain unclear. One objective in better characterizing this relationship is to better understand how (and if) Hsp70 might be used as a putative therapeutic target for these diseases. Specifically, what are the pharmacological relationships between Hsp70’s ATPase activity and polyQ toxicity? Does Hsp70 uniformly protect against toxicity or can its activity actually promote aberrant accumulation of toxic intermediates? These are important questions in understanding Hsp70 biology and, in turn, in learning how to manipulate this chaperone in the context of polyQ expansion disorders.
Here, we employed a pair of newly discovered chemical reagents to tune the ATPase activity of Hsp70 and explore the results of this change on polyQ-related phenotypes in yeast and cultured mammalian cells (PC12). This pair of molecules forms a compelling duo because SW02 stimulates Hsp70s ATPase activity and CE12 inhibits it (Figure 1). Importantly, we found that SW02 and CE12 do not alter Hsp70 expression or stimulate a stress response (Figures 2 and 5) and, therefore, they facilitate insights at physiological chaperone levels.
Surprisingly, we found that stimulating Hsp70 function with SW02 in yeast increased the proportion of more soluble, toxic polyQ aggregates, whereas inhibiting Hsp70’s ATPase with CE12 decreased toxicity. This is an intriguing and somewhat counterintuitive finding because it suggests a possible detrimental role for Hsp70’s activities on the toxicity of aggregated proteins. In other words, “reducing” Hsp70 function with CE12 was protective, while “increasing” its function exacerbated proteotoxicity. At first glance, this result is in contrast with findings from genetic over-expression of Hsp70, in which over-expressing Hsp70 was protective (20, 50). How does one rectify these ideas? One interesting clue might exist in the biochemistry of Hsp70. As mentioned above, the ATP-bound form of Hsp70 has a poor affinity for substrates, while nucleotide hydrolysis improves binding through an allosteric rearrangement (51–53). Thus, some compounds that inhibit ATP turnover would be expected to alter the ratio of free- and bound-substrate by favoring the weak-affinity, ATP-bound form. Based on the biochemistry of the chaperone system, we expect that pharmacological “inhibition” of Hsp70 might, in some cases, produce a phenotype that would be distinct from genetic deletion and, likewise, “activators” would not necessarily phenocopy all aspects of over-expression. More specifically to the case of polyQ proteins, if treatment with SW02 favors accumulation of the ADP-bound form, then it might enhance affinity for the substrate and lead to a net retention of polyQ in early aggregation states. In fact, recent studies suggest that holding other polyQ substrates in the Hsp70-bound form delays proper processing and assembly into aggregates (47). Conversely, inhibition of ATPase activity by CE12 might lead to net release of the substrate, which might facilitate faster progression through the aggregation pathway, thus better bypassing proteotoxic intermediates. Consequently, in this model, there may be a balance between substrate dwell time on Hsp70 and the fate of the substrate. Clearly, many questions about this simplistic model remain unanswered, but we find it intriguing to consider that under specific circumstances, Hsp70 may play a counter-productive role in the processing of polyQ-expansion proteins and possibly also other amyloid-forming proteins.
The toxicity of polyQ-expansion proteins in yeast and other experimental models has been documented to originate from defects in many different cellular functions, such as defects in ER protein quality, defects in endocytosis, defects in protein degradation by the ubiquitin proteasome system, and defects in transcriptional regulation, to name only but a few examples. Future experiments will explore whether SW02 exacerbates and CE12 alleviates all cellular polyQ-induced defects or whether SW02 and CE12 only modulate specific cellular polyQ-induced defects. These experiments may help to determine the cellular mechanisms by which Hsp70 modulates polyQ misfolding and its ensuing toxicity. Yet regardless of the exact molecular and cellular mechanisms that are at work, our results presented here support future translational studies of Hsp70 inhibitors – possibly more so than activators - as potential agents for reducing polyQ toxicity.
Materials and Methods
Chemicals
Tunicamycin was purchased from Sigma-Aldrich.
Synthesis of Hsp70 activators and inhibitors
Compounds SW02 and SW19 were synthesized as described (39). Ethyl 4-(2,4-dichlorophenyl)-2,7,7-trimethyl-5-oxo-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (CE12) was synthesized based on an earlier report (32, 54). To a flame dried round bottom flask (5 mL), 84 mg of dimedone (1.5 eq, 0.6 mmol), 14 mg Yb(OTf)3 (10 mol%), and 0.05 mL ethylacetoacteate (1 eq, 0.4 mmol) were added to 1 mL of acetonitrile, which had been dried over sodium sulfate. The components were stirred at room temperature for 10 minutes, followed by addition of 2,4-dichlorobenzaldehyde (1 eq, 70 mg, 0.4 mmol) and ammonium acetate (1 eq, 39 mg, 0.4 mmol). Reaction proceeded over 3–5 hrs under argon before precipitation by the addition of a water/ice mixture (~2 mL). Precipitation was completed over 1 hour in an ice bath and the resulting precipitate filtered and recrystallized in an ethanol/water system to generate white crystals (87–94% yield). m/z: expected [M+H] 408.11; observed [M+H] 408.1; 1H NMR (Varian 400 MHz; dDMSO): δ 9.184 (s, 1H), 7.388 (d, 1H), 7.319 (s, 1H), 7.10–7.25 (dd, 1H), 5.18 (s, 1H), 3.961–3.986 (m, 2H), 2.287 (s, 3H), 2.13–2.201 (t, 2H), 2.098 (s, 2H), 1.934–1.974 (d, 2H), 1.119 (s, 3H), 1.04 (s, 3H), 0.876 (s, 3H).
ATPase activity assay
The malachite green-based assay for phosphate release by human Hsp72 was carried out as described (55). H. sapiens Hsp72 (HSP1A1) is 74% identical (86% similar) to S. cerevisiae Ssa1p and we used the human protein as a surrogate in the ATPase assays. To improve the malachite green assay signal, Hsp72 (1 μM) was stimulated by human DnaJA2 (0.5 μM). Experiments were performed in triplicate using compounds dissolved in DMSO (final concentration <1% (v/v)).
Yeast strains
Yeast strains in the W303 background (MATa, leu2-3,112, his3-11, trp1-1, ura3-1, can1-100, ade2-1) were used for almost all experiments. The yeast strains expressing the polyQ-expanded Htt fragment were described previously (33, 34, 56, 57). The yeast strain expressing the Ssa1-GFP (BY4743 background) fusion protein was described previously (58). Yeast cells were grown in standard SD media.
Antibodies
The anti-Hsp104 antibody was a kind gift from Susan Lindquist’s laboratory. The anti-Hsp27 antibody was a kind gift from Johannes Buchner’s laboratory. The anti-Hsp70 antibody (3A3) was purchased from Santa Cruz Biotechnology, the anti-Pgk1 antibody was purchased from Molecular Probes/Invitrogen, the anti-GFP antibody was purchased from Sigma-Aldrich, and the anti-tubulin antibody was purchased from Cedarlane.
Growth assay
All growth assays of yeast cells cultures expressing polyQ proteins (spotting assays, growth curves of liquid cultures, and plating assays) were carried out as described previously (59). All assays were performed in at least three independent experiments using three independent yeast colonies to avoid problems with spontaneous suppressors of polyQ toxicity.
Protein extraction and Western blots
Proteins from yeast cells were extracted and Western-blots were carried out as described previously (56). Proteins from PC12 cells were extracted as described previously (57).
β-galactosidase assays
Wild type yeast (W303) was transformed with plasmids including genes expressing the Unfolded Protein Response (UPRE) (44) and Heat Shock Element (HSE) (43) fused to a lacZ reporter. The cells were grown to mid-log phase in selective media followed by dilution to OD600 0.4 and addition of SW02 to a concentration of 100 μM. The cells were grown for another 3.5 h at 30 °C before running a β-galactosidase assay following the manufacturer’s instructions (ThermoFisher Sci.). For the heat shock control, cells were transferred to 39 °C after 2.5 h. and grown at 39 °C for 1 h. before the β-galactosidase assay.
SDD-AGE and Filter Retardation Assay
Protein lysis and the ensuing SDD-AGE experiments were carried out as described previously using an anti-GFP antibody (60). Filter retardation experiments were carried out as described previously using an anti-GFP antibody (59). The SDD-AGE and filter retardation experiments were quantified using NIH Image J software.
Supplementary Material
Acknowledgments
The authors thank Amy Chang for kindly providing the stress reporter constructs. This work was supported by grants from the NIH (NS059690) to J.E.G., predoctoral training grants to A.D.T. (AG354642), C.G.E. (GM008353) and L.N.M. (GM007767), and an AFAR grant and a grant by the Wood foundation to MLD.
Footnotes
This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Soto C, Estrada LD. Protein misfolding and neurodegeneration. Arch Neurol. 2008;65:184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald ME, Gines S, Gusella JF, Wheeler VC. Huntington's disease. Neuromolecular Med. 2003;4:7–20. doi: 10.1385/NMM:4:1-2:7. [DOI] [PubMed] [Google Scholar]
- 3.Bithell A, Johnson R, Buckley NJ. Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington's disease. Biochem Soc Trans. 2009;37:1270–1275. doi: 10.1042/BST0371270. [DOI] [PubMed] [Google Scholar]
- 4.Truant R, Atwal RS, Burtnik A. Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease. Prog Neurobiol. 2007;83:211–227. doi: 10.1016/j.pneurobio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Li XJ, Li S. Proteasomal dysfunction in aging and Huntington disease. Neurobiol Dis. 43:4–8. doi: 10.1016/j.nbd.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voisine C, Pedersen JS, Morimoto RI. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis. 40:12–20. doi: 10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidal R, Caballero B, Couve A, Hetz C. Converging pathways in the occurrence of endoplasmic reticulum (ER) stress in Huntington's disease. Curr Mol Med. 11:1–12. doi: 10.2174/156652411794474419. [DOI] [PubMed] [Google Scholar]
- 8.Gibson GE, Starkov A, Blass JP, Ratan RR, Beal MF. Cause and consequence: mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases. Biochim Biophys Acta. 1802:122–134. doi: 10.1016/j.bbadis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 10.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 11.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 12.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 13.Kettern N, Dreiseidler M, Tawo R, Hohfeld J. Chaperone-assisted degradation: multiple paths to destruction. Biol Chem. 391:481–489. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- 14.Young JC. Mechanisms of the Hsp70 chaperone system. Biochem Cell Biol. 88:291–300. doi: 10.1139/o09-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK) J Biol Chem. 285:21282–21291. doi: 10.1074/jbc.M110.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barral JM, Broadley SA, Schaffar G, Hartl FU. Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, Chalfant MA, Saldanha SA, Hodder P, Tait BD, Garza D, Balch WE, Morimoto RI. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 8:185–196. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trott A, West JD, Klaic L, Westerheide SD, Silverman RB, Morimoto RI, Morano KA. Activation of heat shock and antioxidant responses by the natural product celastrol: transcriptional signatures of a thiol-targeted molecule. Mol Biol Cell. 2008;19:1104–1112. doi: 10.1091/mbc.E07-10-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummings CJ, Sun Y, Opal P, Antalffy B, Mestril R, Orr HT, Dillmann WH, Zoghbi HY. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum Mol Genet. 2001;10:1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- 21.Wacker JL, Huang SY, Steele AD, Aron R, Lotz GP, Nguyen Q, Giorgini F, Roberson ED, Lindquist S, Masliah E, Muchowski PJ. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington's disease. J Neurosci. 2009;29:9104–9114. doi: 10.1523/JNEUROSCI.2250-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem. 53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinwal UK, Miyata Y, Koren J, 3rd, Jones JR, Trotter JH, Chang L, O'Leary J, Morgan D, Lee DC, Shults CL, Rousaki A, Weeber EJ, Zuiderweg ER, Gestwicki JE, Dickey CA. Chemical manipulation of hsp70 ATPase activity regulates tau stability. J Neurosci. 2009;29:12079–12088. doi: 10.1523/JNEUROSCI.3345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata Y, Chang L, Bainor A, McQuade TJ, Walczak CP, Zhang Y, Larsen MJ, Kirchhoff P, Gestwicki JE. High-throughput screen for Escherichia coli heat shock protein 70 (Hsp70/DnaK): ATPase assay in low volume by exploiting energy transfer. J Biomol Screen. 15:1211–1219. doi: 10.1177/1087057110380571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wisen S, Bertelsen EB, Thompson AD, Patury S, Ung P, Chang L, Evans CG, Walter GM, Wipf P, Carlson HA, Brodsky JL, Zuiderweg ER, Gestwicki JE. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem Biol. 5:611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadhwa R, Sugihara T, Yoshida A, Nomura H, Reddel RR, Simpson R, Maruta H, Kaul SC. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000;60:6818–6821. [PubMed] [Google Scholar]
- 27.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fewell SW, Day BW, Brodsky JL. Identification of an inhibitor of hsc70-mediated protein translocation and ATP hydrolysis. J Biol Chem. 2001;276:910–914. doi: 10.1074/jbc.M008535200. [DOI] [PubMed] [Google Scholar]
- 29.Chang L, Miyata Y, Ung PM, Bertelsen EB, McQuade TJ, Carlson HA, Zuiderweg ER, Gestwicki JE. Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem Biol. 18:210–221. doi: 10.1016/j.chembiol.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisen S, Gestwicki JE. Identification of small molecules that modify the protein folding activity of heat shock protein 70. Anal Biochem. 2008;374:371–377. doi: 10.1016/j.ab.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Chang L, Bertelsen EB, Wisen S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 32.Evans CG, Gestwicki JE. Enantioselective organocatalytic Hantzsch synthesis of polyhydroquinolines. Org Lett. 2009;11:2957–2959. doi: 10.1021/ol901114f. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci U S A. 2006;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giorgini F, Muchowski PJ. Exploiting Yeast Genetics to Inform Therapeutic Strategies for Huntington's Disease. Methods Mol Biol. 2009;548:161–174. doi: 10.1007/978-1-59745-540-4_9. [DOI] [PubMed] [Google Scholar]
- 36.Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aiken CT, Tobin AJ, Schweitzer ES. A cell-based screen for drugs to treat Huntington's disease. Neurobiol Dis. 2004;16:546–555. doi: 10.1016/j.nbd.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Evans CG, Jinwal UK, Makley LN, Dickey CA, Gestwicki JE. Identification of dihydropyridines that reduce cellular tau levels. Chem Commun (Camb) 47:529–531. doi: 10.1039/c0cc02253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wisen S, Androsavich J, Evans CG, Chang L, Gestwicki JE. Chemical modulators of heat shock protein 70 (Hsp70) by sequential, microwave-accelerated reactions on solid phase. Bioorg Med Chem Lett. 2008;18:60–65. doi: 10.1016/j.bmcl.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 40.Duerfeldt AS, Brandt GE, Blagg BS. Design, synthesis, and biological evaluation of conformationally constrained cis-amide Hsp90 inhibitors. Org Lett. 2009;11:2353–2356. doi: 10.1021/ol900783m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 42.Amoros M, Estruch F. Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol Microbiol. 2001;39:1523–1532. doi: 10.1046/j.1365-2958.2001.02339.x. [DOI] [PubMed] [Google Scholar]
- 43.Harshman KD, Moye-Rowley WS, Parker CS. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988;53:321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- 44.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 45.Duennwald ML. Polyglutamine misfolding in yeast: Toxic and protective aggregation. Prion. 5 doi: 10.4161/pri.5.4.18071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfe KJ, Cyr DM. Amyloid in neurodegenerative diseases: Friend or foe? Semin Cell Dev Biol. doi: 10.1016/j.semcdb.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter GM, Smith MC, Wisen S, Basrur V, Elenitoba-Johnson KS, Duennwald ML, Kumar A, Gestwicki JE. Ordered assembly of heat shock proteins, Hsp26, Hsp70, Hsp90, and Hsp104, on expanded polyglutamine fragments revealed by chemical probes. J Biol Chem. 286:40486–40493. doi: 10.1074/jbc.M111.284448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arrasate M, Finkbeiner S. Protein aggregates in Huntington's disease. Exp Neurol. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 50.Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 51.Revington M, Zhang Y, Yip GN, Kurochkin AV, Zuiderweg ER. NMR investigations of allosteric processes in a two-domain Thermus thermophilus Hsp70 molecular chaperone. J Mol Biol. 2005;349:163–183. doi: 10.1016/j.jmb.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 52.Slepenkov SV, Witt SN. Kinetic analysis of interdomain coupling in a lidless variant of the molecular chaperone DnaK: DnaK's lid inhibits transition to the low affinity state. Biochemistry. 2002;41:12224–12235. doi: 10.1021/bi0263208. [DOI] [PubMed] [Google Scholar]
- 53.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans CG, Jinwal UK, Makley LN, Dickey CA, Gestwicki JE. Identification of dihydropyridines that reduce cellular tau levels. Chem Commun (Camb) 2011;47:529–531. doi: 10.1039/c0cc02253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang L, Bertelsen EB, Wisén S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 56.Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005;280:23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duennwald ML, Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howson R, Huh WK, Ghaemmaghami S, Falvo JV, Bower K, Belle A, Dephoure N, Wykoff DD, Weissman JS, O'Shea EK. Construction, verification and experimental use of two epitope-tagged collections of budding yeast strains. Comp Funct Genomics. 2005;6:2–16. doi: 10.1002/cfg.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duennwald ML. Monitoring polyglutamine toxicity in yeast. Methods. 53:232–237. doi: 10.1016/j.ymeth.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 60.Halfmann R, Lindquist S. Screening for amyloid aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. J Vis Exp. 2008 doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.