Abstract
Migration of resident dendritic cells (DC) from the skin to local lymph nodes (LN) triggers T cell-mediated immune responses during cutaneous infection, autoimmune disease and vaccination. Here we investigated whether the development and migration of skin resident DC were regulated by interferon regulatory factor 4 (IRF4), a transcription factor that is required for the development of CD11b+ splenic DC. We found that the skin of naïve IRF4−/− mice contained normal numbers of epidermal Langerhans cells (eLC) and increased numbers of CD11b+ and CD103+ dermal DC populations, indicating that tissue DC development and skin residency is not disrupted by IRF4 deficiency. In contrast, numbers of migratory eLC and CD11b+ dermal DC were significantly reduced in the cutaneous LN of IRF4−/− mice, suggesting a defect in constitutive migration from the dermis during homeostasis. Upon induction of skin inflammation, CD11b+ dermal DC in IRF4−/− mice did not express the chemokine receptor CCR7, and failed to migrate to cutaneous LN, while the migration of eLC was only mildly impaired. Thus, while dispensable for their development, IRF4 is crucial for the CCR7-mediated migration of CD11b+ dermal DC, a predominant population in murine and human skin that plays a vital role in normal and pathogenic cutaneous immunity.
Introduction
DC in barrier tissues such as skin are crucial for the initiation of immune responses to infection, vaccination or environmental chemicals that induce allergic contact dermatitis. Cutaneous DC also contribute to the pathology of skin diseases such as psoriasis and atopic dermatitis (1) and cutaneous graft-versus-host disease secondary to allogeneic bone marrow transplantation (2). Inflammation of the skin triggers activation of epidermal and dermal DC, followed by their migration to cutaneous lymph nodes (cLN) where they present peripheral antigens to T cells (3). Tissue DC also constitutively migrate to cutaneous LN during homeostasis (4), and therein play an important role in the induction of tolerance to self or environmental antigens (5). Thus identification of the mechanisms that regulate skin DC migration will provide insight into immunity and tolerance as well as disease pathogenesis.
Multiple subsets of migratory tissue DC have been identified in skin and within the CD11cint MHCIIhi compartment in cutaneous LN [reviewed in (6)]. These include the epidermal langerin+CD11bhiCD103− Langerhans cells (termed eLC), which also may be found in transit in the dermis, and at least three dermal DC (dDC) subsets, the langerin−CD11bhiCD103− DC (termed CD11bhi dDC), langerin−CD11blowCD103− DC (termed CD11blow dDC) and the langerin+CD11blowCD103+ DC (termed CD103+ dDC). The CD11b+ dDC comprise the greatest fraction of dermal DC, while CD103+ dDC are present in minor numbers. These phenotypically defined tissue DC subtypes also are distinguished by their developmental dependence on specific precursors, particular combinations of transcription factors and one or more of the cytokines Flt3 Ligand, GM-CSF or M-CSF (7, 8). eLC require TGFβ, IRF8, ID2, RUNX3 and M-CSF for their development and are derived from self-renewing precursors that seed the skin in fetal life, although they also develop from monocytes during inflammation (8). CD103+ dDC develop from conventional DC (cDC)-restricted precursors (pre-cDC) present in blood, peripheral tissue and lymphoid organs, in a process that requires Flt3 Ligand and GM-CSF and the transcription factors IRF8, ID2 and BATF3 (9–13). In this regard, CD103+ dDC show similar developmental requirements to conventional CD8α+CD11blow DC in the spleen (11).
In contrast, the developmental requirements for the CD11b+ dDC are less well defined. CD11b+ tissue DC depend on Flt3 Ligand, M-CSF and GM-CSF, and adoptive transfer and lineage tracing experiments showed that CD11b+ tissue DC can be derived from both blood monocytes and pre-cDC (9, 14, 15). However, the transcription factors required for their development remain unknown. One candidate was IRF4 since IRF4−/− mice lack conventional CD11bhiCD8α− DC in the spleen, and in this early work, a role of IRF4 in non-lymphoid tissue DC subsets was not investigated (16, 17). Furthermore, in GM-CSF-driven cultures, IRF4−/− BM precursors develop into CD11c+CD11b+ cells that have reduced CIITA mRNA and fail to significantly express MHC class II, suggesting a block in a late developmental stage (16, 17). This prior work led us to test the hypothesis that CD11b+ dermal DC development requires IRF4.
Although most DC subsets have the ability to present antigen and trigger T cell responses, their different developmental requirements and mature phenotypes suggest DC are organized into distinct lineages, which are specialized for particular functional responses. These functional responses may be regulated by the same transcription factors required for DC development, since expression of these factors is usually maintained in mature DC (16, 17). Indeed, both IRF4 and IRF8 regulate inflammatory cytokine production, albeit by distinct mechanisms (18, 19). IRF8 promoted chemokine-induced migration of eLC in vitro and contact hypersensitivity in vivo, although effects of IRF8 deficiency to reduce eLC development or impair migration could not be easily distinguished in vivo (12).
Tissue DC migration to cutaneous LN in homeostasis and inflammation requires the chemokine receptors CCR7 and CXCR4 (4, 20). While eLC employ CXCR4 for migration from the epidermis to the dermis (21), eLC and dDC present in the dermis require CCR7 for entry into the dermal lymphatic vessels and for localization in the T cell zone of LN (4). DC derived from bone marrow of IRF8−/− mice showed reduced levels of CCR7 mRNA, suggesting IRF8 regulates CCR7, consistent with the defect in eLC migration in these mice (12). IRF4 regulates chemokine receptor expression in pre-B cells (22), but a role for IRF4 in regulation of chemokine receptors during skin DC migration has not been reported. Studies of DC migration in contact hypersensitivity (CHS) models of allergic contact dermatitis have revealed roles for migratory langerin+ tissue DC subsets: CD103+ dDC promote but are not absolutely required for CHS, while eLC may suppress CHS (3, 11, 23). However, although they constitute the majority of migrating DC, transcription factors governing the function of CD11b+ dDC during the CHS response have not been identified.
Here we used IRF4−/− mice (24) to determine if IRF4 regulates skin DC development and/or migration during homeostasis and inflammation. We found that naive IRF4−/− mice harbor normal numbers of eLC in the epidermis and increased numbers of CD11b+ and CD103+ DC in the dermis, indicating that the development and skin residency of these DC subsets is not disrupted by IRF4 deficiency. However, numbers of migratory eLC and CD11b+ dDC were significantly reduced in the cutaneous LN of IRF4−/− mice in homeostasis, suggesting a defect in migration from dermis to LN, consistent with the increased numbers of CD11b+ dDC in the dermis. In contrast, the CD103+ dDC subset showed a modest increase in development and migration in IRF4−/− mice. Upon induction of skin inflammation in a CHS assay, CD11b+ dDC in IRF4−/− mice did not express CCR7, and failed to migrate to the draining LN. However, eLC migration after induction of CHS was only mildly impaired in IRF4−/− mice, suggesting that IRF4 governs eLC migration primarily in the absence of inflammation. Taken together, these data show that while not required for their development, IRF4 is crucial for CD11b+ dDC expression of CCR7 and migration from the dermis to the cutaneous LN during homeostasis and skin inflammation.
Materials and Methods
Mice
Heterozygous IRF4+/− on the C57BL/6 background (24) were obtained from Dr. T. Mak (University of Toronto) and bred at OMRF to yield the female and male IRF4+/+ and IRF4−/− littermates analyzed in experiments. Mice were analyzed at 5–6 weeks of age. Female CD45.1+ C57BL/6 mice were obtained from the NCI Animal Production Program. The OMRF IACUC approved the studies.
Cell isolation
Cutaneous (axillary and brachial) LN were digested to a single cell suspension with collagenase type D (1 mg/ml) and DNAse I (0.1 mg/ml) (both from Roche) in Ca2+ and Mg2+-free Hank’s Balanced Salt Solution (HBSS) at 37°C for 30 min. Dermal and epidermal sheets were obtained after treatment of the skin with trypsin (0.25%) in HBSS for 1 hour at 37°C. A dermal cell suspension was obtained upon digestion of dermal sheets in RPMI with 10% FCS, collagenase type D (5 mg/ml), DNAse I (0.2 mg/ml) and hyaluronidase (1.5 mg/ml) (from Worthington) at 37°C for 1 hour as described (25). The epidermal sheets were aspirated up and down in a syringe and then filtered to obtain a single cell suspension. When purified for RNA isolation, epidermal DC were sorted as CD45+ MHCII+ cells.
Flow cytometry
After isolation from tissue, cells were immediately processed for flow cytometry by pre-incubating with anti-CD16/32, and labeling with optimally titered mAbs in FACS buffer (PBS, 5% newborn calf serum, 0.1% sodium azide). LN, spleen, and dermal cells and bone marrow-derived DC were stained with 6–7 parameter combinations of fluorochrome or biotin labeled mAbs specific for CD45.2, CD11c, CD8α, CD11b, CD103, langerin, MHCII, B220, CCR7, CD86 (obtained from BD Biosciences, eBioscience or Biolegend). After surface marker staining, intracellular staining with the anti-langerin Ab was done using a buffer kit from BD Biosciences. Epidermal cells were stained with mAbs specific for CD45.2 and MHCII. Samples were run on an LSRII instrument (BD Biosciences) and data analyzed with FlowJo (TreeStar) software. In the cLN, eLC were gated as CD11c+MHCII+CD8α−langerin+CD11bhiCD103−, CD11bhi dDC were gated as CD11c+MHCII+CD8α−langerin−CD11bhiCD103−, CD11blow dDC were gated as CD11c+MHCII+CD8α−langerin−CD11blowCD103− and CD103+ dDC were gated as CD11c+MHCII+CD8α−langerin+CD11blowCD103+.
Immunohistochemistry
After removal of fat and cartilage, ear skin was incubated in 0.5M EDTA for 1 hr at 37°C to separate the epidermal sheets from the dermis as described (26). The epidermal sheets were fixed in cold acetone for 20 min. and incubated with a PE-conjugated anti-langerin Ab (eBioscience). At least 8 images per sample were captured by AxioVision software after visualization with an Axiovert 200M microscope at 20X magnification. The numbers of eLC/mm2 were calculated.
Induction of contact hypersensitivity
Dibutyl phthalate-acetone (1:1) mixed with a fluorescent cell tracker, chloromethyl fluorescein diacetate (2 mM, CMFDA) (Invitrogen) was applied to the skin of one ear (26). After 24 or 72 hours, DC subpopulations in the dermis, epidermis and the draining auricular LN were analyzed by multi-parameter flow cytometry.
Bone marrow DC cultures
DC were differentiated from bone marrow cells using a GM-CSF-driven culture model as described (27). On day 7, cells were stimulated for 12–18 hr with 100 ng/ml LPS.
Quantitative real time PCR
qPCR of the Ccr7 gene was performed on cDNA generated from the RNA of bone marrow-derived DC and sorted epidermal eLC. Relative expression of genes was determined using the ΔΔCt method with normalization to Gapdh expression. Specific primer sequences were: sense Gapdh 5′ AGGTCGGTGTGAACGGATTTG 3′, antisense Gapdh 5′ TGTAGACCATGTAGTTGAGGTCA 3′; sense Ccr7 5′ CCGTGGCAGACATCCTTTTC 3′, antisense Ccr7 5′ AGGTAGACGCCAAAGATCCAG 3′.
Chemotaxis assay
Bone marrow-derived DC were activated for 12 hr with LPS and 300,000 cells were plated in the upper well of 5μM transwell plates (Costar) in RPMI + 1% BSA. CCL21 (100ng/ml) (Peprotech) was present in the lower chamber. After 3 hours, the number of migrated MHCII+ CD11c+ cells in the lower chamber was determined by flow cytometry.
Adoptive DC transfer
Bone marrow-derived DC from WT and IRF4−/− mice (CD45.2+) were stimulated for 12 hr with LPS and labeled with Cell Trace carboxyfluorescein diacetate, succinimidyl ester (CFSE) or Cell Trace Violet (Invitrogen) according to reagent instructions. 106 cells of each genotype were mixed 1:1 and injected intra-dermally into recipient CD45.1+ C57BL/6 mice as described (4). In independent transfers, WT and IRF4−/− DC were labeled with each cell tracer to ensure that migration was not affected by the cell tracer or labeling procedures. After 36 hrs, popliteal LN cells were analyzed for the presence of CD45.2+ cell tracer-labeled donor DC by flow cytometry.
Statistical analyses
Significant differences between values measured in female and male IRF4+/+ and IRF4−/− mice were determined using the nonparametric Mann-Whitney test, the Wilcoxon matched-pairs signed rank test, the unpaired t test, or a one-way ANOVA with Bonferroni post tests in Prism software as indicated in figure legends. Differences were considered significant when p<0.05. To identify possible sex differences in the effect of genotype on a measured parameter, the data in Fig. 1 also were analyzed using a two-way ANOVA with Bonferroni post tests. The variance in genotype measurements was not due to a genotype x sex interaction, so it was possible to combine female and male data in the Mann-Whitney test.
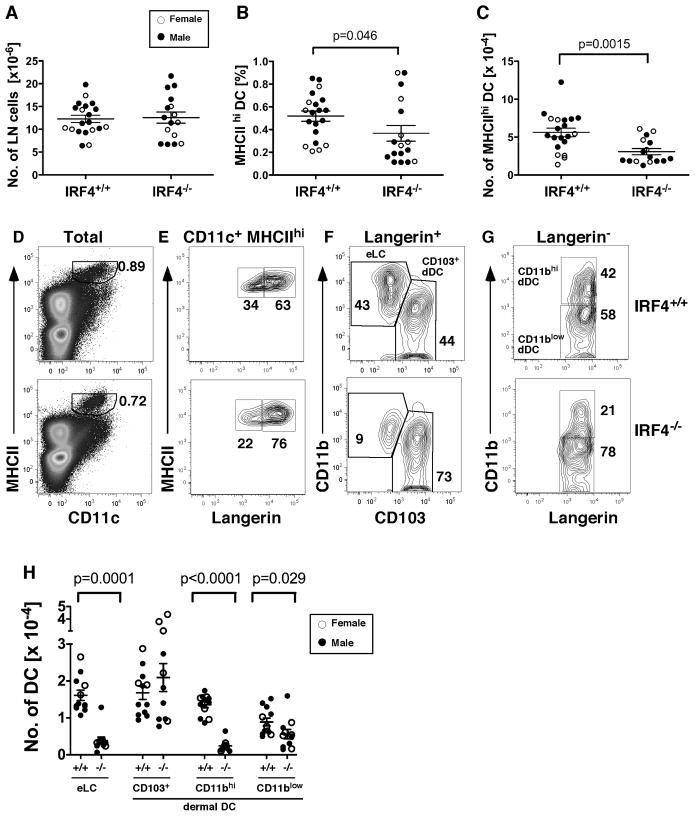
Fig. 1. IRF4−/− cLN have reduced numbers of epidermal Langerhans cells and dermal CD11b+ DC.
(A) Total numbers of inguinal LN cells in IRF4+/+ and IRF4−/− mice. In all graphs, symbols indicate individual female (open circles) and male (closed circles) mice. The mean and SEM are indicated. (B) The fraction of MHCIIhi CD11c+ DC and (C) the numbers of MHCIIhi CD11c+ DC in individual mice were determined by flow cytometry. (D) Shown is the gating of MHCIIhi CD11c+ DC (percentage indicated) within total LN cells. The gating of (E) langerin+ and langerin− DC within the MHCIIhi CD11c+ fraction, (F) CD103+ and CD11bhi DC within the langerin+ fraction, and (G) CD11bhi and CD11blow DC within the langerin− fraction. (H) The numbers of eLC (CD11bhilangerin+CD103−), CD103+ dDC (CD11blowlangerin+CD103+), CD11bhi dDC (CD11bhilangerin−CD103−) and CD11blow dDC (CD11blowlangerin−CD103−) in LN of IRF4+/+ (+/+) and IRF4−/− (−/−) mice. The data were analyzed using a two-way ANOVA with Bonferroni post tests to identify significant differences between sex and genotype. The variance was not due to a genotype x sex interaction. Therefore we used a nonparametric Mann-Whitney test on combined male and female data (n=16–20) to determine significant differences in IRF4+/+ and IRF4−/− genotypes; p values are indicated. A significant sex difference was present only within the CD103+ dDC population in both IRF4+/+ and IRF4−/− mice (two-way ANOVA, p=0.0089).
Results
While the total number of cLN cells is similar in young IRF4+/+ and IRF4−/− mice, the number of migratory skin-resident CD11b+ DC in cLN is significantly reduced in IRF4−/− mice during homeostasis
We used flow cytometry to analyze migratory DC populations in cLN of young ~5 week old mice, in which the total number of cLN cells is similar in IRF4−/− or IRF4+/+ wild type (WT) mice (Fig. 1A). Analysis of young mice was important because IRF4−/− mice develop lymphadenopathy at ~12 weeks of age, due to the accumulation of T and B cells (24).
IRF4−/− mice showed a significant reduction in the percentage and number of skin-derived migratory CD11c+MHCIIhi DC in cLN (Fig. 1, B–D). Within this CD11c+MHCIIhi population of migratory DC, IRF4−/− mice showed a significant reduction in percentage and number of three DC subpopulations, including eLC (langerin+CD11bhiCD103−), CD11bhi dDC (langerin−CD11bhiCD103−) and CD11blow dDC (langerin−CD11blowCD103−) (Fig. 1, E–H). In contrast, the number of CD103+ dDC (langerin+CD11blowCD103+)tended to increase in IRF4−/− mice, but was not significantly different (Fig. 1H). These data led us to suspect that, like conventional splenic CD11b+CD4+ DC (Fig. S1), IRF4 was required for the development of CD11b+ subsets of epidermal and dermal DC.
Previously we showed that IRF4 expression in myeloid progenitors is increased by estrogen receptor alpha signaling during GM-CSF-mediated DC differentiation (27). Thus, in this study we noted the sex of the mice to determine a possible sex bias in IRF4-regulated phenotypes. Statistical analyses showed that the effect of IRF4 deficiency on migratory DC numbers did not differ in males and females. However, we did find that females have greater numbers of CD103+ dDC in cLN in both WT and IRF4−/− mice (p=0.0089) (Fig. 1H).
In IRF4−/− mice, CD11b+ eLC are present in normal numbers in the epidermis, but accumulate in the dermis in homeostasis
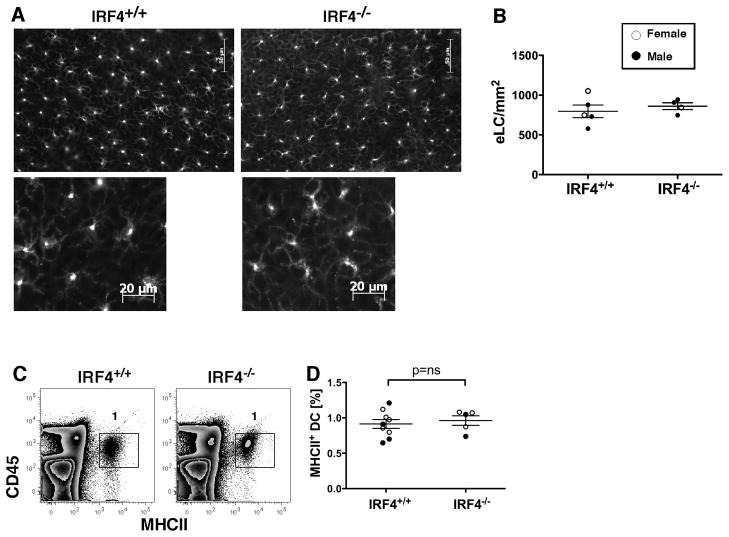
We next determined if IRF4 deficiency led to reduced numbers of resident dermal and epidermal DC. We used fluorescence microscopy with an anti-langerin Ab to identify eLC in epidermal sheets. The number, distribution and morphology of langerin+ DC in the epidermis were not different between WT and IRF4−/− mice (Fig. 2A,B). Similar results were obtained using epidermal cell suspensions and flow cytometry to detect CD45+ MHCII+ eLC (Fig. 2C,D).
Fig. 2. Numbers of epidermal Langerhans cells are similar in IRF4+/+ and IRF4−/− mice in homeostasis.
(A) Epidermal sheets from ear skin of IRF4+/+ and IRF4−/− mice stained with an anti-langerin Ab. Lower panels show DC morphology at a higher magnification. (B) Numbers of eLC/mm2 in individual female (open circles) and male (closed circles) mice (n=4–5 per genotype). The data were analyzed using a Mann-Whitney test. (C) Epidermal eLC within a preparation of ear skin epidermal cells were identified by expression of CD45.2 and MHCII. (D) The percentage of MHCII+ DC in the epidermis of multiple mice was determined (n=5–9).
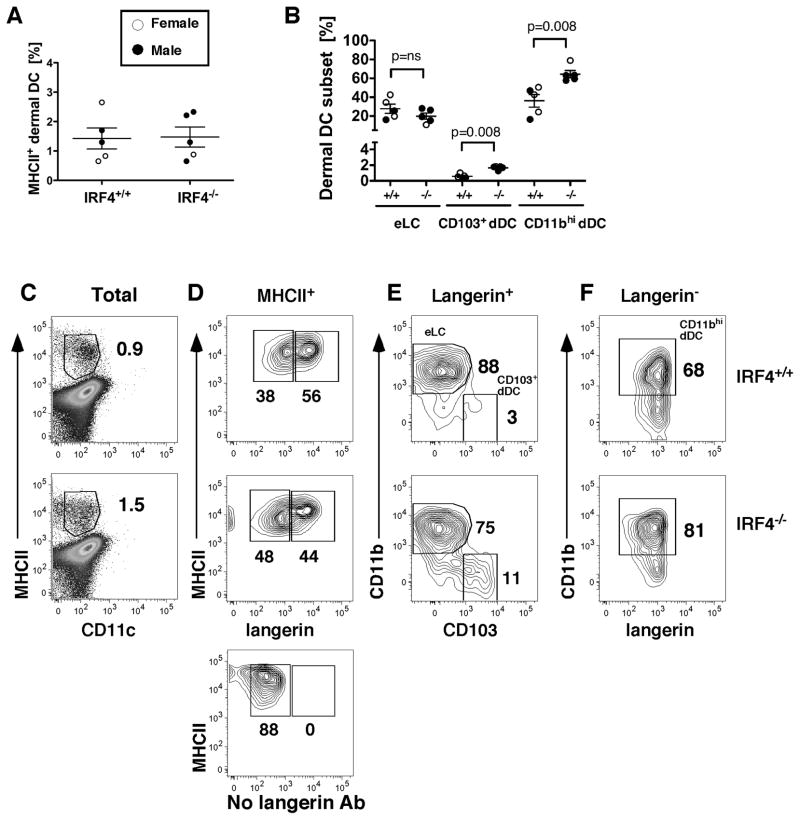
We determined the presence of DC subsets in the dermis by flow cytometry (Fig. 3C–F; Fig. S2). Because accurate dermal cell counts are difficult to obtain, we monitored the fraction of CD45+ cells in the dermis of WT and IRF4−/− mice and determined that it was similar (Fig. S2A); thus it was reasonable to compare the percentages of DC subsets between mice. The fraction of MHCII+CD11c+ cells in the dermis was not significantly different in IRF4−/− mice (Fig. 3A). Importantly, the CD11b+ dDC subset was significantly increased in IRF4−/− mice (Fig. 3B). The CD103+ dDC subset also was increased, while the eLC subset in the dermis was not significantly different (Fig. 3B; Fig. S2). Again we noted no significant differences between males and females.
Fig. 3. The dermis of IRF4−/− mice has an increased proportion of both CD11b+ and CD103+ dDC subsets in homeostasis.
(A) The percentage of MHCII+ cells in the dermis in individual mice. (B) The percentage of each DC subset within the MHCII+ fraction in the dermis of individual mice. (C) Gating of MHCII+ cells (percentage indicated) within a dermal cell suspension. (D) Gating of langerin+ and langerin− populations within the MHCII+ cells of the dermis. The lower panel shows a “fluorescence minus one” staining in which the anti-langerin Ab was omitted. (E) Gating of the CD11b+ eLC and CD103+ dDC within the langerin+ population. (F) Gating of CD11bhi dDC within the langerin− population. In all graphs, females (open circles) and males (closed circles) are indicated. The p values indicate significant differences in IRF4+/+ and IRF4−/− genotypes (males and females combined), determined using a nonparametric Mann-Whitney test, n=5.
These data show that the development of skin-resident eLC and CD11b+ dDC does not require IRF4, and suggests that the reduction in these two DC subsets in IRF4−/− cLN is due to a failure of these cells to migrate from the skin to LN, leading to accumulation of the CD11b+ dDC, but not eLC, in the dermis. Furthermore, the data show that IRF4 deficiency leads to the increased numbers of CD103+ DC in the dermis, although there is no defect in their constitutive migration to LN.
In contact hypersensitivity, CD11b+ dermal DC in IRF4−/− mice accumulate in the dermis and fail to migrate to cutaneous LN
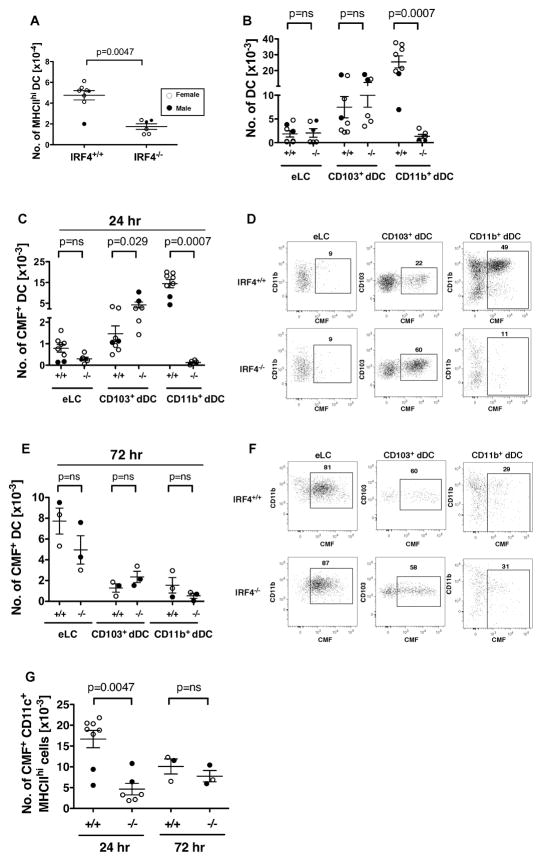
In a commonly used assay for contact hypersensitivity (CHS), increased migration of skin-resident DC into the cLN occurs in response to epicutaneous application of haptens dissolved in chemical sensitizing agents. Prior studies showed that the CD11bhi dDC and CD103+ dDC migrate to the cLN within 24 hr of application of the chemical sensitizing agent, while CD11blow dDC and eLC migrate within 2–4 days (28, 29). To evaluate the role of DC during the sensitization phase of CHS, we painted the skin of one ear in WT and IRF4−/− mice with a mixture of dibutyl phthalate-acetone (1:1) and a fluorescent cell tracker, chloromethyl fluorescein diacetate (CMFDA). After 24 hours, we analyzed DC subpopulations in the draining auricular LN, using the cell tracker to identify newly migrated DC. As during homeostasis, the total number of MHCIIhi DC in LN was reduced in IRF4−/− mice (Fig. 4A). This reduction was primarily due to a ten-fold reduction in the number of CD11b+ dDC (Fig. 4B). At 24 hr post-CHS in WT mice, the majority of migratory cells bearing the cell tracker (CMF+) in the draining LN were within the CD11b+ dDC subset, although some CMF+ eLC and CD103+ dDC also were present in the LN (Fig. 4C,D). In contrast, in IRF4−/− mice, CMF+ CD11b+ dDC (including both CD11bhi and CD11blow subsets) were absent in the draining LN, indicating a profound defect in their migration to the LN (Fig. 4C,D). This defect was also evident when the total number of CMF+ migratory MHCIIhi CD11c+ DC in the cLN at 24 hr post-CHS was counted (Fig. 4G).
Fig. 4. In contact hypersensitivity, CD11b+ dermal DC fail to migrate to cutaneous LN while cell tracker-bearing CD103+ dDC migrate in increased numbers in IRF4−/− mice.
(AD) Dibutyl phthalate-acetone (1:1) and the fluorescent cell tracker CMFDA were applied to ear skin, and cLN cells were harvested after 24 hr. (A) The number of MHCIIhi CD11c+ DC in the draining auricular LN. (B) Relative to eLC and CD103+ dDC, the total number of CD11b+ dDC was significantly reduced in LN of IRF4−/− mice. (C) The number of CMF+ cells within the CD11b+ dDC subset was significantly reduced, and the number of CMF+ cells within the CD103+ dDC subset was significantly increased, 24 hr post-CHS in LN of IRF4−/− mice. (D) Gating of CMF+ cells within the indicated migratory DC subset in cLN at 24 hr. (E–F) Dibutyl phthalate-acetone (1:1) and CMFDA were applied to ear skin, and cLN cells were harvested after 72 hr. (E) The number of CMF+ cells within each migratory DC subset was determined. (F) Gating of CMF+ cells within the indicated migratory DC subset in cLN at 72 hr. (G) The number of CMF+ cells with the gated MHCIIhi CD11c+ fraction (as in Fig. S3A) in IRF4+/+ and IRF4−/− cLN at 24 and 72 hr post-CHS was determined. In all graphs, females (open circles) and males (closed circles) are indicated. The p values indicate significant differences in the IRF4+/+ and IRF4−/− genotypes (males and females combined), determined using a nonparametric Mann-Whitney test, n=3–8.
The migration of CMF+ CD103+ dDC was significantly increased in IRF4−/− mice at 24 hr post-CHS (Fig. 4C), consistent with the increased numbers of this population in the dermis during homeostasis and upon CHS (Fig. 3B, 5B).
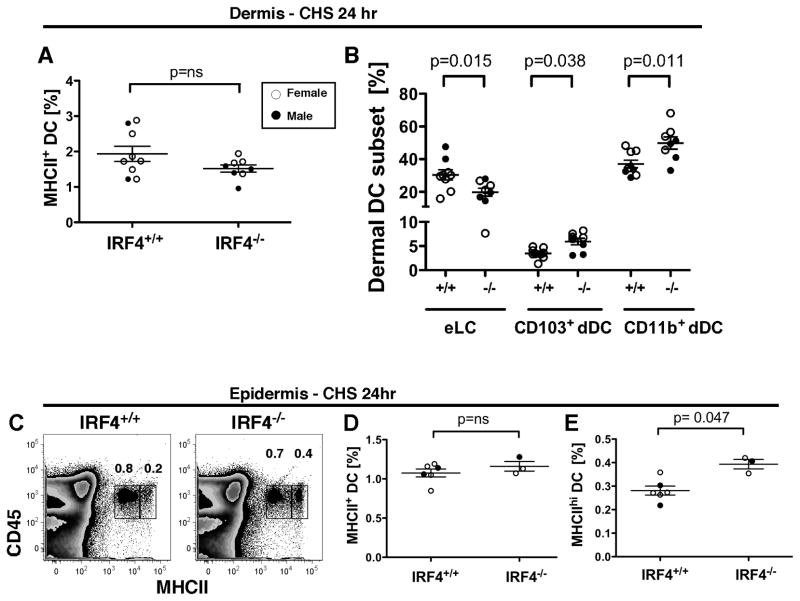
Fig. 5. In contact hypersensitivity, IRF4−/− mice have an increased percentage of CD11b+ and CD103+ dDC subsets in the dermis and MHCIIhi DC in the epidermis.
(A–B) Dibutyl phthalate-acetone (1:1) and the fluorescent cell tracker CMFDA were applied to ear skin, and dermal cells were harvested after 24 hr. Dermal DC subsets were identified by flow cytometry as in Fig. S3. The percentage of (A) total MHCII+ cells and (B) each DC subset within the MHCII+ population in the dermis of multiple mice was determined. (C–E) Dibutyl phthalate-acetone (1:1) and the fluorescent cell tracker CMFDA were applied to ear skin, and epidermal cells were harvested after 24 hr. (C) CD45+ epidermal DC were distinguished by two distinct levels of MHCII. The percentage of (D) total MHCII+ DC and (E) MHCIIhi DC in the epidermis of multiple mice was determined. In all graphs, females (open circles) and males (closed circles) are indicated. The p values indicate significant differences in the IRF4+/+ and IRF4−/− genotypes (males and females combined), determined using a nonparametric Mann-Whitney test, n=3–9.
At 24 hr post-CHS, numbers of migrating CMF+ eLC tended to be reduced in IRF4−/− mice but were not significantly different from eLC numbers in WT mice (Fig. 4C); this may be due to the fact that most eLC migrate at later time points. Therefore, we also determined the migration of skin DC populations 72 hr after application of dibutyl phthalate-acetone/CMFDA. In WT mice at 72 hr post-CHS, the majority of migratory cells bearing the cell tracker (CMF+) in the draining LN were within the eLC subset (Fig. 4E,F). The numbers of migratory CMF+ eLC in cLN tended to be decreased in IRF4−/− mice, but were not significantly different from numbers in WT mice (Fig. 4E). Consistent with this, the total number of CMF+ migratory MHCIIhi CD11c+ DC was not significantly different in WT and IRF4−/− mice at 72 hr post-CHS (Fig. 4G). The numbers of CMF+ CD11b+ dDC in LN of IRF4−/− mice remained very low at 72 hr post-CHS, indicating that the defect in migration of CD11b+ dDC in IRF4−/− mice was not transient (Fig. 4E).
In the dermis, the proportion of MHCII+ DC was similar in WT and IRF4−/− mice at 24 hr post application of the contact sensitizer (Fig. 5A). However, assessment of DC subsets (gated as in Fig. S3) showed that relative to WT mice, the dermis of IRF4−/− mice bore an increased fraction of CD11b+ dDC, and a moderately decreased fraction of eLC (Fig. 5B). In the epidermis at 24 hr post-CHS, in addition to the MHCIIint population observed during homeostasis (Fig. 2C), a second population of CD45+ MHCIIhi eLC was observed (Fig. 5C). Relative to WT mice, IRF4−/− mice showed a moderate increase in this fraction of activated MHCIIhi eLC in the epidermis, suggesting that migration of the most activated eLC was mildly impaired (Fig. 5C–E). Taken together, these analyses of migratory DC in the dermis, epidermis and draining cLN show that in IRF4−/− mice, CD11b+ dDC cannot migrate from dermis to cLN. Furthermore, as manifested by their slight increase in the epidermis and decrease in the dermis, eLC in IRF4−/− mice may have an attenuated ability to migrate from epidermis to dermis during inflammation.
In IRF4−/− mice, dermal CD11b+ DC display reduced levels of CCR7 upon contact sensitivity
CCR7 promotes the migration of skin-resident DC into cLN during homeostasis and during CHS (4, 20). Thus we evaluated the expression of CCR7 on DC subsets in the dermis (gated as in Fig. S3) and epidermis (gated as in Fig. 5C) 24 hr after application of the contact sensitizer. Expression of CCR7 on IRF4−/− CD11b+ dDC and CD103+ dDC correlated with their migratory phenotype. IRF4−/− CD11b+ dDC displayed reduced CCR7 levels, while CD103+ dDC showed normal levels of CCR7 (Fig. 6A).
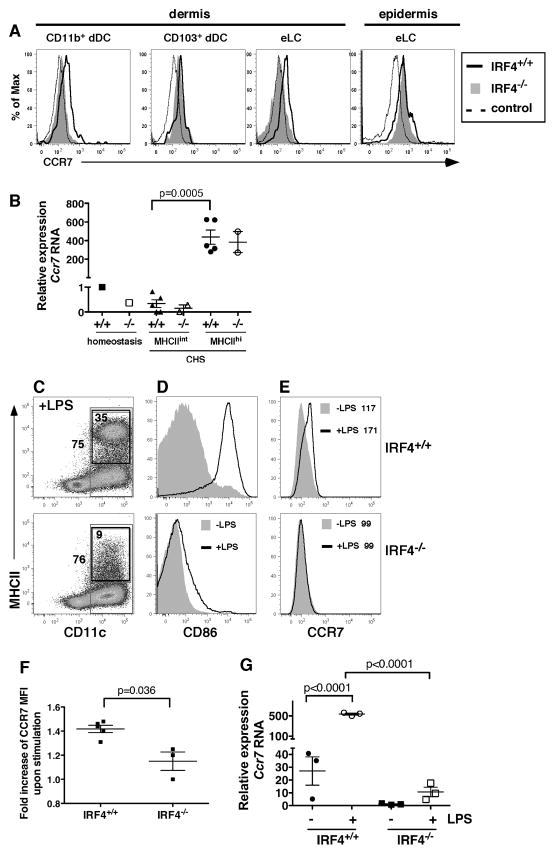
Fig. 6. IRF4−/− DC in the dermis show reduced expression of CCR7.
(A) Dibutyl phthalate-acetone (1:1) and the fluorescent cell tracker CMFDA were applied to ear skin, and dermal and epidermal cells were harvested after 24 hr. The gating of dermal and epidermal DC subsets is shown in Fig. S3 and Fig. 5C. (A) Expression of CCR7 on CD11b+ dDC, CD103+ dDC and eLC subsets in dermis and total MHCII+ eLC in the epidermis of IRF4+/+ (thick solid line) and IRF4−/− (shaded histogram) mice. The dotted line (control) indicates cells stained for DC markers but not CCR7 (“fluorescence minus one” control). The graphs are representative of 3–4 mice of each genotype. (B) CD45+ MHCII+ eLC were sorted from a pooled epidermal cell suspension derived from 3–5 mice during homeostasis (as in Fig. 2C), and separate populations of MHCIIint and MHCIIhi eLC were sorted from the epidermis of individual mice (n=2–5) 24 hr after application of dibutyl phthalate-acetone (1:1) and the fluorescent cell tracker CMFDA (as in Fig. 5C). The relative expression of Ccr7 RNA was determined using qPCR. For the eLC isolated during homeostasis, the data point is the mean of triplicates of the pooled sample (3–5 mice) for the PCR reaction. For the eLC isolated post-CHS, each data point is the mean of triplicates of a sample from a single mouse for the PCR reaction. The significance of the difference between Ccr7 RNA levels in populations of MHCIIint and MHCIIhi eLC in IRF4+/+ mice was evaluated using an unpaired t test. (C–G) DC were differentiated via GM-CSF from bone marrow cells isolated from IRF4+/+ (top panels) and IRF4−/− (bottom panels) mice, and stimulated with LPS for 12–18 hr. (C) In LPS-stimulated cells, the outer box indicates the gating of total CD11c+ cells (75% IRF4+/+ vs. 76% IRF4−/−) and the inset box indicates the gating of CD11c+ MHCII+ DC (35% IRF4+/+ vs. 9% IRF4−/−). The graphs are representative of 3–5 mice of each genotype. (D) The expression of CD86 on resting or LPS-stimulated DC (gated on total CD11c+ cells) is shown. (E) The expression of CCR7 on resting or LPS-stimulated DC (gated on total CD11c+ cells) is shown. The mean fluorescence intensity (MFI) of the overlying histograms is indicated. (F) Fold increase in CCR7 MFI on LPS-stimulated CD11c+ cells (relative to CCR7 MFI on resting cells, as in panel E) in cultures generated from individual mice of each genotype, n=3–5. The significance of these data was evaluated using a Mann-Whitney test. (G) IRF4+/+ and IRF4−/− DC generated from the bone marrow of individual mice were left unstimulated or stimulated for 12 hr with LPS. The relative expression of Ccr7 RNA was determined using qPCR; each data point is the mean of triplicates of an individual sample for the PCR reaction. The significance of these data (n=3) was evaluated using a one-way ANOVA followed by a Bonferroni’s multiple comparison test.
The regulation of CCR7 expression in eLC in IRF4−/− mice is more complex. Consistent with the only minor reduction in eLC migration in IRF4−/− mice, WT and IRF4−/− eLC (total MHCII+ fraction) in the epidermis displayed similar levels of cell surface CCR7 (Fig. 6A). Induced Ccr7 RNA levels were also similar in the activated MHCIIhi eLC isolated from WT and IRF4−/− epidermis during CHS (Fig. 6B). However, despite their relatively minor reduction in migration from dermis to LN, IRF4−/− eLC present in the dermis did show reduced levels of surface CCR7 (Fig. 6A). Unfortunately, we were unable to isolate intact RNA from these dermal DC subsets to assess Ccr7 mRNA levels because distinction of eLC and CD11b+ dDC subsets in the dermis requires intracellular langerin staining, which involves paraformaldehyde fixation. Since the flow cytometric staining of CCR7 is relatively insensitive compared to RNA levels (e.g. compare the magnitude of the activation-induced changes in surface protein expression vs. RNA expression in Fig. 6A,B and 6E–G), it is possible that the apparently reduced CCR7 level on dermal eLC was sufficient to mediate migration of the majority of eLC from the dermis to the LN.
As previously reported, CD11b+ CD11c+ DC in GM-CSF-driven cultures generated from IRF4−/− bone marrow failed to significantly increase MHCII and CD86 expression upon LPS stimulation (Fig. 6C,D). IRF4−/− bone marrow-derived DC also expressed significantly reduced surface CCR7 and Ccr7 mRNA after LPS activation (Fig. 6E–G), suggesting that IRF4 promotes Ccr7 transcription. Taken together, these data show that IRF4 activity promotes the expression of CCR7 in skin-resident CD11b+ dDC and in bone marrow-derived DC.
IRF4−/− DC show an intrinsic defect in migration in vivo and in vitro
To determine if IRF4 deficiency has a cell autonomous effect on DC migration, isolated WT and IRF4−/− bone marrow-derived DC were compared in assays of chemotaxis and in vivo migration. DC were placed into a transwell chemotaxis assay, in which DC migrate across a membrane barrier toward the chemokine CCL21, a ligand of CCR7. While WT DC showed a ~3-fold increase in migration to CCL21 relative to medium alone, IRF4−/− DC failed to migrate toward CCL21 (Fig. 7A).
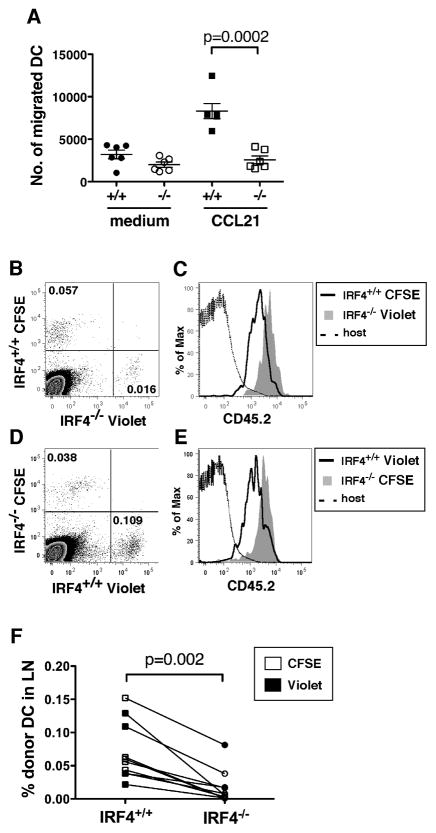
Fig. 7. IRF4−/− DC show an intrinsic defect in migration in vitro and in vivo.
(A) Migration of LPS-activated IRF4+/+ and IRF4−/− bone marrow-derived DC toward the chemokine CCL21 in a transwell chemotaxis assay. Data points represent the number of migrated DC in individual wells; DC are from 2 IRF4+/+ and 2 IRF4−/− mice (3 wells each). Data were evaluated using an unpaired t test. (B–F) LPS-activated IRF4+/+ and IRF4−/− bone marrow-derived DC (CD45.2+) labeled with CFSE or Cell Trace Violet were mixed together in equal numbers and injected intra-dermally into recipient CD45.1+ mice. Inguinal LN were analyzed for the presence of donor DC after 36 hr. (B) Identification of CFSE-labeled IRF4+/+ DC and Cell Trace Violet-labeled IRF4−/− DC among total LN cells. (C) Donor DC populations gated in panel B are CD45.2+. IRF4+/+ DC consistently expressed lower levels of CD45.2 than IRF4−/− DC, independent of the label used. (D) Identification of Cell Trace Violet-labeled IRF4+/+ DC and CFSE-labeled IRF4−/− DC among total LN cells. (E) Donor DC populations gated in panel D are CD45.2+. (F) Compilation of results using DC from 2 IRF4+/+ and 2 IRF4−/− mice each labeled with CFSE (open symbols) or Cell Trace Violet (closed symbols). The percentage of transferred DC among total LN cells is plotted. The significance of these data (n=10) was evaluated using a Wilcoxon matched-pairs signed rank test.
To assess migration in vivo, CD45.2+ WT and IRF4−/− bone marrow-derived DC were separately labeled with CFSE or Cell Trace Violet, mixed 1:1 and transferred via intra-dermal injection into recipient WT CD45.1+ mice. After 36 hr, the relative numbers of donor CD45.2+ WT and IRF4−/− DC in the popliteal LN of each mouse were determined (Fig. 7B–E). Within each mouse, greater numbers of WT than IRF4−/− DC migrated to the LN (Fig. 7F). The reduced ability of IRF4−/− DC to migrate to the LN was independent of the label used (Fig. 7B–E). Taken together, these data show that DC intrinsic IRF4 deficiency results in a reduced ability to migrate toward CCL21 in vitro and to LN in vivo.
Discussion
In this work, we evaluated the role of IRF4 in the development, skin residence and LN-directed migration of migratory tissue DC subsets that are normally present in the epidermis and dermis and also constitute a discrete population of MHCIIhi cells in cutaneous LN. Our data show that IRF4 is not required for the development and skin residence of epidermal and dermal DC, but that it promotes the migration of CD11b+ dDC and eLC from the skin to the LN during homeostasis.
Upon induction of skin inflammation in IRF4−/− mice, the CD11b+ dDC also failed to migrate to the LN. This correlated with their significantly reduced surface CCR7, a chemokine receptor that is crucial for migration of skin DC to cutaneous LN during homeostasis and inflammation. Indeed, IRF4 likely promotes expression of the Ccr7 gene, since LPS-stimulated bone marrow-derived IRF4−/− DC showed a significant reduction in Ccr7 RNA. Activated IRF4−/− CD11b+ bone marrow-derived DC failed to migrate toward the CCR7 ligand CCL21 in a chemotaxis assay, suggesting a DC-intrinsic effect of IRF4 deficiency on migration.
In contrast to CD11b+ dDC, the migration of IRF4−/− eLC from skin to cLN was only minimally impaired during inflammation. This minor defect in migration was reflected in an increased fraction of MHCIIhi eLC in the epidermis and a decreased fraction of eLC in the dermis of IRF4−/− mice. MHCIIhi eLC present in the epidermis of IRF4+/+ and IRF4−/− mice 24 hr post-CHS contained comparable amounts of Ccr7 RNA, indicating that IRF4 is not strictly required for the increased Ccr7 transcription that occurs in activated eLC. However, CCR7 surface expression on eLC did not always correlate with the Ccr7 RNA levels and eLC migration pattern during inflammation. While CCR7 levels on eLC in the epidermis were similar in IRF4+/+ and IRF4−/− mice, eLC in the dermis in IRF4−/− mice did show a reduction in CCR7. Taken together, these data suggest that IRF4-independent chemokine receptors, perhaps regulated by IRF8, may have a stronger effect on eLC migration in an inflammatory environment than during homeostasis.
Our data suggest that the Ccr7 gene may be a direct transcriptional target of IRF4. IRF4 acts alone by binding interferon stimulated response elements (ISRE) or is recruited to composite IRF/PU.1 binding sites on target genes by phosphorylated PU.1 (30). Predicted ISRE and PU.1 binding sites are present upstream of the Ccr7 gene, consistent with direct binding of IRF4. Alternately, IRF4 may indirectly regulate Ccr7 transcription through collaboration with other transcription factors such as NF-κB and AP-1, which do bind to consensus regions in the promoter of Ccr7 gene, a pathway likely to occur secondary to Toll-like receptor signaling in DC (31).
CCR7 may be regulated by both IRF4 and IRF8 since Ccr7 RNA levels were decreased in IRF8−/− bone marrow derived DC (12). Because IRF4−/− CD103+ dDC migrate efficiently, our data suggest that in the absence of IRF4, the putative IRF8 expression in CD103+ dDC was sufficient to promote CCR7 expression, which was previously shown to be required for entry of CD103+ dDC into the cutaneous LN but not the dermis (32).
Our data raise questions about the role of IRF4 in the development of the diverse CD11b+ DC subsets found in non-lymphoid tissue and lymphoid organs. In contrast to the finding that splenic CD11bhi CD4+ cDC do not develop in IRF4−/− mice (16, 17) (Fig. S1), our data show that IRF4 is not required for the initial development of CD11b+ dDC. This is puzzling since splenic CD11b+ cDC and dermal CD11b+ DC can develop from a population of transferred pre-cDC, a process that one might expect to require IRF4 expression in pre-cDC (9, 13). However, lineage tracing and monocyte transfer experiments showed that CD11b+ tissue DC can arise from monocytes in homeostasis (9, 15, 33, 34). Thus it is possible that the dermal CD11b+ DC present in the skin of IRF4−/− mice are derived primarily from monocytes in an IRF4-independent process, while the pre-cDC to CD11b+ DC pathway in dermis and spleen is disrupted by IRF4 deficiency. We also found that MHCIIint CD11b+ resident DC in cutaneous LN are not reduced in IRF4−/− mice in homeostasis (Fig. S1). These data suggest that this resident DC subset also may derive primarily from monocytes in the absence of IRF4.
We also noted a modest increase in CD103+ DC in the dermis in IRF4−/− mice, suggesting that their IRF8-dependent development is enhanced by IRF4 deficiency. This could result from the disruption of the competition between IRF4 and IRF8 for common promoter binding sites or partners such as PU.1 (30). Thus, the absence of IRF4 may increase the efficiency of recruitment of IRF8/PU.1 complexes to composite IRF/PU.1 sites in target genes, thereby enhancing CD103+ dDC development.
DC migration from peripheral tissue to LN is critical for initiation of T cell mediated immune responses that occur in infection, autoimmune disease and vaccination. Our study now places IRF4 in a vital regulatory role during the cutaneous LN-directed migration of CD11b+ dDC, the most abundant DC subset present in murine and human skin (1). Indeed, in humans this IRF4-regulated DC migration pathway is likely to be critical for initiation of adaptive immune responses in skin diseases such as psoriasis and atopic dermatitis, as well as cutaneous infections, graft-versus-host disease, allergic contact dermatitis and dermal vaccination. Furthermore, our study reveals the diverse roles of IRF4 within the complex network of transcription factors that regulates differentiation of DC from precursors as well as mature DC function.
Supplementary Material
Acknowledgments
We thank Dr. Tak Mak for providing IRF4+/− mice. We thank Ms. S. Wilburn and Mr. M. Jones for expert technical assistance, and Drs. Don Capra and José Alberola-Ila for helpful discussions.
Abbreviations
- cDC
conventional dendritic cell
- CHS
contact hypersensitivity
- cLN
cutaneous lymph node
- DC
dendritic cell
- dDC
dermal dendritic cell
- eLC
epidermal Langerhans cell
- IRF4/IRF8
interferon regulatory factor 4 or 8
- qPCR
quantitative real-time reverse transcriptase-polymerase chain reaction
Footnotes
This work was supported by NIH grants AI079616 and AI083715, and the Oklahoma Center for the Advancement of Science & Technology (HR09-055).
Conflict of Interest Disclosures
None.
References
- 1.Johnson-Huang LM, McNutt NS, Krueger JG, Lowes MA. Cytokine-producing dendritic cells in the pathogenesis of inflammatory skin diseases. J Clin Immunol. 2009;29:247–256. doi: 10.1007/s10875-009-9278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merad M, Hoffmann P, Ranheim E, Slaymaker S, Manz MG, Lira SA, Charo I, Cook DN, Weissman IL, Strober S, Engleman EG. Depletion of host Langerhans cells before transplantation of donor alloreactive T cells prevents skin graft-versus-host disease. Nat Med. 2004;10:510–517. doi: 10.1038/nm1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplan DH. In vivo function of Langerhans cells and dermal dendritic cells. Trends Immunol. 2010;31:446–451. doi: 10.1016/j.it.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, Blankenstein T, Henning G, Forster R. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Steinman RM, Nussenzweig MC. Avoiding horror autotoxicus: the importance of dendritic cells in peripheral T cell tolerance. Proc Natl Acad Sci U S A. 2002;99:351–358. doi: 10.1073/pnas.231606698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henri S, Guilliams M, Poulin LF, Tamoutounour S, Ardouin L, Dalod M, Malissen B. Disentangling the complexity of the skin dendritic cell network. Immunol Cell Biol. 2010;88:366–375. doi: 10.1038/icb.2010.34. [DOI] [PubMed] [Google Scholar]
- 7.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helft J, Ginhoux F, Bogunovic M, Merad M. Origin and functional heterogeneity of non-lymphoid tissue dendritic cells in mice. Immunol Rev. 2010;234:55–75. doi: 10.1111/j.0105-2896.2009.00885.x. [DOI] [PubMed] [Google Scholar]
- 9.Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, Stanley ER, Nussenzweig M, Merad M. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King IL, Kroenke MA, Segal BM. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J Exp Med. 2010;207:953–961. doi: 10.1084/jem.20091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelson BT, WKC, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung SS, Murphy TL, Hildner K, Murphy KM. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiavoni G, Mattei F, Borghi P, Sestili P, Venditti M, Morse HCr, Belardelli F, Gabriele L. ICSBP is critically involved in the normal development and trafficking of Langerhans cells and dermal dendritic cells. Blood. 2004;103:2221–2228. doi: 10.1182/blood-2003-09-3007. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu FF, Randolph GJ, Rudensky AY, Nussenzweig M. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kingston D, Schmid MA, Onai N, Obata-Onai A, Baumjohann D, Manz MG. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 2009;114:835–843. doi: 10.1182/blood-2009-02-206318. [DOI] [PubMed] [Google Scholar]
- 15.Jakubzick C, Bogunovic M, Bonito AJ, Kuan EL, Merad M, Randolph GJ. Lymph-migrating, tissue-derived dendritic cells are minor constituents within steady-state lymph nodes. J Exp Med. 2008;205:2839–2850. doi: 10.1084/jem.20081430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki S, Honma K, Matsuyama T, Suzuki K, Toriyama K, Akitoyo I, Yamamoto K, Suematsu T, Nakamura M, Yui K, Kumatori A. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha-dendritic cell development. Proc Natl Acad Sci U S A. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O’Shea JJ, Singh H, Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 18.Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, Matsuyama T, Taniguchi T, Honda K. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc Natl Acad Sci U S A. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HCr, Belardelli F, Gabriele L. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabashima K, Shiraishi N, Sugita K, Mori T, Onoue A, Kobayashi M, Sakabe J, Yoshiki R, Tamamura H, Fujii N, Inaba K, Tokura Y. CXCL12-CXCR4 engagement is required for migration of cutaneous dendritic cells. Am J Pathol. 2007;171:1249–1257. doi: 10.2353/ajpath.2007.070225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouwehand K, Santegoets SJ, Bruynzeel DP, Scheper RJ, de Gruijl TD, Gibbs S. CXCL12 is essential for migration of activated Langerhans cells from epidermis to dermis. Eur J Immunol. 2008;38:3050–3059. doi: 10.1002/eji.200838384. [DOI] [PubMed] [Google Scholar]
- 22.Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Bobr A, Olvera-Gomez I, Igyarto BZ, Haley KM, Hogquist KA, Kaplan DH. Acute ablation of Langerhans cells enhances skin immune responses. J Immunol. 2010;185:4724–4728. doi: 10.4049/jimmunol.1001802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 25.Zimmerli SC, Hauser C. Langerhans cells and lymph node dendritic cells express the tight junction component claudin-1. J Invest Dermatol. 2007;127:2381–2390. doi: 10.1038/sj.jid.5700882. [DOI] [PubMed] [Google Scholar]
- 26.Moodycliffe AM, Shreedhar V, Ullrich SE, Walterscheid J, Bucana C, Kripke ML, Flores-Romo L. CD40-CD40 ligand interactions in vivo regulate migration of antigen-bearing dendritic cells from the skin to draining lymph nodes. J Exp Med. 2000;191:2011–2020. doi: 10.1084/jem.191.11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carreras E, Turner S, Frank MB, Knowlton N, Osban J, Centola M, Park CG, Simmons A, Alberola-Ila J, Kovats S. Estrogen receptor signaling promotes dendritic cell differentiation by increasing expression of the transcription factor IRF4. Blood. 2010;115:238–246. doi: 10.1182/blood-2009-08-236935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kissenpfennig A, Henri S, Dubois B, Laplace-Builhe C, Perrin P, Romani N, Tripp CH, Douillard P, Leserman L, Kaiserlian D, Saeland S, Davoust J, Malissen B. Dynamics and function of Langerhans cells in vivo dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 2005;22:643–654. doi: 10.1016/j.immuni.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Shklovskaya E, Roediger B, Fazekas de St Groth B. Epidermal and dermal dendritic cells display differential activation and migratory behavior while sharing the ability to stimulate CD4+ T cell proliferation in vivo. J Immunol. 2008;181:418–430. doi: 10.4049/jimmunol.181.1.418. [DOI] [PubMed] [Google Scholar]
- 30.Marecki S, Fenton MJ. The role of IRF-4 in transcriptional regulation. J Interferon Cytokine Res. 2002;22:121–133. doi: 10.1089/107999002753452737. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez-Sanchez N, Riol-Blanco L, Rodriguez-Fernandez JL. The multiple personalities of the chemokine receptor CCR7 in dendritic cells. J Immunol. 2006;176:5153–5159. doi: 10.4049/jimmunol.176.9.5153. [DOI] [PubMed] [Google Scholar]
- 32.Ginhoux F, Collin MP, Bogunovic M, Abel M, Leboeuf M, Helft J, Ochando J, Kissenpfennig A, Malissen B, Grisotto M, Snoeck H, Randolph G, Merad M. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J Exp Med. 2007;204:3133–3146. doi: 10.1084/jem.20071733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, Kalchenko V, Geissmann F, Jung S. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med. 2007;204:171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, Stanley ER, Nussenzweig M, Lira SA, Randolph GJ, Merad M. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.