Abstract
A previous report (Biochemistry 46: 2364–2370, 2007) described the application of The Traceable Kinase Method to identify substrates of PKCα in non-transformed human breast MCF-10A cells. Here, a non-radioactive variation of this method compared the phospho-protein profiles of three traceable PKC isoforms (α, δ and ζ) for the purpose of identifying novel, isoform-selective substrates. Each FLAG-tagged traceable kinase was expressed and co-immunoprecipitated along with high affinity substrates. The isolated kinase and its associated substrates were subjected to an in vitro phosphorylation reaction with traceable kinase-specific N6-phenyl-ATP, and the resulting phospho-proteins were analyzed by Western blot with an antibody that recognizes the phosphorylated PKC consensus site. Phospho-protein profiles generated by PKC-α and -δ were similar and differed markedly from that of PKC-ζ. Mass spectrometry of selected bands revealed known PKC substrates and several potential substrates that included the small GTPase-associated effector protein Cdc42 effector protein-4 (CEP4). Of those potential substrates tested, only CEP4 was phosphorylated by pure PKC-α, –δ, and −ζ isoforms in vitro, and by endogenous PKC isoforms in MCF-10A cells treated with DAG-lactone, a membrane permeable PKC activator. Under these conditions, the stoichiometry of CEP4 phosphorylation was 3.2 ± 0.5 (mol phospho-CEP4/mol CEP4). Following knock-down with isoform-specific shRNA-encoding plasmids, phosphorylation of CEP4 was substantially decreased in response to silencing of each of the three isoforms (PKC–α, –δ, or –ζ), whereas testing of kinase-dead mutants supported a role for only PKC-α and –δ in CEP4 phosphorylation. These findings identify CEP4 as a novel intracellular PKC substrate that is phosphorylated by multiple PKC isoforms.
Keywords: phospho-protein, motility, mass spectrometry, Cdc42 effector protein-4, small Rho GTPases
The pleiotropic effects of protein kinase C (PKC) have been documented over the past 30 years but only a few protein substrates that convey the PKC signal for individual pathways have been identified. Previous studies from this laboratory showed that engineered over-expression of PKC-α in non-transformed human breast cells (MCF-10A cells) suppressed cell proliferation rates, while promoting cell movement and altered cell shape (1). That PKC consists of multiple structurally-related isoforms (2) further complicates this model, since each isoform may interact with its unique substrates as well as sharing substrates with other PKC isoforms. The PKC family of ten isoforms consists of three sub-classes, namely the diacylglycerol (DAG)-activated conventional isoforms (α, βI/βII, γ), the DAG-activated novel isoforms (δ, ε, η, θ), and the DAG-unresponsive atypical isoforms (λ/ι and ζ). Additional complexity arises from variation in the isoform expression profiles of different cell types.
In view of the importance of PKC activity to cancer progression, very little is known about PKC substrates that impact cellular transformation. Knowledge of their substrates, whether shared or selected by a single isoform, would clarify whether isoform-specific mechanisms are operating and thereby inform strategies for design of anti-cancer drugs (3, 4). Based on what is known about PKC substrates to date, many are cytoskeletal proteins whose phosphorylation by PKC triggers dynamic changes that drive cell adhesion and migration (5). In light of its central role in producing an aggressive phenotype in breast cancer cells (1, 6, 7), PKC-α continues to be an attractive anti-cancer target. However, in a substantial number of human breast tumors, PKC-α undergoes down-regulation (8) which suggests that the signaling activity of other PKC isoforms, such as PKC-δ, involves some of the same substrates. In human breast cells, PKC-δ promotes migration (like PKC-α), and survival (unlike PKC-α), as well as other aspects of the malignant phenotype (9–13). In contrast, PKC-ζ apparently operates through different signaling pathways since in non-transformed NMuMg human breast cells that are engineered to over-express PKC-ζ, migration is suppressed while partially transforming tumorigenic properties such as clonogenic growth and protease secretion occur that are not promoted by PKC-α (14). Therefore, each of the three PKC isoforms is associated with malignant properties of human breast cancer and may be interacting with some protein substrates that are unique for that isoform, as well as other protein substrates that are shared by multiple isoforms. Dissection of these isoform-directed pathways can be initiated by comparing and analyzing the phosphorylation profiles of individual isoforms.
The present study harnessed an open-ended approach called the “Traceable Kinase Method” to identify novel substrates of PKC-α, -δ and -ζ in human breast cells. This method was introduced in 1997 by K. Shokat who initially used it to identify protein substrates of v-Src (15–18). Several laboratories have since applied this method to determine the substrates of key protein kinases such as JNK (19), ERK2 (20), and Raf-1 (21). The method entails the introduction of a single site mutation at a residue (termed the “gatekeeper”) in the ATP binding site so as to enlarge the space near the N6-amino group of bound ATP. As a result, the mutant, but not the wildtype enzyme, can accommodate an ATP analogue in which the N6-amino bears an aromatic group that occupies the enlarged space of the binding pocket. In the ideal case, productive binding of this analogue by the mutant subsequently supports phosphoryl transfer to the substrate, whereas neither the wildtype enzyme nor any other protein kinase can utilize this sterically-hindered ATP analogue. Therefore, in the presence of this ATP analogue, the mutant is “traceable” since it can be bound productively and used to carry out phosphorylation of protein substrates, while the wildtype protein kinase and presumably other protein kinases remain inactive. In adapting this approach to the study of PKC substrates, our traceable kinase method involves expression followed by co-immunoprecipitation of the mutant or wildtype PKC proteins along with any high affinity substrates bound by the PKC protein (22). Addition of the ATP analogue (N6-phenyl-ATP) to the mutant kinase in the immunopellet generates phosphorylated products that are due to the activity of the traceable kinase. As compared with simple immunoprecipitation of the wildtype enzyme and addition of traditional ATP, use of a traceable kinase restricts phosphotransferase activity primarily to the mutant kinase, greatly reducing background phosphorylation due to other protein kinases that may accompany the traceable kinase during immunoprecipitation.
To address the question of PKC substrates in human breast cells, our laboratory developed a traceable PKC-α for testing in MCF-10A cells as a model system (22). These non-transformed human breast cells express α, δ, and ζ isoforms at low levels (in addition to ε and θ), therefore providing an ideal substrate-rich environment in which to identify their protein substrates in response to diacylglycerol (DAG)-lactone, a membrane-permeable PKC activator. Previously, it was found that engineered over-expression of PKC-α in MCF-10A cells induces migration and alterations in cell morphology while suppressing proliferation (1). By using a traceable mutant of PKC-α, we found that α6-tubulin co-immunoprecipitates with this isoform and is a substrate whose phosphorylation at Ser-165 drives motility of MCF-10A cells (23). This finding provides a plausible mechanism for how engineered over-expression of PKC-α in these non-motile cells induces the motility phenotype. Furthermore, the phosphorylation site in α6-tubulin also serves as a PKC target in highly motile metastatic human breast cells (MDA-MB-468) that do not express any of the conventional isoforms (α, β, γ), but express high levels of PKC-δ. It is therefore possible that α-tubulin is a general PKC substrate recognized by both conventional and non-conventional isoforms. This development opens the possibility that certain PKC isoforms converge on a few shared substrates that consequently promote the motility pathway.
In the present work, traceable kinases were developed for PKC-δ and PKC-ζ isoforms, and their substrate phosphorylation profiles in MCF-10A cells were compared with that of traceable PKC-α. Potential PKC substrates, along with known substrates and binding partners of PKC, were isolated by co-immunoprecipitating the traceable isoform. From these studies, Cdc42 effector protein-4 (CEP4) is identified as a novel substrate of each of the three PKC isoforms being evaluated (α, δ, and ζ).
MATERIALS AND METHODS
Cell culture serum, growth factors, media and DNA mutagenesis and sequencing primers were purchased from Invitrogen, Inc. (Carlsbad, CA). Quik-Change site-directed mutagenesis kit and pCMV4 vector were purchased from Agilent Technologies (Santa Clara, CA). Restriction enzymes were obtained from New England Biolabs (Ipswich, MA), and shRNA-encoding plasmids were purchased from GeneCopoeia (Rockville, MD). Phosphoserine protein phosphatase inhibitor cocktail 3, protease inhibitors, EZ-view anti-FLAG beads, and recombinant PKCζ were acquired from Sigma-Aldrich (St. Louis, MO). Phospho-PKC substrate antibody (product #2261), anti-PAK2, recombinant PKC isoforms (α, δ) and isoform-specific antibodies (α, δ) were obtained from Cell Signaling Technology (Beverly, MA). PKC-ε (N-terminus antibody) was purchase d from BD Biosciences (Franklin Lakes, NJ). The PKC-ε (C-terminus) antibody, PKC-ζ antibody, horseradish peroxidase-conjugated secondary antisera, rabbit IgG-agarose beads, and protein A/G-agarose beads were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). CLASP-1 antibody was obtained from Epitomics, Inc. (Burlingame, CA). Recombinant CEP4 protein was purchased from Novus Biologicals (Littleton, CO) and anti-CEP4 was from Bethyl Laboratories (Montgomery, TX). PolyExpress transfection reagent was purchased from Excellgen, Inc. (Rockville, MD) and Fugene 6 was purchased from Roche Applied Science (Indianapolis, IN). Gelcode Blue and chemiluminescent reagents (Supersignal West Pico) were acquired from Pierce Co. (Rockford, IL). Immobilon-P transfer membranes were purchased from Millipore Corp. (Bedford, MA), and ATP analogues were purchased from Axxora Corp. (San Diego, CA). The DAG-lactone reagent was a gift from Dr. V. Marquez (NCI-Frederick) and either was the pure R-enantiomer (used at 5 µM) or the previously reported racemic mixture (HK654 in ref. 24) (used at 10 µM). Plasmids encoding the kinase-dead forms of human PKC-α, mouse PKC-δ, and rat PKC-ζ (Addgene plasmids 21235, 16389, and 21249, respectively) were obtained from the laboratory of Dr. I.B. Weinstein, and the Trustees of Columbia University, NY, NY.
Construction of Traceable Kinase Mutants for PKC-δ and PKC-ζ
Plasmids encoding human PKC-δ (pEGFP) and mouse PKC-ζ (pCR3) were gifts from Dr. Kiyotsugu Yoshida (Tokyo Medical and Dental University, Japan) and Dr. Marcelo G. Kazanietz (University of Pennsylvania), respectively. The cDNA insert of PKC-δ was subcloned into pCMV4 (restricted at BamH1 and EcoR1) prior to mutagenesis. For construction of each traceable kinase, site-directed mutagenesis was carried out by the Quik-Change Method (Agilent Technologies) at either a methionine (Met) or isoleucine (Ile) residue that corresponded to Met-417 in bovine PKC-α (22). To construct the traceable mutant for PKC-δ, an alanine (Ala) codon was substituted for Met-427 using the following primer: 5’–AAG GAC CAC CTG TTC TTT GTG GCG GAG TTC CTC AAC GGG GGG GAC C-3’. The wildtype and mutated cDNA for PKC-δ were expressed as C-terminally fused FLAG-tagged proteins. To construct the traceable mutant for PKC-ζ, the codon for I330 was substituted with an Ala codon using Quik-Change with the following primer: 5’-CGG TGG TTC CTG GTC GCG GAG TAT GTC AAT GGC GGG- 3’. Expression of wildtype and mutant PKCζ proteins from the pCR3 plasmid conferred a short C-terminal segment from PKCε (ε-tag2) that was fused to the N-terminus of PKCζ. An antibody recognizing the C-terminal fragment of PKC-ε (Santa Cruz Biotechnology, Inc.) was used to immunoprecipitate and to characterize the PKCζ mutant. For each construct, the entire opening reading frame was verified by DNA sequencing (Macrogen Corp. USA, Rockville, MD).
Cell Culture and Transfection
Mid-passage MCF-10A cells were obtained from the Barbara Ann Karmanos Cancer Institute. The cells were cultured in DMEM/F-12 medium (1:1) supplemented with 15% equine serum, insulin (10 µg/ml), epidermal growth factor (20 ng/ml), cholera toxin (100 ng/ml), and hydrocortisone (0.5 µg/ml), as previously described (25). MDA-MB-231 cells were cultured in Iscove’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum. Each cell line was maintained with penicillin (100 units/ml), streptomycin (100 µg/ml), and fungizone (0.5 µg/ml). Cells were plated into 10-cm plates one day prior to transfection in order to achieve 60–80% confluence. Unless otherwise indicated, plasmid DNA was combined with Fugene 6 transfection reagent in a 1:2 ratio and the mixture was added to the cells cultured in 4 ml serum-free medium. The cells were incubated with serum-free medium for 6 h and followed by addition of complete medium.
Cell Lysis and Western Blotting
For preparing whole cell lysates for western blotting, MCF-10A cells transfected with PKC-δ or PKC-ζ plasmids were lysed by sonication (3 X 10 s) in 0.5 ml lysis buffer [1% TX-100, 20 mM TRIS (pH 7.4), 2 mM MgCl2, and 2 mM EGTA]. All buffers contained protease inhibitors (1 mM phenylmethanesulfonyl fluoride, 10 ng/mL leupeptin, and 10 ng/mL soybean trypsin inhibitors) and phosphatase inhibitors. After centrifugation at 5700 × g for 10 min, SDS sample buffer was added to the lysates, and the samples were heated for 5 min at 95°C prior to 8% SDS-PAGE. After transfer, the membrane was blocked in 5% milk for 1 h at room temperature, followed by incubation for 2 h at room temperature with either anti-FLAG (1:1000) for detection of PKC-α and PKC-δ, or anti-PKC-ε (C-terminal) (1:1000) for PKC-ζ. The blot was washed with TBS containing 0.1% Tween-20, incubated with secondary antibody for 1h, and developed by chemiluminescence.
Immunoprecipitation
For MCF-10A cells expressing PKC-α or PKC-δ, cells were lysed in hypotonic, detergent-free buffer [20 mM TRIS (pH 7.4), 2 mM MgCl2, 2 mM EGTA and 1 mM dithiothreitol (DTT), 10 µM bis-indoleylmaleimide, protease inhibitors, and phosphatase inhibitors] followed by immunoprecipitation with anti-FLAG EZview beads. Sixty microliters of anti-FLAG beads (pre-equilibrated in cold PBS) were added to a lysate and rotated for 2 h at 4°C. The beads were centrifuged at 8200 × g for 30 sec and washed twice with 500 µl PBS and a third time with 0.5 ml 2X kinase buffer (50 mM TRIS (pH 7.4), 20 mM magnesium acetate, 1 mM EGTA and 1 mM DTT).
Cells transfected with PKC-ζ were lysed in 0.5 ml TBS containing 0.5% NP-40 with protease inhibitors and phosphatase inhibitors, as detailed above. Because PKC-ζ contained the ε-tag (corresponding to the C-terminus of PKC-ε), a pre-clearing step was used to remove endogenous PKC-ε protein. For this purpose, anti-PKC-ε (N-terminal) (8 µg) was added to each lysate for 2 h at 4°C and the immuno-complexes containing endogenous PKC-ε were collected by addition of protein A/G beads (25 µl) and rotation for 1 h at 4°C, followed by centrifugation at 3000 × g for 5 min. After transfer of the supernatants to fresh tubes, WT PKC-ζ or its traceable mutant was immunoprecipitated by addition of 8 µg of anti-PKC-ε (C-terminal) for 2 h at 4°C and 50 µl protein A/G-agarose beads for 1 h at 4°C. Following centrifugation, the pellets were washed twice, as described above.
Assay of Analogue Specificity with Myelin Basic Protein as Substrate
WT PKCs (α, δ, or ζ) or their corresponding traceable mutant kinases were expressed in MCF-10A cells. Following immunoprecipitation, the pellets were washed twice in lysis buffer and a final wash with 2X kinase buffer (50 mM TRIS (pH 7.4), 20 mM magnesium acetate, 1 mM EGTA and 2 mM DTT). An equal volume of each enzyme was aliquotted into each tube in a set of five eppendorf tubes. To each tube was added either ATP or the specified ATP analogue (N6-phenyl-ATP, N6-benzyl-ATP, N6-phenethyl-ATP or N6-(3-methyl)-benzyl-ATP). The final reaction medium consisted of 25 mM TRIS (pH 7.4), 0.5 mM Ca2+ (for PKC-α) or 0.5 mM EGTA (for PKC-δ and PKC-ζ), 10 mM magnesium acetate, 1 mM DTT with protease inhibitors and phosphatase inhibitors, 0.1 mg/ml phosphatidylserine, 10 µM DAG-lactone (PKCα, δ) or 1 µM ceramide (for PKCζ), 50 µg myelin basic protein (MBP), and at t = 0, 100 µM ATP or ATP analogue was added to initiate the reaction. The reaction was carried out for 10 min at 30°C and quenched by adding 5X SDS sample buffer. The samples were heated for 5 min at 95°C and analyzed by 12% SDS-PAGE. The best analogue for each mutant was determined by assay of MBP phosphorylation (18 kDa band) by Western blot using the phospho-PKC substrate antibody.
Phosphorylation Profile of PKC-α, -δ, and -ζ in MCF-10A Cells
PKCα and PKCδ and their corresponding traceable mutants were expressed in MCF-10A cells. Cells cultured on 15-cm plates were lysed in 0.5 ml hypotonic, detergent-free buffer containing protease inhibitors and phosphatase inhibitors. Immunoprecipitation with either anti-FLAG-EZview beads for FLAG-tagged PKCα and δ enzymes, or anti-PKCε for ε-tagged PKCζ was carried out for 2h at 4°C, followed by two washes with PBS and a third wash with 2X kinase buffer. To each immunopellet was added 1X kinase buffer containing Ca2+ (for PKCα) or EGTA (for PKCδ/ζ) and activating phospholipid [PS (1.0 mg/ml) for PKCα/δ, or ceramide (1 µM) for PKCζ, plus protease inhibitors and phosphatase inhibitors. The reaction was carried out for 1 h at 30°C and quenched by adding SDS sample buffer and heating at 95°C for 5 min. Approximately 25% of each sample was loaded into one half of a 7.5 % SDS-PAGE gel (0.75 mm) for Western blot analysis, and the remainder of the sample was loaded into the other half of the gel for Gelcode Blue staining. Following electrophoresis, the gel was cut in half with a scalpel and staining or transfer to a PDVF membrane was performed. To locate bands of interest using Western blot analysis, the membrane was probed with phospho-PKC substrates antibody. Bands of interest were excised from the gel and submitted for analysis by mass spectrometry.
Protein Identification by Mass Spectrometry
Identification of proteins was performed at the Keck Foundation Mass Spectrometry Resource Laboratory at Yale Cancer Center. Gel-resolved proteins were digested in situ with trypsin, phospho-peptides were enriched by TiO2, and the resulting peptides were analyzed by LC MS/MS on a LTQ Orbitrap mass spectrometer. All MS/MS spectra were searched against the NCBI database with a probability or significance threshold of p< 0.05 using the automated MASCOT algorithm. Identification required that two or more MS/MS spectra matched the same protein entry in the database, and that matched peptides correspond to tryptic peptides in the protein.
Motility Assay
MCF-10A cells were plated onto a 10-well slide through a cell sedimentation manifold (CSM, Inc., Phoenix, AZ) as concentric circles followed by incubation at 37% and 5% CO2. After removal of the manifold (t=0), images were recorded with a camera attached to an inverted Nikon Diaphot microscope. After 16 hours, during which cells radiated outwardly, images were taken again and motility was calculated by the change in total area (in µm2) occupied by the cells (Motic Image Plus 2.0). Each reported value is the average of triplicate measurements and the corresponding standard deviation.
RESULTS
Preparation and characterization of traceable mutants of PKC-δ and PKC-ζ
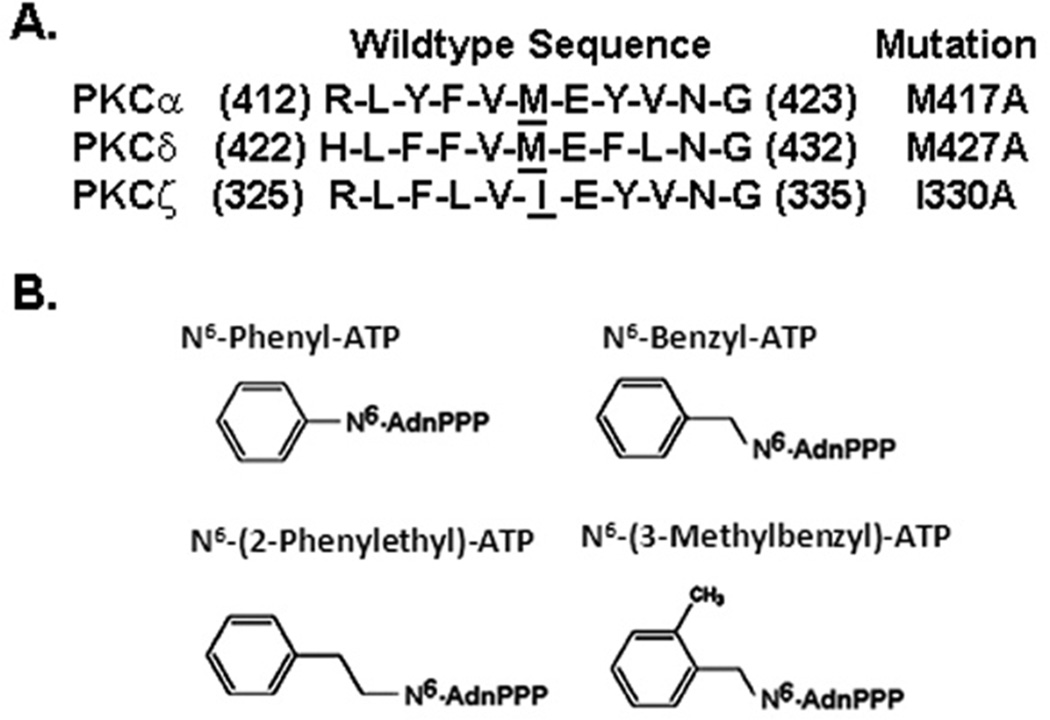
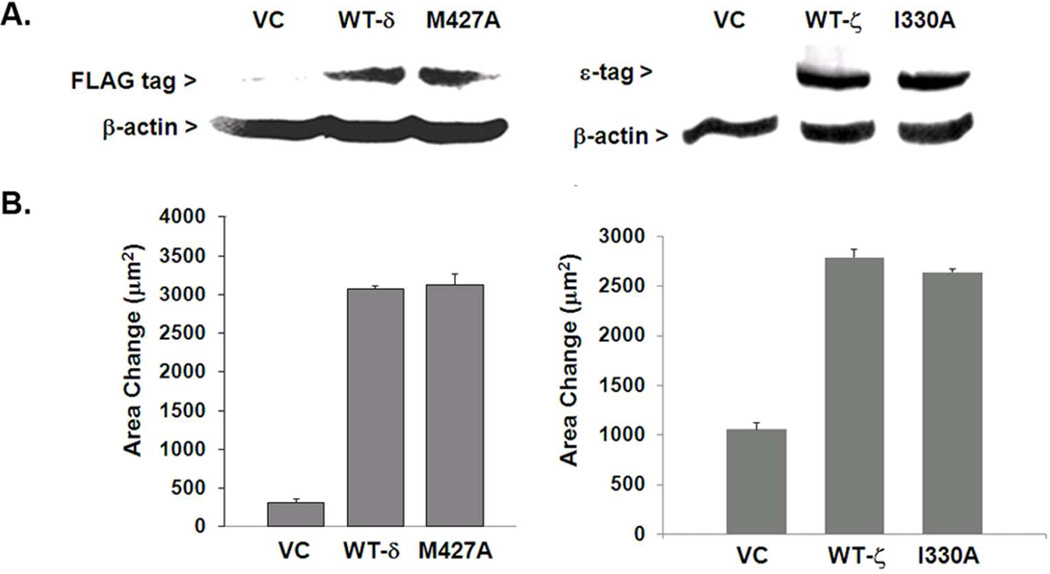
The traceable kinases M427A for PKC-δ and I330A for PKC-ζ were prepared by substituting the gatekeeper residue (Met or Ile) with Ala by site-directed mutagenesis. Each site was chosen based on its alignment with Met-417 in PKC-α (Figure 1A). As predicted from the X-ray crystal structure of PKA, this position is closest to the N6-amino group in ATP when it is bound at the active site, as previously described (22). Wildtype (WT) PKC-δ or its corresponding traceable mutant was sub-cloned into a pCMV4 vector that confers a FLAG tag at the C-terminus. The cDNA encoding the WT PKC-ζ or its corresponding traceable mutant was expressed from a pCR3 vector that confers a short sequence of the carboxyl terminus of PKC-ε (ε-tag) that subsequently proved useful both for immunoprecipitation and detection in western blotting. Following transient transfection, expression levels were assessed with either anti-FLAG (for WT or mutant PKC-δ) or PKC-ε antibody (for ε-tagged WT and mutant PKC-ζ) (Figure 2A). WT and mutant forms for PKC-δ were expressed to equivalent levels as 75-kDa proteins, while PKC-ζ with its ε-tag was expressed as a 72-kDa protein.
Figure 1.
Chemical and genetic tools used for developing and testing traceable PKC isoforms. A. Primary sequences for PKC-δ and -ζ that align with M417 of PKC-α. Remodeling of the ATP binding sites in PKC-δ and PKC-ζ was carried out by site-directed mutagenesis of the cDNA corresponding to the underlined residue to an Ala codon (designated to the right of each sequence). B. Structures of the ATP analogues tested in this study.
Figure 2.
Expression and phenotypic analysis of traceable mutants and wildtype PKC isoforms. (A) Following transfection of a plasmid that conferred a FLAG epitope on the expressed protein, wildtype (WT) PKCδ and its corresponding traceable kinase were expressed in MCF-10A cells, resolved by SDS-PAGE (75 µg total protein per lane), and detected by Western blot analysis. WT and mutant PKCδ proteins (75 kDa) were detected with anti-FLAG (1:1000), whereas the PKCζ proteins (72 kDa) were detected with anti-ε tag (1:1000). For both isoforms, equivalent sample loading was confirmed by staining with anti-β-actin (1:5000). (B) Expression of either WT or traceable mutant for PKCδ (left panel) or PKCζ (right panel) produced increased motility of MCF-10A cells (PKCδ) or MDA-MB-231 cells (PKCζ). Triplicate measurements of motility were performed as described in the ‘Methods’ and the results were averaged. Each experiment was carried out at least twice with similar results.
To determine whether the expressed mutant selects the same intracellular substrates as the WT, the mutant should reproduce the same phenotype, such as motility, that is engendered by the WT enzyme in a cell-based assay. Increased motility of MCF-10A cells which are weakly motile, was induced following expression of the wildtype or traceable mutant of PKCα, as previously reported (22). This observation was consistent with retention of substrate specificity by the traceable mutant. Similar to traceable PKCα, WT PKC-δ and its corresponding traceable mutant were also observed to increase the motility of MCF-10A cells by almost 5-fold relative to the vector control (Figure 2B, left panel). In contrast, both WT PKC-ζ and its corresponding traceable mutant showed neither activating nor suppressive effects on MCF-10A cell motility (not shown). However, both WT-PKCζ and its traceable mutant exhibited a similar ability to stimulate motility of metastatic MDA-MB-231 cells (Figure 2B, right panel) thereby demonstrating that each mutant possesses the necessary substrate specificity to produce the motility phenotype. That elevated motility by overexpression of either PKC-α (22) or –δ occurs without addition of DAG-lactone implies that each enzyme is being endogenously activated in MCF-10A cells, and that PKCζ is also being activated by an unknown mechanism in MDA-MB-231 cells that is not operating in MCF-10A cells. As has been discussed elsewhere (23), this effect in MCF-10A cells was attributed to a high enzyme dosage (that titrates a putative inhibitor) and elevated intracellular DAG levels that is generated as a consequence of the EGF requirement in the MCF-10A cell culture medium (25). It is noted that because PKC-ζ is an atypical isoform, it is not responsive to endogenous DAG. Regardless of their modes of activation, the traceable kinases for PKC-δ and -ζ produced the same effect on motility as their corresponding WT enzyme. It is concluded that, similar to PKC-α (22), the mutation at the ATP binding site did not impair the ability of PKC-δ and PKC-ζ to bind endogenous ATP and to select the correct substrates to engender the motility phenotype.
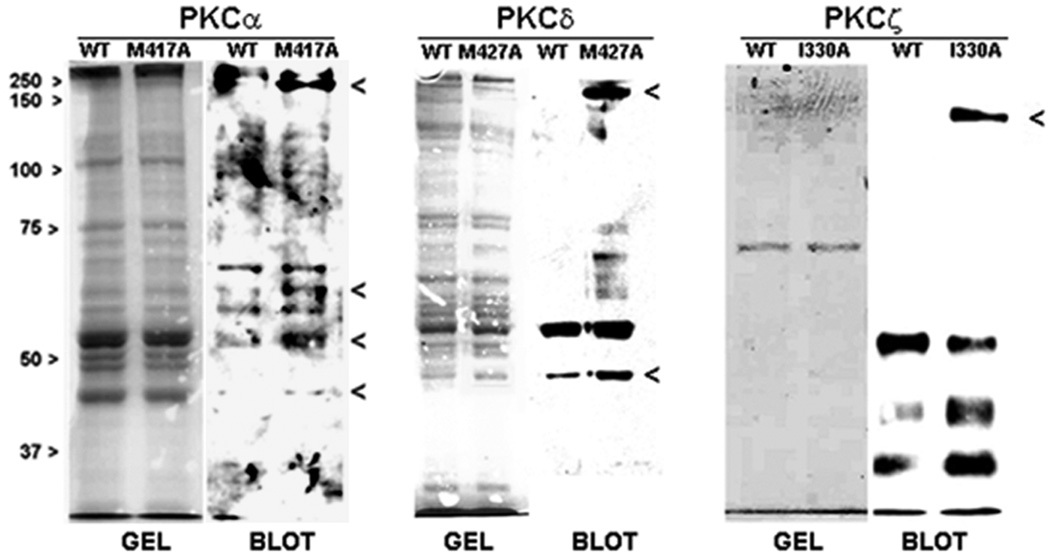
The next step was to identify an ATP analogue that best supported the kinase activity of the traceable mutant while being relatively non-reactive with the WT enzyme. A series of ATP analogues derivatized at the N6-amino group of the adenosine moiety were chosen that included the N6-phenyl, N6-benzyl, N6-phenylethyl, and N6-(3-methyl)-benzyl derivatives (Figure 1B). To select the best ATP analogue, a kinase reaction with myelin basic protein (MBP) as artificial substrate was carried out with each ATP analogue and compared with ATP itself. WT and mutant enzymes were expressed in MCF-10A cells, immunoprecipitated, and aliquotted into multiple tubes along with MBP. The specified ATP analogue or ATP itself was added to initiate the phosphorylation of MBP. The level of phospho-MBP (18 kDa band) of each sample was determined by Western Blotting with an antibody that specifically recognizes the phosphorylated consensus site in a PKC substrate (PKC substrate antibody). The results obtained for the WT isoforms (Figure 3) indicated that unlike PKC-α, the WT form for PKC-δ was reactive with the benzyl derivatives of ATP. In contrast, WT PKC-ζ was the most permissive isoform since it displayed substantial reactivity with every analogue except phenyl-ATP. This finding with PKC-ζ may arise from subtle differences in the glycine-rich region (GXGXXG) of the ATP binding loop of PKC-ζ (2). Experiments performed with the traceable mutants demonstrated that N6-phenyl-ATP supported the highest level of phospho-MBP production while being minimally reactive with the corresponding WT PKC. Because this ATP analogue produced the highest differential in phospho-MBP levels between mutant and WT enzymes, it was selected for use in subsequent experiments.
Figure 3.
Analysis of WT and traceable PKC isoform activities with different ATP analogues. FLAG-tagged WT isoforms or traceable kinase mutants for PKCα (A), PKCδ (B), and PKCζ (C) were immunoprecipitated with anti-FLAG and assayed with myelin basic protein (MBP) as protein substrate. Each immunoprecipitated enzyme (WT or mutant) was distributed in equal volume into tubes for each set of reactions that were carried out with either ATP or the specified ATP analogue. The abbreviations refer to the modification at the N6-amino group in adenosine of each analogue: N6-phenyl (Phen), N6-benzyl (Benz), N6-phenethyl (Phen-Et), N6-3-(methyl)-benzyl (3Me-Benz). Phosphorylated MBP standard (P-MBP) indicates the position of the 18 kDa phospho-protein product of the reaction. Details of the assay are given in the ‘Methods’. The experiment was carried out twice with similar results.
Phosphorylation profiles of PKC-α, -δ, and -ζ in MCF-10A cells and identification of potential substrates
To identify direct substrates of PKC-α, −δ, and -ζ in MCF-10A cells, phosphorylation profiles were produced from the high affinity substrates that co-immunoprecipitated with WT or mutant enzymes under detergent-free conditions that preserved protein-protein interactions. Any high-affinity substrates pulled-down by the enzymes were phosphorylated in vitro following addition of N6-phenyl-ATP to the immunopellet to initiate the phosphorylation reaction. Products were resolved by SDS-PAGE, and phosphorylated proteins were detected by Western blot using the PKC substrates antibody. As compared with the WT enzyme, bands unique to the mutant were observed in the reaction products for each traceable protein kinase, as shown in Figure 4. The traceable mutant for PKCα produced unique bands at 43 kDa, 53 kDa, 62 kDa, and 200 kDa, while that for the PKC-δ mutant produced unique bands at 200-kDa and 43-kDa. The corresponding Gelcode Blue-stained gels for the same samples showed detectable protein bands at these positions for the traceable mutants.
Figure 4.
Phospho-protein profiles of WT and traceable PKC isoforms. MCF-10A transfectants expressing FLAG-tagged WT or traceable mutant of each PKC isoform were lysed in detergent-free medium and each lysate was immunoprecipitated with anti-FLAG, as described in the ‘Methods’. This step facilitated the isolation of high affinity substrates of each PKC isoform. Following addition of N6-phenyl-ATP to each immunoprecipitated enzyme under activating conditions (phosphatidylserine for PKCα and δ, ceramide for PKCζ), phosphorylated products were resolved on SDS-PAGE and stained with Gelcode Blue. Western blot analysis of an aliquot of each sample (representing 25% of the total sample) was carried out with a duplicate gel and the resulting blot was probed with PKC substrates antibody (1:1000). Each WT/traceable mutant pair was aligned with the corresponding Gelcode blue-stained gel that serves as the loading control. Bands excised for MS/MS analysis are indicated with a caret (>). Each phospho-protein profile is representative of three independent experiments.
In contrast with PKC-α and –δ, the profile produced by WT PKC-ζ and its corresponding mutant revealed a strong 130-kDa band in the Western blot for the mutant that was absent with the wildtype enzyme, and that corresponded to a weakly detectable band in the stained gel. Mass spectrometry of a gel slice excised from this region revealed the presence of a few proteins such as mitochondrial NAD(P) transhydrogenase and elongation factor 1 (EF1)-α 2. It is notable that EF1 is already known to be a PKC substrate, the consequence of which is the stimulation of GDP/GTP exchange in this protein (26). Furthermore, a related isoform, EF1-α1, was also detected by mass spectrometry in a band produced by traceable PKCα (22). In the phospho-protein profile produced by PKCζ, a major protein band was detected at approximately 43 kDa. However, in the corresponding Gelcode blue-stained gel, there were no detectable bands in this region in either the wildtype or mutant samples.
MS/MS analysis of selected phosphoprotein bands produced by traceable PKC-α and –δ revealed the presence of several proteins that are related to the small GTPases, Rho, Cdc42 and Rac1 (Table 1). Small GTPase-related molecules included Rho kinase (ROCK-1), cdc42 effector protein-4 (CEP4), and p21 activating kinase-2 (PAK-2) all of which serve as downstream effectors for the activated small GTPases. Other proteins detected included α- and β-tubulin, microtubule-associated proteins (MAP4, MAPKKK7, dynein), as well as proteins that bind specifically to the growing ends (“plus ends”) of microtubules (CLASP, CLIP-1). Previous work from this laboratory identified and characterized α6-tubulin as a substrate whose phosphorylation by endogenous PKC promotes motility of MCF-10A cells (23). Several other proteins already known to be PKC substrates (Table 1) included IQGAP, VASP, and MAP4 which co-immunoprecipitated with PKC and are known to associate with small GTPases. In addition, the PKC binding protein plectin was detected. It is important to emphasize that unless otherwise known to be a PKC substrate, identification of a protein by MS/MS merely indicates its co-immunoprecipitation with PKC during the experiment and therefore its substrate status requires further verification.
TABLE 1.
Proteins that co-immunoprecipitate with traceable PKC isoforms.
| A. | ||||
|---|---|---|---|---|
| Protein associated with: |
||||
| Co-immunoprecipitating Protein | PKC-α | score | PKC-δ | score |
| *Cdc42 effector protein-4 (CEP4) | 43 kDa | 193 | 43 kDa | 196 |
| β-Actin | 43 kDa | 1060 | 43 kDa | 906 |
| MAPKK1 | 43 kDa | 53 | ||
| Dynein-1 | 53 kDa | 133 | ||
| LIM protein 3 | 53 kDa | 38 | ||
| *p21-activated kinase-2 (PAK2) | 62 kDa | 58 | ||
| Ras GTPase activating protein-binding protein 1/2 | 62 kDa | 84/103 | ||
| MAPKKK7-interacting protein | 62 kDa | 303 | ||
| AMP-activated protein kinase | 62 kDa | 47 | ||
| Desmoglein | 62 kDa | 118 | 43 kDa | 76 |
| *CLIP-associated protein-1 (CLASP-1) | 200 kDa | 81 | ||
| *Rho-activated kinase-1 and 2 (ROCK-1/2) | 200 kDa | 823/114 | 200 kDa | 868 |
| ARFGAP with Rho-GAP domain (ARAP2) | 200 kDa | 66 | ||
| *Rho/cdc42/Rac activating protein-1 (RICS) | 200 kDa | 51 | 200 kDa | 49 |
| Brefeldin A-inhibitor of GTP-exchange protein-2 (ARFGEF2) | 200 kDa | 318 | ||
| B. | |||||||
|---|---|---|---|---|---|---|---|
| Protein associated with: |
|||||||
| Ref. | PKC-α | score | PKC-δ | score | PKC-ζ | score | |
| VASP (Ser-157) | 41 | 43 kDa | 271 | 43 kDa | 275 | ||
| α-Tubulin (Ser-165) | 23 | 53 kDa | 65 | ||||
| PACSIN 2 | 42 | 62 kDa | 84 | ||||
| EF-1α | 26 | 130 kDa | 57 | ||||
| IQGAP-1 (Ser-1443) | 40 | 200 kDa | 64 | 200 kDa | 249 | ||
| Plectin | 33 | 200 kDa | 3022 | 200 kDa | 37 | ||
| MAP4 (Ser-815) | 43 | 200 kDa | 60 | ||||
A. Proteins were identified by MS/MS, as described in the ‘Methods’. The Mascot protein score is indicated next to each detected protein. Those proteins that contain one or more potential PKC phosphorylation sites at either partial or full consensus sequences are signified by (*).
B Proteins identified by MS/MS that had been previously identified as either a substrate or binding protein of PKC, are shown along with the site of PKC phosphorylation (given in parentheses).
In vitro and intracellular phosphorylation of potential substrates
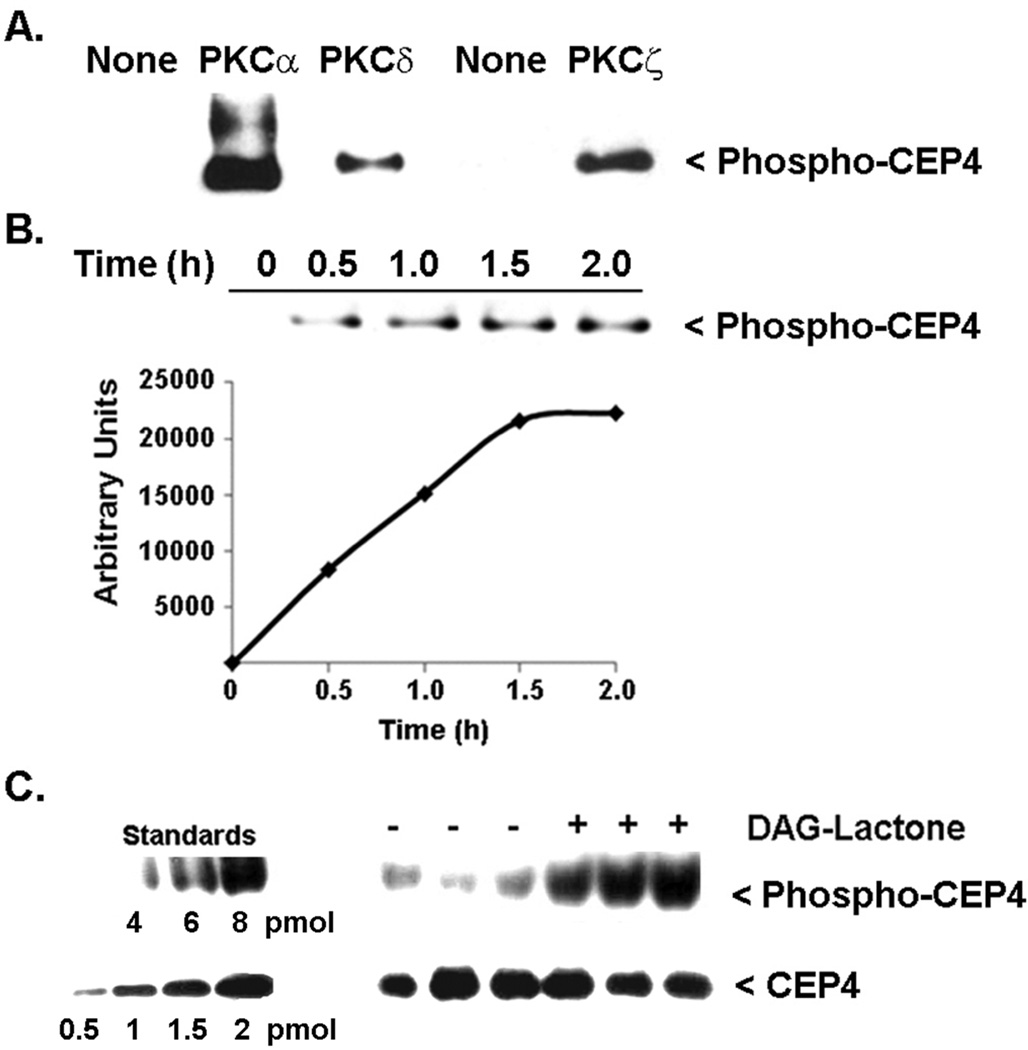
Four proteins that co-immunoprecipitated with either PKCα and/or PKCδ, namely CEP4 (43 kDa band), PAK2 (63 kDa band), ROCK-1 (200 kDa band) and CLASP-1 (170 kDa), were evaluated for intracellular phosphorylation in MCF-10A cells. These proteins were selected because they contain either partial or complete consensus sites for PKC phosphorylation, and are known to serve roles that relate to the cytoskeleton, cell morphology, or cell invasion. CEP4, ROCK1 and PAK2 are effector proteins of the small GTPases (27. 28), whereas CLASP-1 is phosphorylated by glycogen synthase kinase-β3 and thereupon mediates downstream interactions between microtubules and Rac1 (29). Each of the four candidate substrates was analyzed for intracellular phosphorylation following treatment of MCF-10A cells with DAG-lactone to stimulate endogenous activity of conventional and novel PKC isoforms. The prospective substrate was immunoprecipitated with a protein-specific antibody and the pellets were analyzed by Western blot using the PKC substrates antibody. Of these proteins, only CEP4 (48 kDa) was phosphorylated in response to DAG-lactone (Figure 5C), whereas ROCK1, CLASP-1 and PAK2 did not undergo DAG-lactone stimulated intracellular phosphorylation (data not shown). In an in vitro assay with pure, recombinant PKC (α, δ, or ζ) proteins, CEP4 underwent direct phosphorylation by each isoform (Figure 5A), thereby documenting that a direct interaction between each PKC isoform and CEP4 is indeed possible.
Figure 5.
CEP4 is a substrate of PKC in vitro and in MCF-10A cells. (A) Pure recombinant CEP4 (750 ng) was tested in vitro with pure, recombinant PKCα (170 ng), PKCδ (100 ng), or PKCζ (166 ng). The reaction medium carried out in 1X kinase buffer contained 0.1 mg/ml phosphatidylserine plus either 5 µM DAG-lactone (for PKCα, δ), or 1 µM ceramide (for PKC-ζ). The reaction was initiated by addition of 100 µM ATP and incubated at 30°C. After 30 min, each reaction was quenched with sample buffer and analysis of phosphorylation was carried out by Western blot with anti-PKC substrates antibody (1:1000). The results are representative of three independent experiments. (B) Phosphorylation of pure, recombinant CEP4 (1 µg) was carried out with pure, recombinant PKCα (0.74 µg) (Sigma-Aldrich), as in described in (A), and was complete after 2 h. Each time point consisted of 200 ng (4 pmol) CEP4 protein. (C) Intracellular phosphorylation of CEP4 was tested in MCF-10A cells cultured in 60-mm plates and treated for 1 h with 10 µM DAG-lactone or DMSO (0.1% v/v) as vehicle control. Whole cell lysates were prepared in 0.5 ml of 50 mM TRIS-HCl, 150 mM NaCl, 5 mM EGTA, 5 mM EDTA, 1% Triton-X, 10 µM bis-indoleylmaleimide, phosphatase inhibitors and protease inhibitors. Lysates were pre-cleared with a 1:1 combination of rabbit IgG-agarose and protein A/G-agarose, and CEP4 protein was immunoprecipitated with 3 µg CEP4 antibody and protein A/G-agarose. Western blotting of the immunopellets was used to assess CEP4 phosphorylation with the PKC substrates antibody (1:1500). To demonstrate equivalent protein loading, a duplicate blot was probed with anti-CEP4 (1:2000). The results are representative of two independent experiments. The ratio of intracellular phospho-CEP4 relative to total CEP4 in each sample was determined by Image J software and by reference to known standards for phospho-CEP4 or CEP4. Standard phospho-CEP4 was that produced by pure, recombinant PKCα in (B) and represents the maximally phosphorylated protein.
The stoichiometry of phosphorylation of intracellular CEP4 relative to total CEP4 was determined by reference to known amounts of pure, recombinant CEP4 standard that was phosphorylated in vitro by pure, recombinant PKCα. A time course of in vitro phosphorylation (Figure 5B) demonstrated that maximal phosphorylation of pure, recombinant CEP4 was achieved after 2.0 h. This fully phosphorylated product was subsequently used as a standard for quantitating the phosphorylation of intracellular CEP4 (Figure 5C). The experiment consisted of one Western blot containing known picomole amounts of standard phospho-CEP4 for comparison with immunoprecipitated CEP4 from MCF-10A cells treated for 1 h with DAG-lactone, and a duplicate blot with identical samples and known amounts of pure, recombinant unphosphorylated CEP4. Each blot was probed with either the PKC substrates antibody or CEP4 antibody to detect phospho-CEP4 or total CEP4 protein, respectively. Quantitation of phospho-CEP4 or total CEP4 in triplicate samples was performed with Image J software by comparing the samples with known amounts of standard CEP4 or phospho-CEP4 protein present in the same blot (Figure 5C). The ratio of phospho-CEP4 to total CEP4 in DAG-lactone-treated cells was calculated to be 3.2 ± 0.5 (pmol/pmol), the average of two independent experiments (each consisting of triplicate measurements). This finding implies that in response to DAG-lactone, PKC isoforms phosphorylate an average of three to four sites in CEP4.
Interference of intracellular CEP4 phosphorylation by shRNA-encoding plasmids and kinase-defective mutants of PKC isoforms
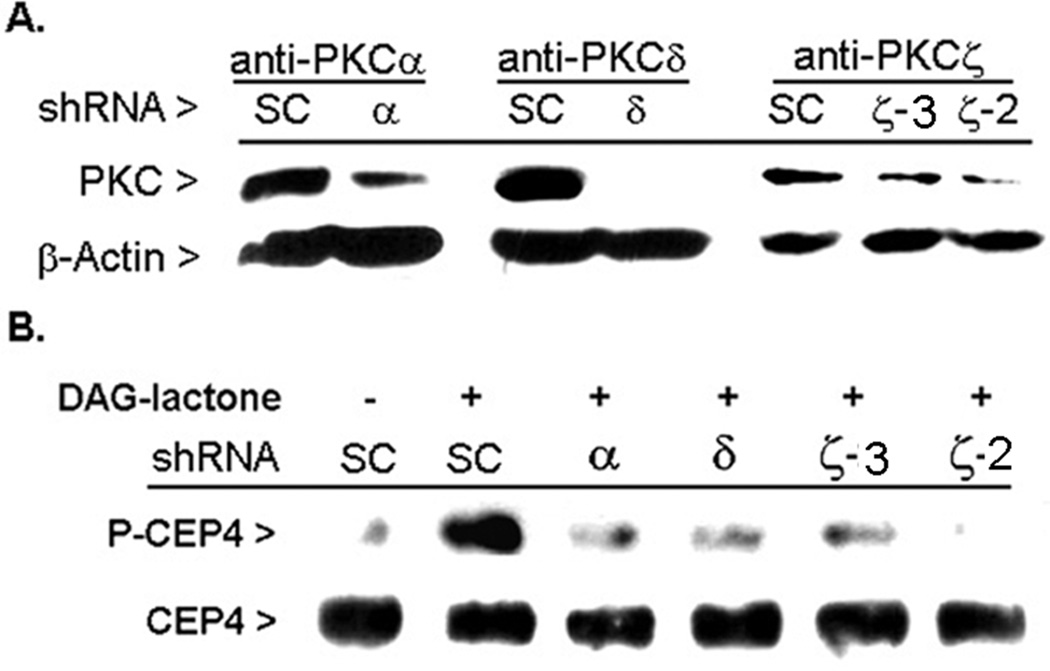
To evaluate whether CEP4 serves as an intracellular substrate for one or more PKC isoforms, MCF-10A cells were transfected with individual shRNA-encoding plasmids for α, δ, or ζ and their interference in CEP4 phosphorylation was evaluated under conditions of DAG-lactone stimulation. Each shRNA-encoding plasmid featured co-expression of GFP so that the transfection efficiency could be determined. Following a 48-h incubation period, the transfection efficiency was found to be 70% or higher. However, a period of 96 h was required to produce maximal knock-down of each PKC isoform. The single plasmids used for PKC-α and –δ had been validated by the manufacturer for their ability to knock-down their intended target and were effective at knocking down expression of these isoforms by 60–100% in MCF-10A cells (Figure 6A). Of four constructs obtained for PKCζ, two (ζ-2, ζ-3) were effective in achieving knockdown of PKCζ (by 80–90%) in MCF-10A cells. In DAG-lactone treated cells, silencing of each individual isoform led to a substantial inhibition of CEP4 phosphorylation relative to the scrambled control (SC) shRNA plasmid. When compared with SC-treated cells, inhibition of CEP4 phosphorylation averaged 60% with either the PKCα or PKCδ shRNA reagent, and >85% with PKCζ shRNA (ζ-2), where each value represents the average of three independent experiments. These findings suggest that each of the three PKC isoforms phosphorylate CEP4 in the intracellular environment.
Figure 6.
Knock-down of individual PKC isoforms blocks CEP4 phosphorylation. MCF-10A cells were cultured in 6-well plates and transiently transfected with a 3:1 mixture of PolyExpress reagent (µl) to a plasmid (µg) encoding shRNA that targets a single PKC isoform (α, δ, or ζ). Following incubation of the cells at 37°C for 96 h, lysates were prepared and normalized for total protein. An aliquot of each lysate (30 µg) was removed for demonstration of knock-down of each isoform using Western blot by use of an isoform-specific antibody, as shown in (A). The remaining portion of each lysate (100 µg protein) was immunoprecipitated with anti-CEP4 (3 µg) and the resulting immunopellets were analyzed in duplicate Western blots for either phosphorylated CEP4 (PKC substrates antibody, 1:1500 dilution), or for total CEP4 protein (anti-CEP4, 1:2000 dilution), as shown in (B). The results are representative of three independent experiments.
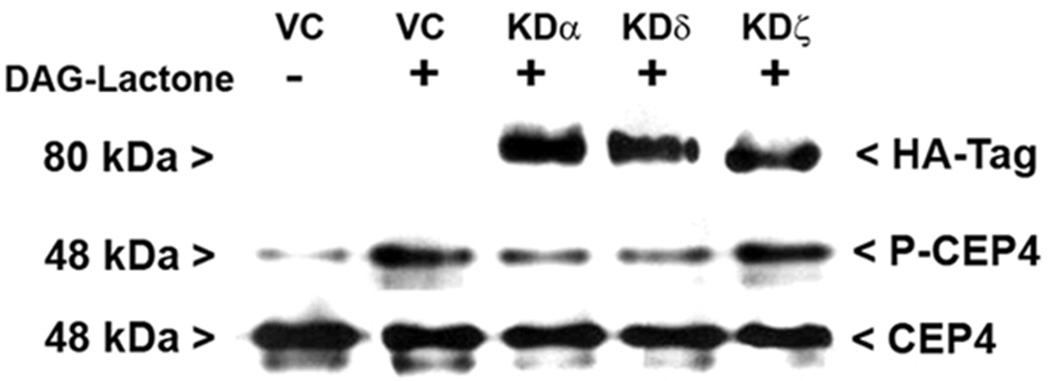
In a complementary approach, MCF-10A cells were transfected with individual kinase-dead (KD) hemagglutinin (HA)-tagged mutants for α (K368R), δ (K376R), or ζ (K281M) and their interference in CEP4 phosphorylation was evaluated under conditions of DAG-lactone stimulation. In each lysate, the expression of a kinase-dead mutant was verified by Western blot with an HA-tag antibody (Figure 7). Lysates were immunoprecipitated with anti-CEP4 and the resulting immunopellets were analyzed for phosphorylated CEP4 and for total CEP4 protein by Western blot. The results, shown in Figure 7, reveal a substantial dominant-negative effect on CEP4 phosphorylation by the KD mutants of PKC-α and -δ averaging 40–70% for three independent experiments. However, no significant interference (<10%) was observed with KD-PKCζ and may have had limited access (see “Discussion”). The agreement of the shRNA and kinase-dead mutant experiments for PKCα and –δ is further evidence that CEP4 is an intracellular substrate of these DAG-sensitive isoforms.
Figure 7.
Kinase-dead mutants for PKC-α and –δ interfere with phosphorylation of CEP4. MCF-10A cells were cultured on 60-mm plates and transfected with a 3:1 mixture of PolyExpress reagent (µl) to plasmid (µg) encoding the HA-tagged, kinase-dead (KD) mutant for each PKC isoform (KDα, KDδ, or KDζ) or the vector control (VC). Expression of HA-tagged KD mutants was verified in each whole cell lysate (50 µg/lane) by probing with anti-HA tag (1:1000). CEP-4 was immunoprecipitated from each cell lysate with anti-CEP4 and the pellets were evaluated for phosphorylated CEP4 (P-CEP4) by Western blot with the PKC substrates antibody (1:1500). Equivalent CEP4 content for all samples was established by probing immunopellets in a duplicate blot with anti-CEP4 (1:2000). The results are representative of three independent experiments.
DISCUSSION
In this study we employed the Traceable Kinase Method to define substrate profiles of three PKC isoforms in MCF-10A human breast cells. Traceable kinases for PKC-α, PKC-δ, and PKC-ζ were compared with respect to their phospho-protein profiles that were generated from high affinity binding proteins co-immunoprecipitating with each traceable kinase. Among the co-immunoprecipitated proteins for PKC-α and –δ were proteins known to serve regulatory or effector roles associated with the small GTPases. These results indicated that there is some redundancy of co-immunopreciptating proteins/substrates for PKC-α and -δ. In contrast, PKC-ζ produced a very different phosphorylation pattern with only one detectable phospho-protein that was unique to the traceable kinase.
Preparation of the traceable isoforms for PKC-δ and PKC-ζ was carried out in a manner similar to our earlier work with PKC-α (22). An important departure from that initial study was the use of a non-radioactive approach to detect phospho-protein products. This innovation was made possible by the availability of a PKC substrates antibody (Cell Signaling Technology) that specifically recognizes a phosphorylated Ser in the PKC consensus motif of unknown proteins. Judging from the number and M.W. range of the resulting profiles, the background is much lower and the sensitivity is comparable to what was observed with the radioactive method (22). The observation of background signals by the WT enzyme may indicate that there are additional protein kinases in the immunopellet able to utilize the ATP analogue (present at 100 µM) and therefore contribute to product formation during the in vitro reaction. In this regard, we observed background reactivity even in experiments that employed the radiolabelled analogue (22), signifying that it is not simply the outcome of the antibody-based method employed in the present study. Importantly, detection of several previously identified PKC substrates among proteins co-immunoprecipitating with the traceable mutants (Table 1B) supports the validity and effectiveness of the methods used here.
Our experiments focused on Cdc42 effector protein-4 (CEP4) as a newly identified PKC substrate. CEP4 was identified by MS/MS in the 43-kDa band in phospho-protein profiles generated by both PKC-α and -δ. In vitro reactions with pure, recombinant isoforms showed that each isoform can phosphorylate pure, recombinant CEP4. In intact MCF-10A cells, phosphorylation of CEP4 in response to DAG-lactone was carried out by PKCα or PKCδ, as shown by interference of CEP4 phosphorylation by individual isoform-specific shRNA reagents or kinase-dead mutants. The loss of DAG-stimulated CEP4 phosphorylation by PKCζ knockdown was surprising because it implied that this isoform, which is insensitive to DAG, phosphorylates CEP4 only in DAG-treated control cells. The possibility was considered that, in addition to PKCζ, the two PKCζ shRNA reagents also silenced DAG-sensitive PKC isoforms. However, no effect on either PKCα or PKCδ expression accompanied PKCζ knockdown (data not shown). A plausible explanation is that DAG is known to cause recruitment of PKCζ to the plasma membrane via DAG-stimulated PKC isoforms that consequently activate the MAP kinase cascade, as recently reported for bronchial epithelial cells stimulated with phorbol esters (44). It is further noted that blockade of DAG-stimulated CEP4 phosphorylation by silencing of native PKCζ (Figure 6) is inconsistent with the lack of interference by the kinase-dead PKCζ mutant (Figure 7). A reasonable explanation is that, unlike the native enzyme, the kinase-dead mutant had limited access to CEP4. For example, kinase-dead PKCζ may not have been able to bind CEP4 because its substrate binding site was already occupied by a high affinity substrate whose release depended upon being phosphorylated. Nevertheless, the outcome produced by silencing native PKCζ by each of two different shRNA reagents clearly demonstrated the role of this isoform in CEP4 phosphorylation. The efforts taken here to establish which of the three PKC isoforms phosphorylate CEP4 in the intracellular environment, lead us to conclude that CEP4 serves as a substrate for all of them.
CEP4 is a member of a family of Cdc42 effector proteins (also known as BORG proteins) that upon binding the activated GTP-bound form of Cdc42 with high affinity, promotes increased actin stress fiber and pseudopodia formation, and leads to loss of E-cadherin from adherens junctions (30, 31). Judging from the strong signals we observed for immunoprecipitated CEP4 in Western blots, this protein is highly abundant in both MCF-10A cells (Figure 5) and human breast tumor cells (MDA-MB-231 cells) (unpublished observations). It is notable that there are five structurally-related human CEP isoforms (31), of which CEP4 is the only member to possess a canonical PKC consensus site at Ser-18 (two pairs of cationic residues flanking a serine or threonine residue). CEP4 was phosphorylated in a stoichiometric ratio of almost 4:1 (mol phosphoCEP4/mol CEP4) in DAG-lactone-stimulated MCF-10A cells (Figure 5C). Therefore, it is likely that additional sites are being modified by DAG-stimulated PKC isoforms. Inspection of the human CEP4 primary sequence shows that in addition to a full consensus site centered at Ser-18, there are three partial consensus sites (two cationic residues on one side of a serine residue) located at Ser-77, Ser-80, and Ser-86. The significance of these proposed sites of phosphorylation to CEP4 function provides an avenue for further investigation.
In addition to CEP4, our investigation considered ROCK1, PAK2, and CLASP-1 as substrates of PKC by testing for their intracellular phosphorylation following DAG-lactone treatment of non-transformed MCF-10A cells. None of these proteins exhibited detectable phosphorylation in MCF-10A cells under conditions in which only DAG-responsive PKC isoforms were stimulated (data not shown). Nonetheless, their co-immunoprecipitation with PKC-α and δ isoforms in MCF-10A cell lysates implies that they are physically associated either directly or indirectly with each isoform. It is possible that these proteins are components of a hub that includes other PKC substrates (e.g. MAP4, VASP, and IQGAP) and thereby organizes PKC-related signaling components. That plectin was also observed among the co-immunoprecipitated proteins for both PKCα and –δ (Table 1) supports this idea since it is known to sequester RACK (Receptor for Activated C Kinase) and to provide a cytoskeletal scaffold for binding and directing PKC signaling activity at the plasma membrane and cytoskeleton (5, 32, 33). In view of the array of GTPase-related proteins that co-immunoprecipitated with PKC, this putative hub may organize certain PKC isoforms, the small GTPases (Rho, Cdc42, Rac1), their respective downstream effectors (ROCK1, PAK, CEP4), and their associated regulatory enzymes (RICS, IQGAP, ARFGEF2, ARFGAP).
There is growing recognition that the RhoA family of GTPases (RhoA, Cdc42, and Rac 1) and their regulatory proteins and effectors, have mechanistic significance to human breast cancer (27, 28, 34–36). An earlier study from this laboratory demonstrated that Rac1 was functionally involved in the motility phenotype produced by PKC-α expression in MCF-10A cells (1). More recent studies from other laboratories showed that ErbB2-mediated transformation of human breast cancer epithelial cells requires the interaction of Rac1 with its downstream effector PAK (37), and that PAK1/PAK2 mediate invasiveness of human breast cancer cells (38). However, while analyzing potential substrates, we found that PKC-α and -δ phosphorylated PAK2 in vitro, but found no evidence that PKC phosphorylates this enzyme in response to DAG-lactone treatment of MCF-10A cells (data not shown). Other studies have indicated that PKC influences the binding activity of inhibitory proteins that regulate the activation state of the Rho family of GTPases. In this regard, a guanine nucleotide dissociation inhibitor was shown to undergo phosphorylation by PKCα, causing it to dissociate and thereby promote nucleotide exchange and activation of RhoA GTPase (39). In another example, PKC-mediated phosphorylation of IQGAP increased its affinity for nucleotide-depleted, inactive Cdc42 relative to the Cdc42-GTP active form, and was proposed as a means to sequester Cdc42 in its inactive form (40). A common theme in these examples is that PKC-mediated phosphorylation provides a mechanism by which to modulate the affinity of a regulatory protein for a small Rho GTPase. An avenue for further investigation will be to determine if PKC-mediated phosphorylation of CEP4 alters its binding affinity for activated Cdc42, thereby modulating CEP4/Cdc42 signaling pathways and producing one or more phenotypes previously attributed to PKC in human breast cells.
ACKNOWLEDGMENTS
This research was supported by grant CA125632 from the National Cancer Institute (to S.A.R.).
Plasmids encoding bovine PKC-α, human PKC-δ, and mouse PKC-ζ were gifts from Dr. Peter J. Parker (Cancer Research UK London Research Institute, London, U.K.), Prof. K. Yoshida (Tokyo Medical and Dental University, Japan) and Prof. M. G. Kazanietz (University of Pennsylvania), respectively. The DAG-lactone reagent was provided by Dr. V. Marquez (NCI-Frederick, NIH). Mass spectrometry was performed at the Keck Foundation Mass Spectrometry Resource Laboratory at Yale Cancer Center. The authors thank Professors Corinne Michels, Karl Fath, Cathy Savage-Dunn (Queens College), Jill Bargonetti (Hunter College), and Jeffrey Segall (Albert Einstein College of Medicine) for helpful discussions throughout the course of this research.
ABBREVIATIONS
- PKC
protein kinase C
- WT
wildtype
- VC
vector control
- CEP4
Cdc42 effector protein-4PS, phosphatidylserine
- DAG-lactone
diacylglycerol-lactone
- MBP
myelin basic protein
- shRNA
small hairpin RNA
- GFP
green fluorescent protein
REFERENCES
- 1.Sun X-g, Rotenberg SA. Over-expression of PKCα in MCF-10A human breast cells engenders dramatic alterations in morphology, proliferation and motility. Cell Growth Differ. 1999;10:343–352. [PubMed] [Google Scholar]
- 2.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol. Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teicher BA. Protein kinase C as a therapeutic target. Clin. Cancer Res. 2006;12:5336–5345. doi: 10.1158/1078-0432.CCR-06-0945. [DOI] [PubMed] [Google Scholar]
- 4.Konopatskaya O, Poole AW. Protein kinase Cα: disease regulator and therapeutic target. Trends Pharmacol. Sci. 2010;31:8–14. doi: 10.1016/j.tips.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsson C. Protein kinase C and regulation of the actin cytoskeleton. Cell. Signalling. 2006;18:276–284. doi: 10.1016/j.cellsig.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Ghoul A, Serova M, Benhadji KA, Cvitkovic E, Faivre S, Philips E, Calvo F, Lokiec F, Raymond E. Protein kinase C α and δ are members of a large family of high potential for anticancer targeted therapy. Targ. Oncol. 2006;1:42–53. [Google Scholar]
- 7.Lonne GK, Cornmark L, Zahirovic IO, Landberg G, Jirstrom K, Larsson C. PKCalpha expression is a marker for breast cancer aggressiveness. Mol. Cancer. 2010;9:76–90. doi: 10.1186/1476-4598-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerfoot C, Huang W, Rotenberg SA. Immunohistochemical analysis of advanced human breast carcinomas reveals down-regulation of protein kinase Cα. J. Histochem. Cytochem. 2004;52:419–422. doi: 10.1177/002215540405200314. [DOI] [PubMed] [Google Scholar]
- 9.Kiley SC, Clark KJ, Goodnough M, Welch DR, Jaken S. Protein kinase delta involvement in mammary tumor cell metastasis. Cancer Res. 1999;59:3230–3238. [PubMed] [Google Scholar]
- 10.McCracken MA, Miraglia LJ, McKay RA, Strobl JS. Protein kinase C δ is a pro- survival factor in human breast tumor cell lines. Mol. Cancer Ther. 2003;2:273–281. [PubMed] [Google Scholar]
- 11.Kruger JS, Reddy KB. Distinct mechanisms mediate the initial and sustained phases of cell migration in epidermal growth factor receptor-overexpressing cells. Mol. Cancer Res. 2003;1:801–809. [PubMed] [Google Scholar]
- 12.McKiernan E, O’Brien K, Grebenchtchikov N, Geurts-Moespot A, Sieuwerts AM, Martens JW, Magdolen V, Evoy D, McDermott E, Crown J, Sweep FC, Duffy MJ. Protein kinase Cdelta expression in breast cancer as measured by real-time PCR, western blotting, and ELISA. Brit. J. Cancer. 2008;18:1644–1650. doi: 10.1038/sj.bjc.6604728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonne GK, Masoumi KC, Lennartsson J, Larsson C. Protein kinase Cdelta supports survival of MDA-MB-231 breast cancer cells by suppressing the ERK1/2 pathway. J. Biol. Chem. 2009;284:33456–33465. doi: 10.1074/jbc.M109.036186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urtreger AJ, Grossoni VC, Falbo KB, Kazanietz MG, Bal de Kier Joffe ED. Atypical protein kinase C-ζ modulates clonogenicity, motility, and secretion of proteolytic enzymes in murine mammary cells. Mol. Carcinogenesis. 2005;42:29–39. doi: 10.1002/mc.20066. [DOI] [PubMed] [Google Scholar]
- 15.Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ting AY, Witte K, Shah K, Kraybill B, Shokat KM, Schultz PG. Phage-display evolution of tyrosine kinases with altered nucleotide specificity. Biopolymers. 2001;60:220–228. doi: 10.1002/1097-0282(2001)60:3<220::AID-BIP10035>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 17.Shah K, Shokat KM. A chemical genetic screen for direct v-src substrates reveals ordered assembly of a retrograde signaling pathway. Chem. Biol. 2002;9:35–47. doi: 10.1016/s1074-5521(02)00086-8. [DOI] [PubMed] [Google Scholar]
- 18.Witucki LA, Huang X, Shah K, Liu Y, Kyin S, Eck MJ, Shokat KM. Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phosphoacceptor specificity of the wildtype enzyme. Chem. Biol. 2002;9:25–33. doi: 10.1016/s1074-5521(02)00091-1. [DOI] [PubMed] [Google Scholar]
- 19.Habelhah H, Shah K, Huang L, Burlingame AL, Shokat KM, Ronai Z. Identification of new JNK substrate using ATP pocket mutant JNK and a corresponding ATP analogue. J. Biol. Chem. 2001;276:18090–18095. doi: 10.1074/jbc.M011396200. [DOI] [PubMed] [Google Scholar]
- 20.Eblen ST, Kumar NV, Shah K, Henderson MJ, Watts CKW, Shokat KM, Weber MJ. Identification of novel ERK2 substrates through use of an engineered kinase and ATP analogs. J. Biol. Chem. 2003;278:14926–14935. doi: 10.1074/jbc.M300485200. [DOI] [PubMed] [Google Scholar]
- 21.Hindley AD, Park S, Wang L, Shah K, Wang Y, Hu X, Shokat KM, Kolch W, Sedivy JM, Yeung KC. Engineering the serine/threonine protein kinase Raf-1 to utilize an orthogonal analogue of ATP substituted at the N6 position. FEBS Lett. 2004;556:26–34. doi: 10.1016/s0014-5793(03)01352-8. [DOI] [PubMed] [Google Scholar]
- 22.Abeyweera TP, Rotenberg SA. Design and characterization of a traceable protein kinase C-α. Biochemistry. 2007;46:2364–2370. doi: 10.1021/bi0622017. [DOI] [PubMed] [Google Scholar]
- 23.Abeyweera TP, Chen X, Rotenberg SA. Phosphorylation of α6-tubulin by protein kinase Cα activates motility of human breast cells. J. Biol. Chem. 2009;284:17648–17656. doi: 10.1074/jbc.M902005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Bermejo ML, Leskow FC, Fuji T, Wang Q, Blumberg PM, Ohba M, Kurok T, Han KC, Lee J, Marquez VE, Kazanietz MG. Diacylglycerol (DAG)-lactones, a new class of protein kinase C (PKC) agonists, induce apoptosis in LNCaP prostate cancer cells by selective activation of PKCalpha. J. Biol. Chem. 2002;277:645–655. doi: 10.1074/jbc.M107639200. erratum in (2004) J. Biol. Chem. 279, 23846. [DOI] [PubMed] [Google Scholar]
- 25.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 26.Peters HI, Chang YW, Traugh JA. Phosphorylation of elongation factor 1 (EF-1) by protein kinase C stimulates GDP/GTP-exchange activity. Eur. J. Biochem. 1995;234:550–556. doi: 10.1111/j.1432-1033.1995.550_b.x. [DOI] [PubMed] [Google Scholar]
- 27.Bouzahzah B, Albanese C, Ahmed F, Pixley F, Lisanti MP, Segall JD, Condeelis J, Joyce D, Minden A, Der CJ, Chan A, Symons M, Pestel RG. Rho family GTPases regulate mammary epithelium cell growth and metastasis through distinguishable pathways. Mol. Med. 2001;7:816–830. [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Y, Olufemi L, Wang MT, Nie D. Role of rho GTPases in breast cancer. Front. Biosci. 2008;13:759–776. doi: 10.2741/2718. [DOI] [PubMed] [Google Scholar]
- 29.Wittman T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3β in migrating epithelial cells. J. Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirsch DS, Pirone DM, Burbelo PD. A new family of cdc42 effector proteins, CEPs, function in fibroblast and epithelial cell shape changes. J. Biol. Chem. 2001;276:875–883. doi: 10.1074/jbc.M007039200. [DOI] [PubMed] [Google Scholar]
- 31.Joberty G, Perlungher RR, Macara IG. The Borgs, a new family of cdc42 and TC10 GTPase-interacting proteins. Mol. Cell. Biol. 1999;19:6585–6597. doi: 10.1128/mcb.19.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaken S, Parker PJ. Protein kinase C binding partners. BioEssays. 2000;22:245–254. doi: 10.1002/(SICI)1521-1878(200003)22:3<245::AID-BIES6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 33.Osmanagic-Myers S, Wiche G. Plectin-RACK1 (receptor for activated C kinase-1) scaffolding: a novel mechanism to regulate protein kinase C activity. J. Biol. Chem. 2004;279:18701–18710. doi: 10.1074/jbc.M312382200. [DOI] [PubMed] [Google Scholar]
- 34.Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Brit. J. Cancer. 2002;87:635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burbelo P, Wellstein A, Pestell RG. Altered rho GTPase signaling pathways in breast cancer cells. Breast Cancer Res. Treat. 2004;84:43–48. doi: 10.1023/B:BREA.0000018422.02237.f9. [DOI] [PubMed] [Google Scholar]
- 36.Zuo Y, Wu Y, Chakraborty C. Cdc42 negatively regulates intrinsic migration of highly aggressive breast cancer cells. J. Cell Physiol. 2012;227:1399–1407. doi: 10.1002/jcp.22853. [DOI] [PubMed] [Google Scholar]
- 37.Arias-Romero LE, Villamar-Cruz O, Pacheco A, Kosoff R, Huang M, Muthuswamy SK, Chernoff J. A Rac-Pak signaling pathway is essential for ErbB2-mediated transformation of human breast epithelial cancer cells. Oncogene. 2010;29:5839–5849. doi: 10.1038/onc.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coniglio SJ, Zavarella S, Symons MH. Pak1 and Pak2 mediate tumor cell invasion through distinct signaling mechanisms. Mol. Cell. Biol. 2008;28:4162–4172. doi: 10.1128/MCB.01532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta D, Rahman A, Malik AB. Protein kinase C-α signals rho-guanine nucleotide dissociation inhibitor phosphorylation and rho activation and regulates the endothelial cell barrier function. J. Biol. Chem. 2001;276:22614–22620. doi: 10.1074/jbc.M101927200. [DOI] [PubMed] [Google Scholar]
- 40.Grohmanova K, Schlaepfer D, Hess D, Gutierrez P, Beck M, Kroschewski R. Phosphorylation of IQGAP1 modulates its binding to cdc42, revealing a new type of Rho-GTPase regulator. J. Biol. Chem. 2004;279:48495–48504. doi: 10.1074/jbc.M408113200. [DOI] [PubMed] [Google Scholar]
- 41.Chitaley K, Chen L, Galler A, Walter U, Daum G, Clowes AW. Vasodilator-stimulated phosphoprotein is a substrate for protein kinase C. FEBS Lett. 2004;556:211–215. doi: 10.1016/s0014-5793(03)01435-2. [DOI] [PubMed] [Google Scholar]
- 42.Ritter B, Modregger J, Paulsson M, Plomann M. PACSIN 2, a novel member of the PACSIN family of cytoplasmic adapter proteins. FEBS Lett. 1999;454:356–362. doi: 10.1016/s0014-5793(99)00830-3. [DOI] [PubMed] [Google Scholar]
- 43.Mori A, Aizawa H, Saido TC, Kawasaki H, Mizuno K, Murofushi H, Suzuki K, Sakai H. Site-specific phosphorylation by protein kinase C inhibits assembly-promoting activity of microtubule-associated protein 4. Biochemistry. 1991;30:9341–9346. doi: 10.1021/bi00102a029. [DOI] [PubMed] [Google Scholar]
- 44.Xiao H, Bai XH, Wang Y, Kim H, Mak AS, Liu M. MEK/ERK pathway mediates PKC activation-induced recruitment of PKCζ and MMP-9 to podosomes. J. Cell Physiol. 2012 doi: 10.1002/jcp.24146. (Epub ahead of print). [DOI] [PubMed] [Google Scholar]