Abstract
Sialidases hydrolytically remove sialic acids from sialylated glycoproteins and glycolipids. Sialidases are widely distributed in nature and sialidase-mediated desialylation is implicated in normal and pathological processes. However, mechanisms by which sialidases exert their biological effects remain obscure, in part because sialidase substrate preferences are poorly defined. Here we report the design and implementation of a sialidase substrate specificity assay based on chemoselective labeling of sialosides. We show that this assay identifies components of glycosylated substrates that contribute to sialidase specificity. We demonstrate that specificity of sialidases can depend on structure of the underlying glycan, a characteristic difficult to discern using typical sialidase assays. Moreover, we discovered that S. pneumoniae sialidase NanC strongly prefers sialosides containing the Neu5Ac form of sialic acid, versus those that contain Neu5Gc. We propose using this approach to evaluate sialidase preferences for diverse potential substrates.
Introduction
Sialic acid refers to a family of nine-carbon α-keto acids typically located at the non-reducing termini of many mammalian glycoproteins and glycolipids. Various sialic acids occur in nature,(1) including N-acetylneuraminic acid (Neu5Ac), the most common sialic acid in humans, N-glycolylneuraminic acid (Neu5Gc), produced by many non-human mammals, and deaminated sialic acid (2-keto-3-deoxy-D-glycero-D-galacto-nononic acid; KDN), produced by lower vertebrates and bacteria. In mammals, sialylated glycoconjugates, also called sialosides, play roles in physiological processes including cell differentiation, proliferation, apoptosis, and the immune response. Additionally, sialosides facilitate non-host interactions by serving as receptors for pathogens and secreted toxins.(2) The extent of glycoconjugate sialylation is regulated by sialyltransferases, which add sialic acid to glycoconjugates, and by sialidases, also called neuraminidases, which remove sialic acid from glycoconjugates. Sialidases hydrolytically cleave the ketosidic bond between sialic acid and an underlying glycan. These enzymes can regulate cell signaling through their ability to rapidly and dramatically change glycosylation of cell surface adhesion molecules, a transformation that can impact responses to extracellular stimuli.
Genes encoding sialidases are found in viral, bacterial, and eukaryotic genomes. Viral and bacterial sialidases play roles in pathogen infection of human hosts. In mammals, sialidase activities are critical to normal physiology, and changes in sialidase expression levels are associated with cancer, both in primary tumor samples and in cancer cell lines.(3) However, mechanistic link(s) between sialidase activity and cellular responses remain obscure. Improved knowledge of the substrate specificity of sialidases could reveal clues as to the regulatory processes that are normally controlled by sialidases and those that are disrupted by sialidase activity in infection and in cancer.
Routine measurements of sialidase activity are conducted using methylumbelliferyl-N-acetyl-α-D-neuraminic acid (4-MU-NANA), an artificial substrate consisting of a proto-fluorophore linked to Neu5Ac. Hydrolysis of the ketosidic bond by a sialidase liberates the fluorophore, enabling facile detection of sialidase activity by continuous fluorescence measurement. However, the 4-MU-NANA reagent cannot provide information about the sialidase’s specificity for sialic acid linkage or the underlying glycan to which the sialic acid is attached. Specificity studies require examination of sialidase activity on a variety of sialoside substrates. One approach is to quantify the amount of sialic acid released, either by direct measurement (4) or by coupled enzyme assays,(5) but the availability of defined substrates and the quantity of material required limit the scope of these approaches. Another strategy to studying sialidase specificity involves the use of lectin binding to detect changes in the sialylation state of immobilized glycans.(6, 7) However, low specificity and variable affinity of lectins limits their utility. As an alternative, two groups reported disaccharide substrates that are tagged with para-nitrophenyl or methylumbelliferone groups at the reducing terminus. Removal of sialic acid by a sialidase frees a tagged monosaccharide that can be hydrolyzed by excess glycosidase, and the liberated tag produces a quantitative UV or fluorescent signal.(8, 9) This approach yielded information about the substrate specificity of both bacterial and mammalian sialidases,(10, 11) but remains limited to disaccharide substrates.
Here we describe a simple sialidase specificity assay that circumvents limitations of existing assays. By employing a chemoselective reaction that labels any sialoside, this assay enables analysis of sialidase activity on a wide variety of sialosides, including relatively complex glycans. Implementing this assay, we demonstrate that the activities of a human cytosolic sialidase and a previously uncharacterized pneumococcal sialidase are influenced by multiple structural elements contained within the sialic acid and the underlying glycan.
Results and Discussion
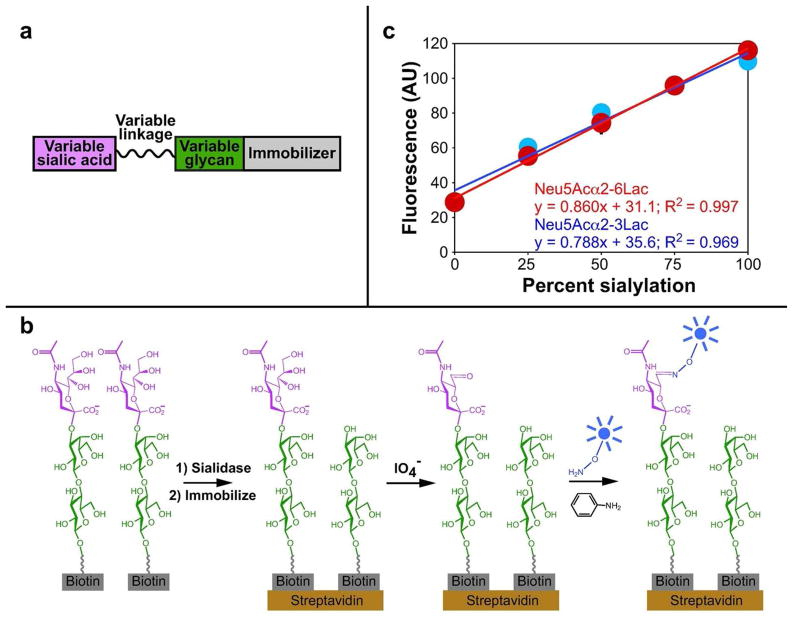
We sought a simple and reliable method to assess the sialylation level of a variety of glycans. Because the primary limitation of existing sialidase specificity assays is access to suitable substrates, the ideal method should be readily applicable to existing glycans that have been prepared to study the specificity of glycan-binding proteins.(12) Immobilized glycan arrays are widely used to rapidly assess the specificity of carbohydrate binding proteins(13–15). Potential hits are then validated by solution-based binding measurements. We envisioned an analogous approach to studying sialidase specificity: immobilized arrays of sialylated glycans could be subjected to sialidase treatment and the relative desialylation of potential substrates measured. Interesting trends in substrate specificity could be confirmed by further solution-based kinetics measurements. To measure the extent of desialylation, we turned to the periodate oxidation and aniline catalyzed ligation (PAL) method, which capitalizes on the unique reactivity of the sialic acid polyhydroxyl side chain.(16) Mild oxidation conditions selectively oxidize the C7 position of sialic acid to an aldehyde. Subsequent oxime formation by reaction with an aminooxy reagent is catalyzed by aniline, yielding a labeled sialic acid. This detection method is ideally suited to specificity studies because it is independent of the glycan to which sialic acid is attached and of most modifications to the sialic acid itself, although modifications to the C7 and C8 positions of sialic acid preclude its use. We reasoned that we could employ the PAL method to detect the ability of sialidases to desialylate a variety of biotinylated sialosides in which the sialic acid structure, sialic acid linkage, and structure of the underlying glycan were varied (Figure 1, panel a).
Figure 1. PAL method adapted to 96-well plate format.
(a) The sialidase specificity assay accommodates a variety of sialosides. The form of sialic acid, the nature of the linkage, and structure of the underlying glycan can be varied. (b) Glycans are incubated with sialidase, then immobilized. Sialic acid-containing glycoconjugates are fluorescently labeled by the periodate oxidation and aniline catalyzed ligation (PAL) method and detected. (c) The PAL method quantitatively detects sialylation. Biotinylated sialylated and unsialylated glycans were mixed in ratios indicated and immobilized. Sialylated glycans were labeled by periodate oxidation followed by aniline-catalyzed oxime ligation to Alexa Fluor 488. Error bars represent standard deviation of three trials.
First, we adapted the PAL reaction(16) for use in a 96-well format (Figure 1, panel b). We used glycans conjugated to biotin at the reducing end via a spacer unit (Sp). We immobilized each glycan in a single well of a streptavidin-coated plate, then oxidized by sodium periodate treatment. Oxidized glycans were reacted with an aminooxy detection reagent in the presence of aniline catalyst, using acidic conditions (pH 4.5) to promote the reaction.(17) Of the fluorophores examined, Alexa Fluor 488 provided the best sensitivity, yielding a 3- to 4-fold increase in fluorescence for sialylated glycans relative to unsialylated glycans (Supplementary Figure 1). In the absence of periodate oxidation, there was no difference in fluorescence signal between sialylated and unsialylated glycans, and in the absence of aniline the difference was greatly reduced (Supplementary Figure 2). Taken together, these results indicate that the detection method reliably discriminates sialylated and unsialylated glycans.
Next, we identified conditions where the fluorescence signal accurately reflected the amount of sialylated glycan. We prepared defined mixtures of Neu5Acα2-3Lac-Sp-biotin and Lac-Sp-biotin. We immobilized the mixtures in individual wells of a 96-well streptavidin-coated plate and detected sialylated molecules using the PAL method. In this format, achieving signal that accurately reflected the degree of sialylation required use of milder oxidation conditions (0.1 mM NaIO4) than used previously (1 mM NaIO4) for live cell labeling (Supplementary Figure 1).(16) Additionally, while introducing a step where the fluorescence signal was enzymatically amplified enhanced the fluorescence change, it resulted in a non-linear response (Supplementary Figure 1, panel d), so we chose to rely on direct detection with an aminooxyfluorophore conjugate. With optimized conditions, observed fluorescence signal related linearly to the fraction of sialylated glycan present (Figure 1, panel c). Sialic acid linkage did not affect the fluorescence signal and the signal was proportional to the fraction of glycan that was sialylated. Subsequent data are reported as “percent sialylation,” calculated for each experimental observation based on fluorescence signals of sialylated and unsialylated glycans (Supplementary Figure 3).
We evaluated two approaches to sialidase treatment, either preceding or following glycan immobilization (Supplementary Figure 4). We observed robust and, in some cases, complete desialylation of glycans when the glycans were sialidase-treated before immobilization, while using the same amount of sialidase after immobilization resulted in less desialylation. Furthermore, sialidase treatment prior to immobilization yielded a more proportional relationship between amount of sialidase added and the loss of fluorescence signal. We speculate that some aspect of the immobilization process sterically occludes enzyme access to glycan. Thus, all experiments were conducted with sialidase treatment of glycans preceding immobilization.
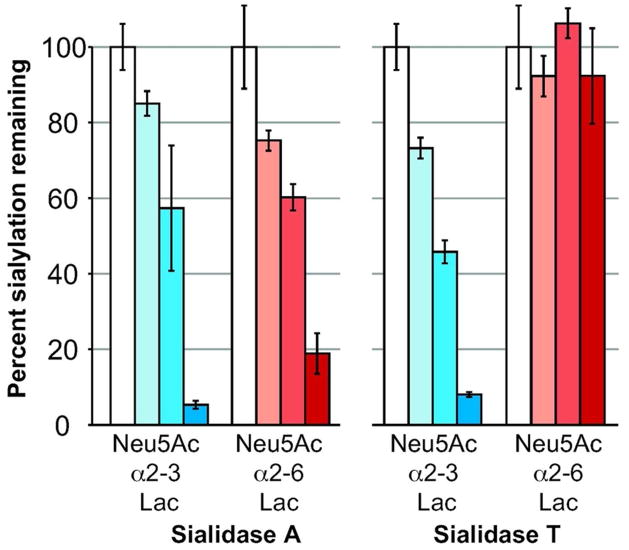
We investigated whether we could accurately detect sialidase specificity by examining two well-characterized bacterial sialidases. Sialidase A from Arthrobacter ureafaciens hydrolytically cleaves both α2-3- and α2-6-linked sialic acids.(18) Sialidase T from Salmonella typhimurium exhibits a strong preference for sialic acids in the α2-3-linkage.(19) Using Neu5Acα2-3Lac-Sp-biotin and Neu5Acα2-6Lac-Sp-biotin, we observed desialylation of both α2-3- and α2-6-linked sialosides by sialidase A, while sialidase T displayed a strong preference for α2-3-linked sialic acid (Figure 2).
Figure 2. Sialidase assay accurately characterizes specificity of bacterial sialidases.
Biotinylated Neu5Acα2-3Lac and Neu5Acα2-6Lac were incubated with Arthrobacter ureafaciens sialidase A (0, 0.55, 1.66, or 5 mU, bars left to right) or Salmonella typhimurium sialidase T (0, 6.4, 19.3, or 58 μU, bars left to right) for 2 h at 37 °C. The PAL method was used to determine the fraction of sialylated glycan remaining after sialidase treatment. Error bars represent standard deviation of three trials.
Next, we examined the human sialidase NEU2, also known as the cytosolic sialidase or the soluble sialidase.(20) Mammalian NEU2 is highly expressed in skeletal muscle, where it is implicated in myoblast differentiation,(21) and also present in other tissues. In vitro assays have demonstrated that NEU2 accepts glycoprotein, glycolipid, and oligosaccharide substrates,(22, 23) but its physiological substrates remain unknown. We measured the pH dependence of GSTNEU2 activity toward Neu5Acα2-3Lac-Sp-biotin (Figure 3, panel a). Consistent with prior reports, we found that GST-NEU2 exhibits optimal activity at pH 5.6.(10, 23) We also tested the effects of varying the buffer composition (Figure 3, panel b). GST-NEU2’s activity toward Neu5Acα2-3Lac-Sp-biotin did not depend on divalent ions. On the other hand, GST-NEU2 was inhibited by sialidase inhibitors 2-deoxy-2,3-didehydro-N-acetylneuraminic acid (DANA) and zanamivir (Relenza), but was not affected significantly by oseltamivir (Tamiflu), consistent with previous findings.(24)
Figure 3. Activity of GST-NEU2 depends on buffer composition.
(a) GST-NEU2 (1.13 mU) was incubated with Neu5Acα2-3Lac in buffer containing 200 mM sodium phosphate, 100 mM sodium acetate, 100 mM NaCl at the indicated pH, for 1 h at 37 °C. Error bars represent standard deviation of three trials. (b) GST-NEU2 activity (1.13 mU) was measured in 100 mM NaOAc, pH 5.6, 100 mM NaCl at 37 °C for 1 h. Buffer was varied by presence of 1 mM CaCl2, MgCl2, EDTA, zanamivir, DANA, or oseltamivir, as indicated. Error bars represent standard deviation of three trials.
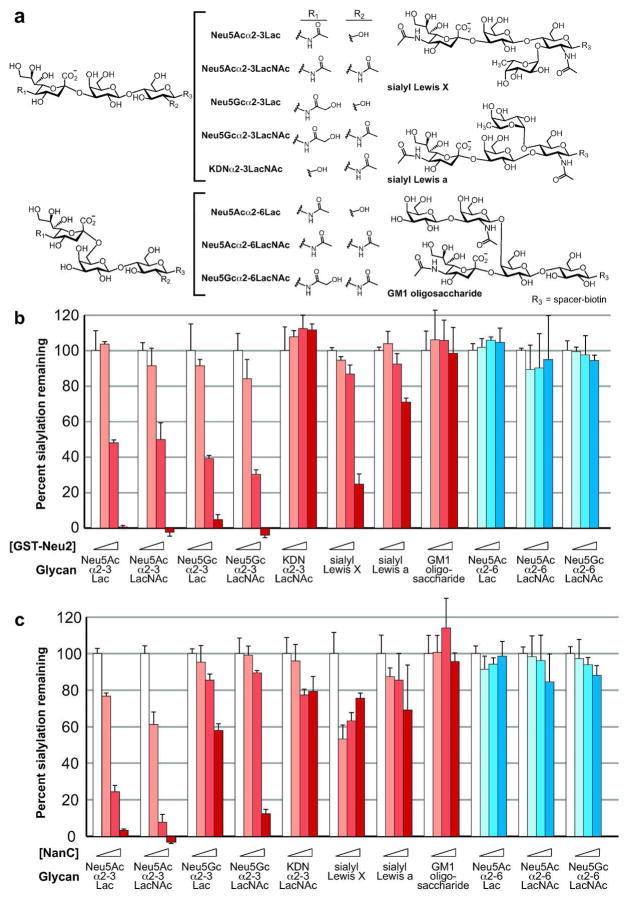
We examined the sialoside specificity of NEU2 using a small panel of potential substrates, with variable sialic acid linkage, sialic acid form, and underlying glycan structure (Figure 4, panel a). An early report describing the purification of cytosolic sialidase activity indicated that it prefers the α2-3-linkage,(25) and subsequent studies using recombinant NEU2 support that finding.(10, 23) However, using lysates from cells overexpressing mouse NEU2, Koda et al. observed activity toward both α2-3- and α2-6-linked sialosides.(26) Our data are consistent with the prevailing view, that NEU2 exhibits a strong preference for α2-3-linked sialic acids; we observed only minor activity toward the α2-6-linked sialosides (Figure 4, panel b). We examined NEU2’s tolerance of different forms of sialic acid. Consistent with the findings of others,(10) we observed that GST-NEU2 readily hydrolyzes both Neu5Ac and Neu5Gc, but not the deaminated sialic acid, KDN. We examined how sialidase activity depends on the structure of the underlying glycoconjugate and found that GST-NEU2 did not strongly discriminate between Lac- or LacNAc-based glycans. Although GST-NEU2 readily accepted Neu5Acα2-3LacNAc-Sp-biotin, it showed diminished activity toward sialyl Lewis X, in which fucose is attached to the N-acetylglucosamine (GlcNAc) residue. Furthermore, sialyl Lewis a and the GM1 oligosaccharide, both of which contain Neu5Acα2-3Gal, were also poor substrates, suggesting that the enzyme is sensitive to glycan structure. The lack of NEU2 activity on the GM1 oligosaccharide is consistent with other studies,(23) but sialyl Lewis X and sialyl Lewis a had not been examined previously. We validated our finding by monitoring glycan desialylation by thin layer chromatography (Supplementary Figure 5). NEU2 hydrolyzes sialyl Lewis X more slowly than Neu5Acα2-3Lac, consistent with the specificity assay data.
Figure 4. Human and bacterial sialidases exhibit sialoside specificity dependent on sialic acid linkage, sialic acid form, and underlying glycan structure.
(a) Sialylated glycans examined. (b) GST-NEU2 activity detected by PAL method. GST-NEU2 (0, 0.76, 2.3, or 6.9 mU, bars left to right) was incubated with 1 μM of glycan in 100 mM NaOAc, pH 5.6, and 100 mM NaCl for 2 h at 37 °C, followed by immobilization and PAL labeling. (c) NanC (0, 0.8, 2.4, or 7.2 mU, bars left to right) was incubated with 1 μM of glycan in 100 mM NaOAc, pH 5.6, and 100 mM NaCl for 2 h at 37 °C, followed by immobilization and PAL labeling. Error bars represent standard deviation of three trials.
We examined the substrate specificity of a Streptococcus pneumoniae sialidase, NanC (Figure 4, panel c). NanC is expressed by approximately 50% of S. pneumoniae isolates and is enriched in cerebrospinal fluid.(27) As suggested previously,(28) NanC prefers α2-3- over α2-6-linked sialic acids. NanC also displayed a dramatic preference for Neu5Ac over Neu5Gc, a result we confirmed using a p-nitrophenol activity assay (Supplementary Figure 6). This result may help explain S. pneumoniae’s preference for the human host and may also relate to its ability to infect the brain. Humans are incapable of Neu5Gc synthesis and although dietary Neu5Gc can be incorporated into cellular glycoconjugates, it seems to be excluded from the brain.(29, 30) NanC also preferred LacNAc- over Lac-containing substrates. S. pneumoniae encodes a β-galactosidase and an N-acetylglucosaminidase that cleave LacNAc on N-glycans, resulting in resistance to opsonophagocytic killing.(31) Removal of LacNAc-linked sialic acids may enable increased bacterial colonization by exposing LacNAc substrate. NanC also exhibited a preference for non-fucosylated glycans and was essentially inactive toward the GM1 oligosaccharide. These results provide insight into NanC specificity, and represent a step toward defining NanC’s mechanistic roles in infection.
In summary, we report a method for evaluating sialidase specificity. We adapted the PAL method to monitor desialylation of immobilized glycoconjugates. Then, we examined how the activities of mammalian and bacterial sialidases depend on multiple substrate features, including sialic acid linkage, sialic acid identity, and structure of the underlying glycan. In the future, immobilized glycoproteins and glycolipids could be examined or the method could be adapted to interrogate larger arrays of robotically printed glycans.(32) Knowledge of sialidase specificity will provide insight into sialidase roles in normal physiology and in disease.
Methods
PAL method in 96-well format
Biotinylated glycans at a final concentration of 1 μM were combined with sialidase in a volume of 100 μL and incubated at 37 °C. For Sialidase A, 0.55, 1.66, or 5 mU (unit = 1 μmol of sialic acid released per minute) of enzyme in 50 mM sodium phosphate, pH 6.0, was incubated with glycan for 1 h. For Sialidase T, 6.4, 19.3, or 58 μU of enzyme in 50 mM sodium citrate, pH 6.0, containing 100 mM sodium chloride and 100 μg mL−1 BSA was incubated with glycan for 1 h. GST-NEU2 (0.76, 2.3, or 6.9 mU) or NanC (0.8, 2.4, or 7.2 mU) were incubated with 1 μM glycan in standard buffer (100 mM NaOAc, pH 5.6, containing 100 mM NaCl) for 2 h at 37 °C. For pH dependence analysis of GST-NEU2 activity, 1.13 mU of GST-NEU2 was incubated with glycan for 1 h in buffer of the indicated pH. Buffers were composed of 200 mM sodium phosphate, 100 mM sodium acetate, 100 mM NaCl and titrated to the appropriate pH with HCl or NaOH. To immobilize glycans, 96-well streptavidin-coated plates (Fisher 15503) were incubated with 90 μL of the sialidase/glycan mixture at 4 °C for 1 h and then washed three times with PBS, pH 7.4, containing 0.05% (v/v) Tween (PBST).
For PAL detection, glycans were oxidized with 0.1 mM NaIO4 in PBS for 30 min at 4 °C. Oxidation was quenched by 1 M glycerol in PBS pH 7.4. Wells were washed three times with PBST, then reacted with 10 μM Alexa Fluor 488-hydroxylamine (Invitrogen, A-30629) and 10 mM aniline in 100 mM NaOAc, pH 4.5, for 2 h at 4 °C. Wells were washed three times with PBST. Fluorescence was measured on a Spectramax M5 plate reader with 488 nm excitation and 515 nm emission.
Supplementary Material
Acknowledgments
We thank K. Brown for use of the fluorescence plate reader and acknowledge support from the National Institutes of Health (R01GM090271) and the Welch Foundation (I-1686). Biotinylated glycans were from the NIH-funded Consortium for Functional Glycomics (U54GM62116). J. J. K. is an Alfred P. Sloan Research Fellow.
Footnotes
Detailed descriptions of method optimization, protein expression and purification, and specificity validation are provided in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Yu H, Chen X. Carbohydrate post-glycosylational modifications. Org Biomol Chem. 2007;5:865–872. doi: 10.1039/b700034k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest. 2007;87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyagi T, Yamaguchi K. Mammalian sialidases: Physiological and pathological roles in cellular functions. Glycobiology. 2012;22:880–896. doi: 10.1093/glycob/cws057. [DOI] [PubMed] [Google Scholar]
- 4.Corfield AP, Higa H, Paulson JC, Schauer R. The specificity of viral and bacterial sialidases for α(2-3)- and α(2-6)-linked sialic acids in glycoproteins. Biochim Biophys Acta. 1983;744:121–126. doi: 10.1016/0167-4838(83)90080-8. [DOI] [PubMed] [Google Scholar]
- 5.Cabezas JA, Calvo P, Eid P, Martin J, Perez N, Reglero A, Hannoun C. Neuraminidase from influenza virus A (H3N2) - Specificity towards several substrates and procedure of activity determination. Biochim Biophys Acta. 1980;616:228–238. doi: 10.1016/0005-2744(80)90141-2. [DOI] [PubMed] [Google Scholar]
- 6.Rogerieux F, Belaise M, Terzidistrabelsi H, Greffard A, Pilatte Y, Lambre CR. Determination of the sialic acid linkage specificity of sialidases using lectins in a solid phase assay. Anal Biochem. 1993;211:200–204. doi: 10.1006/abio.1993.1257. [DOI] [PubMed] [Google Scholar]
- 7.Onodera S. A microplate assay for sialidase activity using plant lectin binding to N-acetyllactosamine. Biol Pharm Bull. 1994;17:29–33. doi: 10.1248/bpb.17.29. [DOI] [PubMed] [Google Scholar]
- 8.Chokhawala HA, Yu H, Chen X. High-throughput substrate specificity studies of sialidases by using chemoenzymatically synthesized sialoside libraries. Chembiochem. 2007;8:194–201. doi: 10.1002/cbic.200600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Indurugalla D, Watson JN, Bennet AJ. Natural sialoside analogues for the determination of enzymatic rate constants. Org Biomol Chem. 2006;4:4453–4459. doi: 10.1039/b613909d. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Cao H, Yu H, Chen Y, Lau K, Qu J, Thon V, Sugiarto G, Chen X. Identifying selective inhibitors against the human cytosolic sialidase NEU2 by substrate specificity studies. Mol Biosys. 2011;7:1060–1072. doi: 10.1039/c0mb00244e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao HZ, Li YH, Lau K, Muthana S, Yu H, Cheng JS, Chokhawala HA, Sugiarto G, Zhang L, Chen X. Sialidase substrate specificity studies using chemoenzymatically synthesized sialosides containing C5-modified sialic acids. Org Biomol Chem. 2009;7:5137–5145. doi: 10.1039/b916305k. [DOI] [PubMed] [Google Scholar]
- 12.Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR, Paulson JC. Sialoside specificity of the Siglec family assessed using novel multivalent probes. J Biol Chem. 2003;278:31007–31019. doi: 10.1074/jbc.M304331200. [DOI] [PubMed] [Google Scholar]
- 13.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manimala JC, Li ZT, Jain A, VedBrat S, Gildersleeve JC. Carbohydrate array analysis of anti-Tn antibodies and lectins reveals unexpected specificities: Implications for diagnostic and vaccine development. Chembiochem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- 15.Fukui S, Feizi T, Galustian C, Lawson AM, Chai WG. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 16.Zeng Y, Ramya TNC, Dirksen A, Dawson PE, Paulson JC. High-efficiency labeling of sialylated glycoproteins on living cells. Nat Meth. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirksen A, Dawson PE. Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling. Biocong Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchida Y, Tsukada Y, Sugimori T. Enzymatic properties of neuraminidases from Arthrobacter ureafaciens. J Biochem. 1979;86:1573–1585. doi: 10.1093/oxfordjournals.jbchem.a132675. [DOI] [PubMed] [Google Scholar]
- 19.Hoyer LL, Roggentin P, Schauer R, Vimr ER. Purification and properties of cloned Salmonella typhimurium LT2 sialidase with virus-typical kinetic preference for sialyl α2‚3 linkages. J Biochem. 1991;110:462–467. doi: 10.1093/oxfordjournals.jbchem.a123603. [DOI] [PubMed] [Google Scholar]
- 20.Monti E, Preti A, Rossi E, Ballabio A, Borsani G. Cloning and characterization of NEU2, a human gene homologous to rodent soluble sialidases. Genomics. 1999;57:137–143. doi: 10.1006/geno.1999.5749. [DOI] [PubMed] [Google Scholar]
- 21.Fanzani A, Giuliani R, Colombo F, Zizioli D, Presta M, Preti A, Marchesini S. Overexpression of cytosolic sialidase Neu2 induces myoblast differentiation in C2C12 cells. FEBS Lett. 2003;547:183–188. doi: 10.1016/s0014-5793(03)00709-9. [DOI] [PubMed] [Google Scholar]
- 22.Burg M, Müthing J. Characterization of cytosolic sialidase from Chinese hamster ovary cells: Part I: Cloning and expression of soluble sialidase in Escherichia coli. Carb Res. 2001;330:335–346. doi: 10.1016/s0008-6215(00)00294-9. [DOI] [PubMed] [Google Scholar]
- 23.Tringali C, Papini N, Fusi P, Croci G, Borsani G, Preti A, Tortora P, Tettamanti G, Venerando B, Monti E. Properties of recombinant human cytosolic sialidase hsNEU2. J Biol Chem. 2004;279:3169–3179. doi: 10.1074/jbc.M308381200. [DOI] [PubMed] [Google Scholar]
- 24.Hata K, Koseki K, Yamaguchi K, Moriya S, Suzuki Y, Yingsakmongkon S, Hirai G, Sodeoka M, von Itzstein M, Miyagi T. Limited inhibitory effects of oseltamivir and zanamivir on human sialidases. Antimicrob Agents Chemother. 2008;52:3484–3491. doi: 10.1128/AAC.00344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warner TG, Chang J, Ferrari J, Harris R, McNerney T, Bennett G, Burnier J, Sliwkowski MB. Isolation and properties of a soluble sialidase from the culture fluid of Chinese hamster ovary cells. Glycobiology. 1993;3:455–463. doi: 10.1093/glycob/3.5.455. [DOI] [PubMed] [Google Scholar]
- 26.Koda T, Kijimoto-Ochiai S, Uemura S, Inokuchi J. Specific expression of Neu2 type B in mouse thymus and the existence of a membrane-bound form in COS cells. Biochem Biophys Res Comm. 2009;387:729–735. doi: 10.1016/j.bbrc.2009.07.100. [DOI] [PubMed] [Google Scholar]
- 27.Pettigrew MM, Fennie KP, York MP, Daniels J, Ghaffar F. Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect Immun. 2006;74:3360–3365. doi: 10.1128/IAI.01442-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu GG, Kiefel MJ, Wilson JC, Andrew PW, Oggioni MR, Taylor GL. Three Streptococcus pneumoniae sialidases: Three different products. J Amer Chem Soc. 2011;133:1718–1721. doi: 10.1021/ja110733q. [DOI] [PubMed] [Google Scholar]
- 29.Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, Varki A, Muchmore E. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc Natl Acad Sci U S A. 2003:12045–12050. doi: 10.1073/pnas.2131556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muchmore EA, Diaz S, Varki A. A structural difference between the cell surfaces of humans and the great apes. Am J Phys Anthropol. 1998;107:187–198. doi: 10.1002/(SICI)1096-8644(199810)107:2<187::AID-AJPA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 31.Dalia AB, Standish AJ, Weiser JN. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect Immun. 2010;78:2108–2116. doi: 10.1128/IAI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens J, Blixt O, Paulson JC, Wilson IA. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Micro. 2006;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.