Abstract
TonB-dependent transporters (TBDTs), which transport iron-chelating siderophores and vitamin B12 across the outer membrane of gram negative bacteria, share a conserved architecture of a 22-stranded beta-barrel with an amino-terminal plug domain occluding the barrel. We previously reported that we could induce TBDTs to reversibly open in planar lipid bilayers via the use of urea and that these channels were responsive to physiological concentrations of ligands. Here we report that in the presence of urea, trypsin can cleave the amino-terminal 67 residues of the plug of the TonB-dependent transporter FhuA, as assessed by gel shift and mass spectrometry assays. On the bilayer, trypsin treatment in the presence of urea resulted in the induced conductance no longer being reversed upon removal of urea, suggesting that urea opens intact FhuA channels by pulling the plug at least partly out of the barrel, and that removal of the urea then allows reinsertion of the plug into the barrel. When expressed separately, the FhuA plug domain was found to be a mostly unfolded structure that was able to occlude isolated FhuA beta-barrels inserted into the membrane. Thus, although folded in the barrel, the plug need not be folded upon exiting the barrel. The rate of insertion of the beta-barrels into the membrane was tremendously increased in the presence of an osmotic gradient provided by either urea or glycerol. Negative staining electron microscopy showed that FhuA in a detergent solution formed vesicles, thus explaining why an osmotic gradient promoted the insertion of FhuA into membranes.
Gram negative bacteria use special outer membrane receptors, called TonB-dependent transporters (TBDTs), to acquire vitamin B12 and iron-bound siderophores. These TBDTs, found across many genera including Escherichia, Yersinia and Salmonella, all possess a conserved architecture consisting of a 22-stranded antiparallel beta-barrel with short periplasmic turns and long extracellular loops, along with a roughly 160 residue amino terminus, called the plug domain, which folds into and occludes this very large barrel (for a review, see(1)).
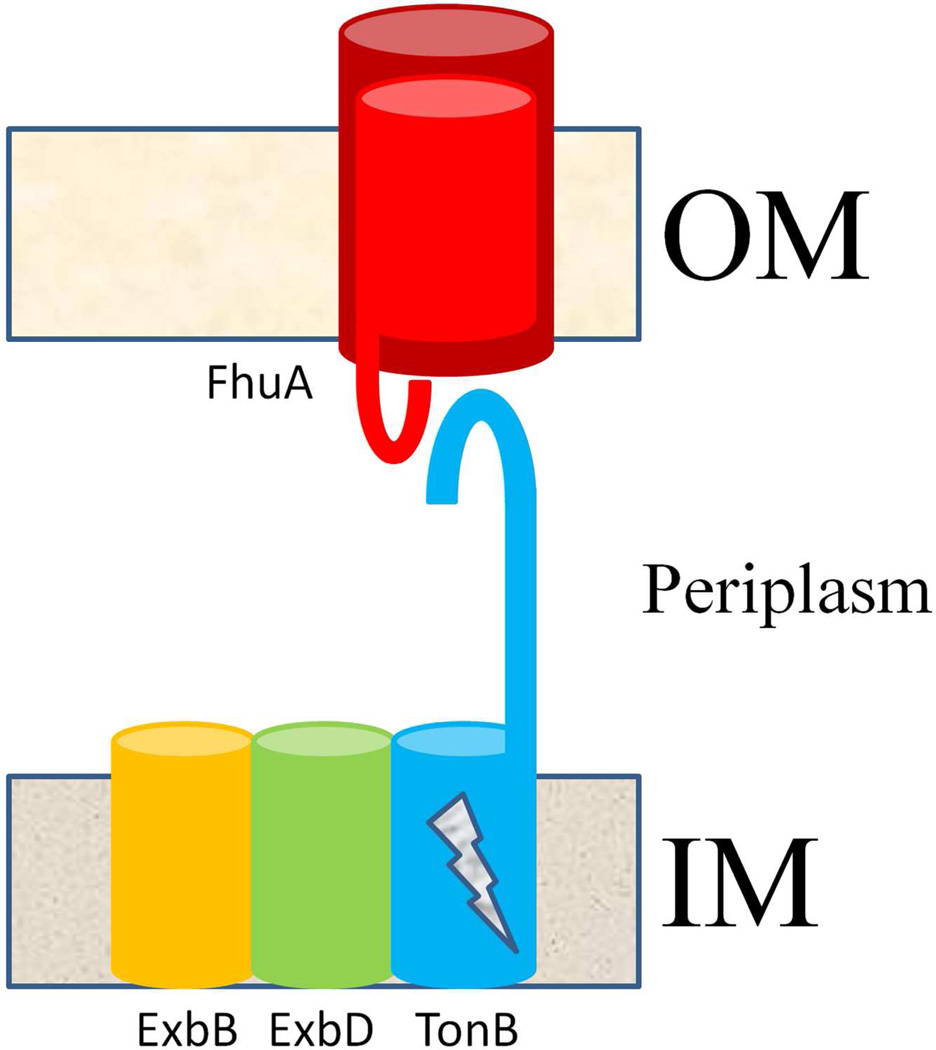
Genetic experiments have shown that transport of the siderophore across the outer membrane via the TBDTs requires the presence of, and interaction with, the TonB protein of the inner membrane TonB complex (Figure 1, reviewed in(2)). About five residues (called the TonB box) near the very beginning of the amino terminus of the TBDTs were found to be very well conserved and essential for TonB interaction and TBDT function. The site of interaction with TonB was localized to TonB’s carboxy terminus, and the nature of the interaction between TonB and TBDTs was more clearly defined when co-crystal structures of the carboxy terminus of TonB with FhuA(3) and BtuB(4) were solved. These structures showed the carboxy terminus of TonB forming a beta sheet, with the TonB box of the TBDT extending out of the barrel to form another strand of the beta sheet, a so-called strand-exchange interaction.
Figure 1.
Cartoon of spatial arrangement of the siderophore outer membrane permeation machinery. Siderophore receptors (FhuA is depicted in the figure) are TBDTs that sit in the outer membrane (OM) of gram negative bacteria and possess a conserved beta-barrel (dark red) architecture. Upon siderophore binding to the receptor, the plug domain of the receptor (light red) extends its TonB box (red hook), located near the amino terminus of the polypeptide, into the periplasm to interact with the carboxy terminus (blue hook) of the TonB protein of the inner membrane (IM) TonB complex, consisting of ExbB, ExbD and TonB. In an energy-dependent manner (indicated by a lightning bolt on TonB), TonB’s interaction with the receptor’s TonBbox somehow allows a pathway to form within the lumen of the receptor’s barrel, through which the siderophore diffuses.
Co-crystal structures of TBDTs and several known ligands have also been solved. For example, Ferric-Hydroxamate Uptake protein A (FhuA), with its cognate siderophore, ferrichrome, was solved in 1998(5). It came as a surprise to many that there were not major conformational changes within the protein, specifically, within the plug domain of the protein, when the siderophore was bound. This left unanswered one of the key questions in the field: how does the siderophore move through an ostensibly occluded barrel to gain access to the periplasm of the cell?
The current hypothesis posits that upon binding of the siderophore to the TBDT, the TonB box of the TBDT extends into the periplasm to interact with TonB, and that this interaction, in an energy-dependent manner, allows the formation of a pathway for the siderophore to move through the lumen of the TBDT. This of course begs the question of what happens to the plug domain during siderophore translocation.
The Coulton(6), and later the van der Helm(7) laboratories, using engineered disulfide bonds to fix the plug to the barrel domain, or engineered intra-plug disulfide linkages, concluded that the plug domain remains mostly within the barrel domain during siderophore translocation. However, based on accessibility of fluorescein maleimide to cysteines within the barrel domain of the TBDT FepA, the Klebba laboratory concluded that the plug domain moves completely out of the barrel(8). The Postle laboratory(9), using accessibility of BMCC to cysteines on the plug and within the barrel, as well as proteinase K accessibility to the plug in the periplasm, concluded that at least 51 residues of the plug domain exited the barrel in the presence of colicin B, another ligand of FepA. Because of the patterns of the result, however, it was suggested that the plug may come completely out of the barrel, but be protected from BMCC and proteinase K by another periplasmic protein.
To this extensive body of in vivo evidence, we add data from our in vitro studies, in the hope of shedding further light on the issue of the location of the plug domain during siderophore translocation. We found that trypsin could cleave approximately 40% of the plug domain of FhuA, but ony if 4 M urea was present. The barrel domain forms constitutively open channels in planar lipid bilayers, and these open channels can, in fact, be occluded by separately purified plug domain. And finally, the plug domain is mostly unfolded when in solution.
Materials and Methods
Protein expression and purification
E. coli strain AW740(10) has deletions of the genes for the major outer membrane proteins OmpF and OmpC. A fhuA mutant derivative of AW740(11) bearing the pBR322-based plasmid pHX405, encoding FhuA with a hexahistidine tag inserted after codon 405 of FhuA(12), was the kind gift of Dr. James Coulton. pHX405 was isolated from this strain and the Stratagene QuikChange Kit (Agilent Technologies, Santa Clara, CA) was used to delete residues 6–160 of fhuA, to give FhuA Δ6–160 (FhuA barrel). Strain KJ740 was created by curing the plasmid from the above-described AW740, by extended growth in 5 µg/ml coumermycin A1 (Promega, Fitchburg, Wisconsin), as described(13). KJ740 is thus ΔompF ΔompC fhuA31. DNA sequences encoding FhuA residues 1–160 were amplified from pHX405 by PCR (FhuA plug) and inserted into pET52-b (Novagen, Darmstadt, Germany), which creates an HRV 3C-cleavable Strep II tag at the amino terminus and a thrombin-cleavable His10 tag at the carboxy terminus of the FhuA plug sequence. FhuA barrel was expressed and purified from KJ740 bearing the plasmid FhuA Δ6–160 essentially as described for whole FhuA(14). FhuA plug was expressed in BL21 (DE3) cells and purified on both Strep-Trap and His-Trap columns (GE Healthcare, Piscataway, NJ) as specified by the manufacturer.
Concerned about the contribution of the double tags of the plug to plug behavior, we decided to remove these tags to examine the behavior of a “tagless plug” on the system. To this end, we attempted to use thrombin and HRV 3C protease to cleave the C-terminal His10 and N-terminal Strep II tags, respectively, from the plug. Gel shift assays suggested that HRV 3C cleaved the Strep II tag as expected, but exposure of the plug to thrombin resulted in degradation of the plug (data not shown). To overcome this problem, a new plug was engineered with a C-terminal TEV site replacing the thrombin cleavage site (and still retaining the N-terminal HRV 3C cleavage site), and was then purified as described above. Exposure of this plug to HRV 3C (Novagen, Darmstadt, Germany) and AcTEV™ (Invitrogen, Carlsbad,Ca) in a buffer containing 100 mM NaCl, 1 mM DTT and 20 mM sodium phosphate, pH 7.2, for 3 hours at room temperature (~23 °C) yielded the correctly sized fragment as assessed by SDS-PAGE. This reaction was then incubated with Ni-NTA resin (Qiagen, Valencia, Ca) to remove both the free His10 and the endoproteases (both possess a poly-histidine tag), and the supernatant from this incubation was used directly in bilayer experiments. This tagless plug construct exhibited similar behavior to that of the doubly-tagged plug (data not shown).
Reagents
Stock solutions of whole FhuA (0.33 or 2.2 mg/mL in 50 mM ammonium acetate, pH 8.0, 250 mM NaCl, 70 mM imidazole, 0.1% lauryldimethylamine-oxide [LDAO]) and FhuA barrel (1.6 mg/mL in 50 mM Tris-Cl, pH 8.0, 500 mM NaCl, 70 mM imidazole, 0.1% LDAO) were stable when kept at 4 °C for over eighteen months; FhuA plug (0.3 mg/mL, 100 mM NaCl, 20 mM NaPO4, pH 7.2) was stored at −80 °C and the batch used for all experiments reported here was stable when kept at 4°C for at least six months. However, several other batches, prepared with identical protocols exhibited degradation with a half time of several days if kept at 4°C. Tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Worthington Biochemical Corp., Lakewood, NJ) was dissolved in 1 mM HCl and frozen at −20 °C until the day of use. For inhibited trypsin, powders of trypsin and soybean trypsin inhibitor (SBTI) (Worthington Biochemical Corp., Lakewood, NJ) were dissolved in 100 mM KCl, 1 mM EDTA, 5 mM CaCl2, 20 mM HEPES, pH 7.2, at a ratio of 7.5:1 SBTI:trypsin by weight (according to the Worthington documentation, 1 g of SBTI inhibits 2.1 g trypsin), allowed to incubate 5 minutes at room temperature, and then frozen at −20 °C until the day of use.
Planar Lipid Bilayer Experiments
Planar lipid bilayers were formed at room temperature by the brush technique(15) across a 0.5 mm hole in a 125 µm thick Teflon partition. Membranes separated two Lucite compartments, each containing 3 ml of 100 mM KCl, 1 mM EDTA, 5 mM CaCl2, 20 mM HEPES, pH 7.2, which could be stirred by small magnetic stir bars. The membrane-forming solution was 3% diphytanoylphosphatidylcholine (Avanti Polar Lipids, Alabaster, AL) in n-decane. After the membrane formed, either whole FhuA (1.25 nM) or FhuA barrel (2–20 nM) was added to the cis compartment, which also contained 4 M urea. The final concentrations of FhuA plug, trypsin, and SBTI were 45 nM, 100 µg/mL and 750 µg/mL, respectively, in those experiments in which they were added. Perfusions of each compartment were performed for 8 minutes via a Polystaltic Pump (Buchler Instruments, Kansas City, MO) at a flow rate of 2 mL/min. All experiments were done under voltage-clamp conditions; voltages are those of the cis solution (to which the whole FhuA or barrel was added) with respect to that of the trans solution, which was held at virtual ground. Current responses were filtered at 10 Hz and recorded concurrently on a Narco physiograph chart recorder (Houston, TX) and an NI6211 Data Acquisition Board (National Instruments, Austin, TX) and stored digitally. Each of the experiments reported here was performed at least 3 times with similar results.
Proton NMR
A Proton 1D NMR Spectrum of FhuA plug was obtained on a Varian 600 Instrument (Palo Alto, CA) at 25 °C. The analyzed solution was: 0.3 mg/mL FhuA plug in 100 µM 2,3,5-triisopropylbenzenesulfonyl chloride, 10% D2O, 100 mM NaCl, 20 mM NaPO4, pH 7.2.
Circular Dichroism
A circular dichroism spectrum consisting of 3 averaged scans was acquired from a Jasco J-815 instrument (Easton, MD) analyzing the FhuA plug solution (0.3 mg/mL in 100 mM NaCl, 20 mM NaPO4, pH 7.2). The circular dichroism spectrum was fed into the K2D2 program(16) for secondary structure estimation.
Trypsin Digest for SDS-PAGE
A reaction buffer of 0.05% LDAO, 100 mM KCl, 5 mM CaCl2, 1 mM EDTA, 20 mM HEPES, pH 7.2 containing 100 µg/mL whole FhuA (and, where indicated, 4 M urea), 100 µg/mL trypsin or preincubated trypsin (100 µg/mL) with SBTI (750 µg/mL) was allowed to incubate at room temperature (~23 °C) for 20 minutes. The reaction was then treated with reducing sample buffer, boiled for 1 minute, and resolved on a 4–15% acrylamide gradient SDS gel, followed by visualization with Coomassie blue staining.
Trypsin Digest for Mass Spectrum Analysis
The sample for mass spectrum analysis was prepared as follows: a reaction buffer of 4 M urea, 100 mM KCl, 5 mM CaCl2, 1 mM EDTA, 0.05% LDAO, 20 mM HEPES, pH 7.2 containing 800 µg/mL whole FhuA and 400 µg/mL trypsin was incubated at 37 °C for 120 minutes, and then at room temperature for an additional 60 minutes, before being quenched with 0.1 % trifluoroacetic acid. The deconvoluted masses, along with enzyme identity and FhuA sequence (from PDB file 1QKC) were then entered into Expasy’s FindPept (http://web.expasy.org/findpept/), ProtParam (http://web.expasy.org/cgi-bin/protparam/protparam) and PeptideCutter (http://web.expasy.org/peptide_cutter/) programs to infer possible trypsin cleavage sites.
Electron Microscopy
Solutions of whole FhuA were diluted 10 fold in 100 mM KCl, 5 mM CaCl2, 1 mM EDTA, 20 mM HEPES, pH 7.2 and used to perform negative staining.
Results
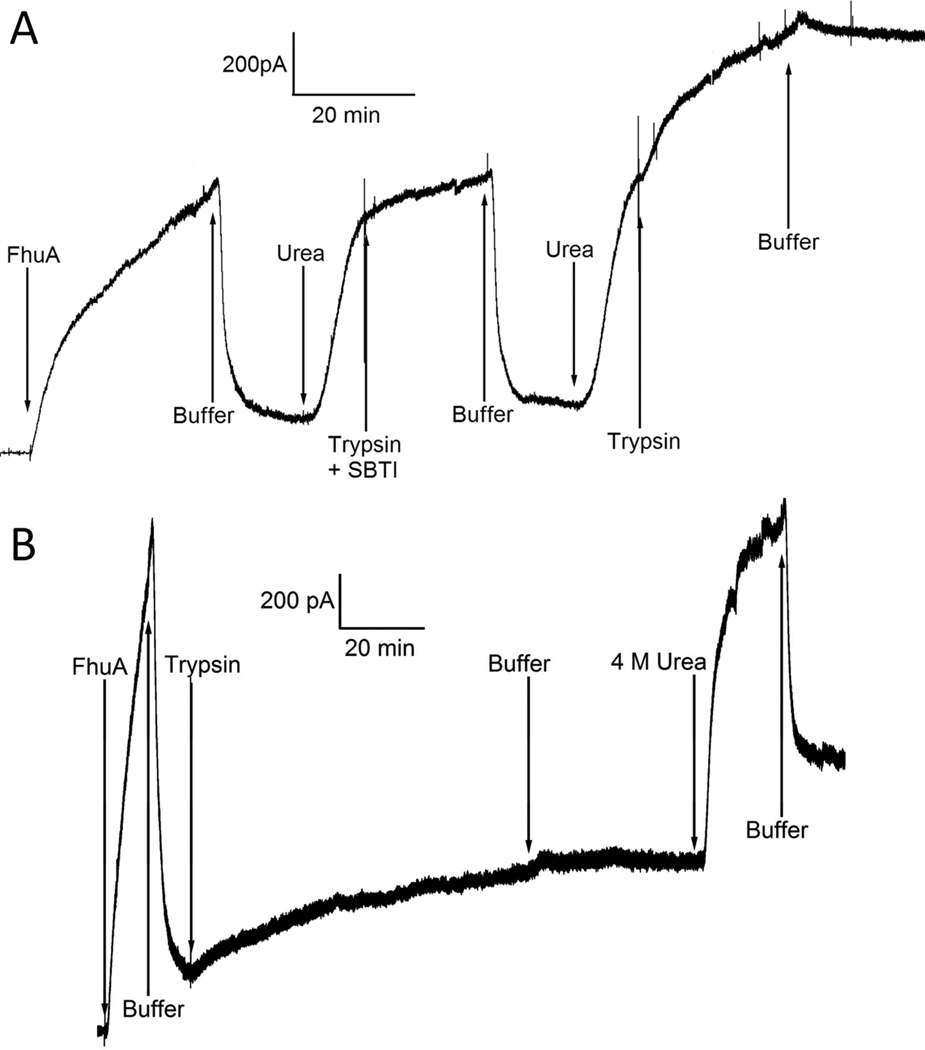
With FhuA in planar lipid bilayers, we normally find a reversible conductance increase and decrease corresponding to introduction and removal of 4 M urea in the cis solution (a urea cycle)(17). However, if the FhuA-containing membrane was exposed to both 4 M urea and trypsin at the same time, we found not only an increase in conductance beyond that seen with 4 M urea alone, but also little conductance loss upon removal of urea (Fig. 2A). (It is known that even in 10 mM CaCl2 and 5 M urea, trypsin retains ~75 percent of its activity after 1 hour at 37 °C(18).) In contrast, a FhuA-containing membrane exposed to trypsin in the absence of urea subsequently displayed a urea cycle response comparable to that seen without trypsin exposure (Fig. 2B) (The exposure to trypsin did produce a small conductance increase.). Thus, in the absence of urea, the sites of trypsin action were not readily accessible to trypsin.
Figure 2.
Trypsin in the presence of urea, but not in its absence, resulted in loss of reversibility of conductance. Both current traces begin with 4 M urea in the cis compartment and the voltage held at −20 mV throughout the experiment. A) Addition of FhuA (1.25 nM) to the cis compartment and removal of urea via buffer perfusion (labeled “Buffer”) resulted in the standard rise and fall, respectively, of conductance. Subsequent exposure of the membrane to cis trypsin (100 µg/mL) with bound soybean trypsin inhibitor (750 µg/mL) (labeled “Trypsin + SBTI”) in the presence of urea did not alter the conductance reversal level when the urea (plus trypsin and SBTI) were perfused out. However, addition of cis trypsin (100 µg/mL), without the trypsin inhibitor, in the presence of urea resulted in a conductance increase beyond that seen without trypsin, and, more importantly, there was little fall in conductance upon buffer perfusion of the compartment. B) Addition of FhuA (1.25 nM) to the cis compartment and removal of urea via buffer perfusion resulted in the standard rise and fall, respectively, of conductance. When typsin was added to the cis compartment (in the absence of urea) there was a small, slow rise of conductance that terminated after the trypsin was removed via perfusion with buffer. More significant, however, was the fact that now when urea was cycled in and out of the cis compartment, the response was similar to that seen when no trypsin was added. Contrast the final buffer perfusion in Figure 2B with that in Figure 2A.
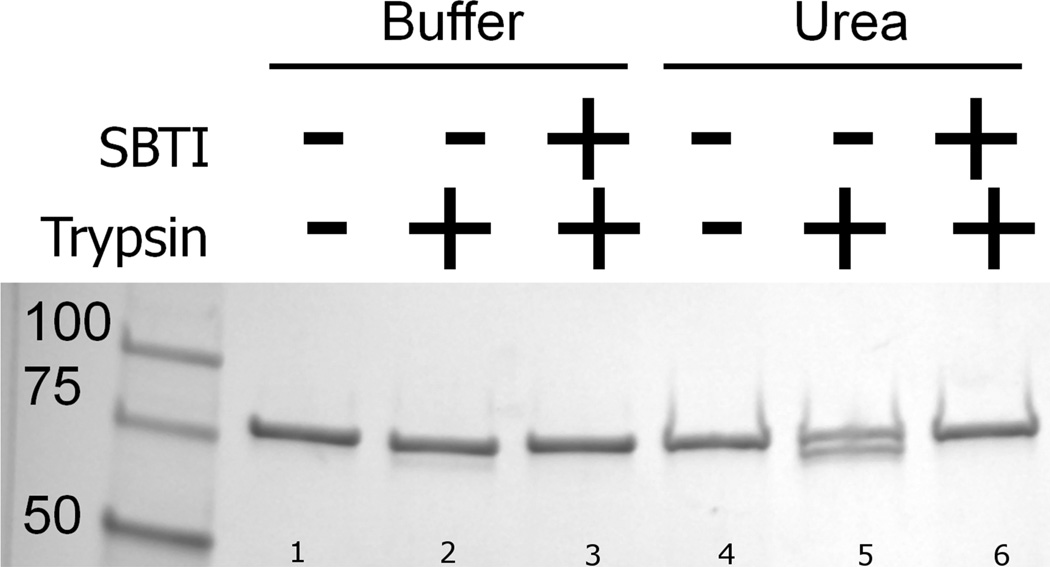
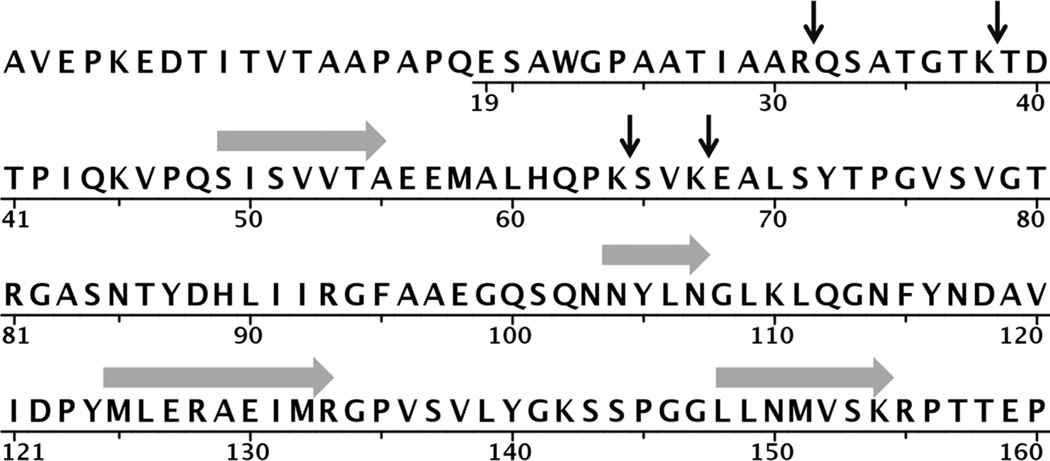
In an effort to identify the sites of trypsin action, we incubated FhuA (in its LDAO-containing solution) with trypsin in the presence and in the absence of urea (see Materials and Methods) and resolved the products on SDS-PAGE. The Coomassie-visualized proteins shown in Figure 3 reveal that in the absence of urea, trypsin had a very slight effect on the FhuA band, whereas in the presence of urea, two protein bands appeared, one migrating at the normal position and another faster-migrating band, seemingly several kilodaltons less in size. We then performed the same reaction to completion (See Materials and Methods), quenched with trifluoroacetic acid and did a mass spectrum analysis. Using the deconvoluted masses with tools from Expasy’s website (see Materials and Methods), we found several logical sites for trypsin action on FhuA. Entities corresponding to 1–31, 32–64 and 39–64 were found. The mass spectrometer also found entities of mass that corresponded to the carboxy terminus of the FhuA polypeptide after trypsin cleavage at sites R31, K64 and K67. Together, these results suggest a total of four trypsin cleavage sites (Fig. 4), with K67 being the furthest from the amino terminus in the primary sequence.
Figure 3.
Trypsin treatment of whole FhuA in the presence of 4 M urea resulted in a faster-migrating band on SDS-PAGE (see Materials and Methods for details of the trypsin treatment). Whole FhuA protein migrates at ~80 kDa when boiled in either buffer or urea, as shown in lanes 1 and 4 (numbers at bottom of figure). In the presence of trypsin alone (lane 2), there may be a very faint band representing a faster migrating species, whereas trypsin and urea together (lane 5) converted about half of the 80 kDa band to the faster migrating species. A preincubation of trypsin with soybean trypsin inhibitor resulted in no faster migrating species (lanes 3 and 6).
Figure 4.
Results of Mass Spectrum Analysis of whole FhuA incubated with trypsin in 4 M urea (see Materials and Methods). Three small amino-terminal peptides from whole FhuA were identified, corresponding to trypsin cleavage sites R31, K38 and K64. Carboxy-terminal polypeptides were also identified corresponding to trypsin cleavage sites at R31, K64 and K67. The black vertical arrows indicate the trypsin cleavage sites while the gray horizontal arrows indicate the residues that form the beta strands composing the central beta sheet of the plug (PDB file 1QKC; residues 49–55,104–107,125–133 and 148–154). The first 18 residues of FhuA in this PDB structure were disordered, denoted by the lack of an underline for those residues.
Because it seems that in 4 M urea the plug of whole FhuA is somewhat exposed to solution, we decided to separately express the barrel and plug domains of FhuA and look for interactions on the bilayer. We found that if the FhuA barrel was exposed to the membrane in the presence of either 3 M glycerol or 4 M urea, we could induce robust channel conductance across the membrane (Fig. 5). This is due to the formation of ~100 nm vesicles vesicles in the FhuA-detergent solution (Fig. 6). As has been shown(19), an osmotic gradient across the membrane greatly facilitates the fusion of vesicles to that membrane. It should be noted that the vesicles only formed in detergent solution with FhuA present, and not in detergent alone.
Figure 5.
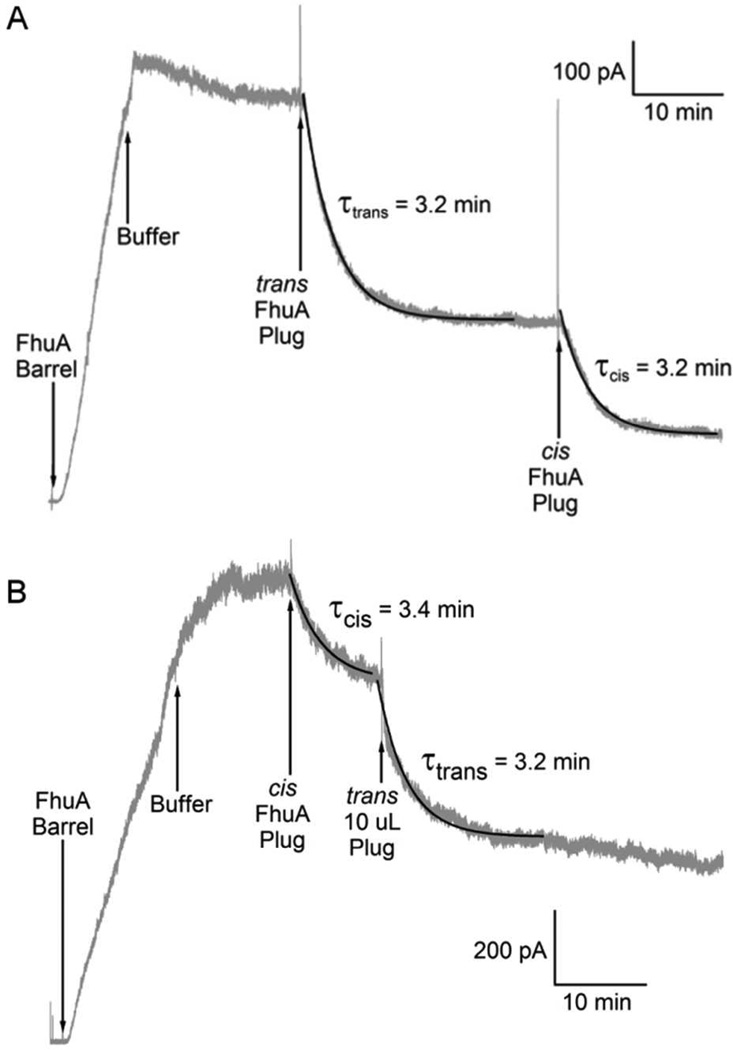
FhuA plug occluded FhuA barrel. Current traces begin with either 4 M urea (A) or 3 M glycerol (B) in the cis compartment, and the voltage held at −20 mV throughout the experiment. Conductance was induced by the addition of FhuA barrel (9 nM) to the cis solution, and unlike whole FhuA, this conductance persisted even when urea was removed. (Also unlike whole FhuA, conductance was induced by 3 M glycerol.) Furthermore, the addition of FhuA plug (45 nM) to either side of the membrane resulted in some loss of conductance. (Addition of cis plug resulted in a 25% and 22% loss of conductance in panels A and B, respectively; addition of trans plug resulted in a 50% and 40% loss of conductance in panels A and B, respectively).
Figure 6.
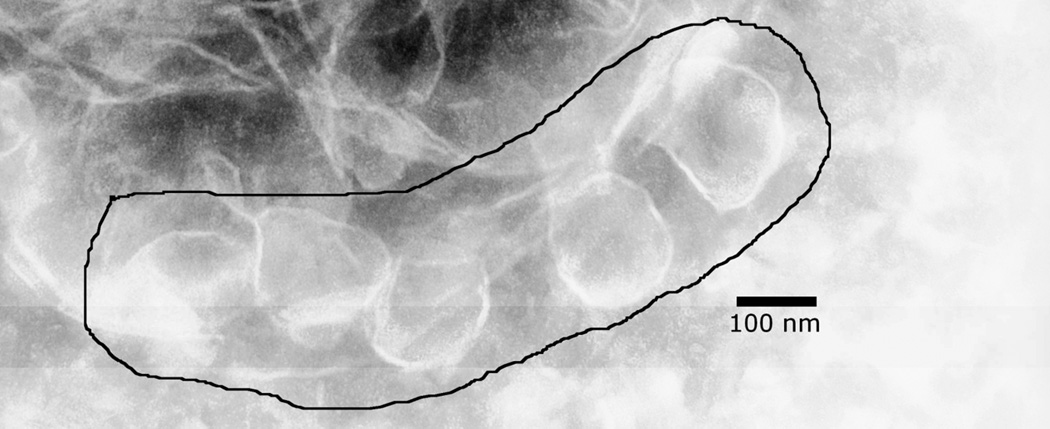
FhuA forms vesicles in detergent solution. Shown here is a negative stained electron microscopy image at 50,000 magnification of whole FhuA after it had been diluted 10-fold into the buffer used in the bilayer experiments. (This resulted in a 0.03mg/mL FhuA protein concentration and a 0.01% LDAO detergent concentration.) The black line encloses several ~100 nm vesicles. No vesicles were observed in solutions with detergent alone (not shown).
In contrast to the results with whole FhuA, the conductance induced by the barrel was not reversed upon removing the urea. (Also unlike whole FhuA, significant conductance was induced in the presence of 3 M glycerol.) This was to be expected, if the other effect of urea (besides its effect on the fusion of the vesicles with the membrane) is disruption of the plug domain. The addition of the FhuA plug to either the cis or trans compartment resulted in a loss of conductance ranging from 55–90% of total conductance (Fig. 5), with 15–30% conductance loss due to plug action from the cis compartment and 40–60% loss due to plug action from the trans compartment. The conductance block by the plug followed exponential kinetics with a time constant from the cis and trans sides of 4.69 ± 1.05 minutes (n = 8) and 4.45 ± 1.45 minutes (n = 13) respectively. Unexpectedly, the subsequent addition of urea did not cause the channels to reopen (data not shown) (see Discussion).
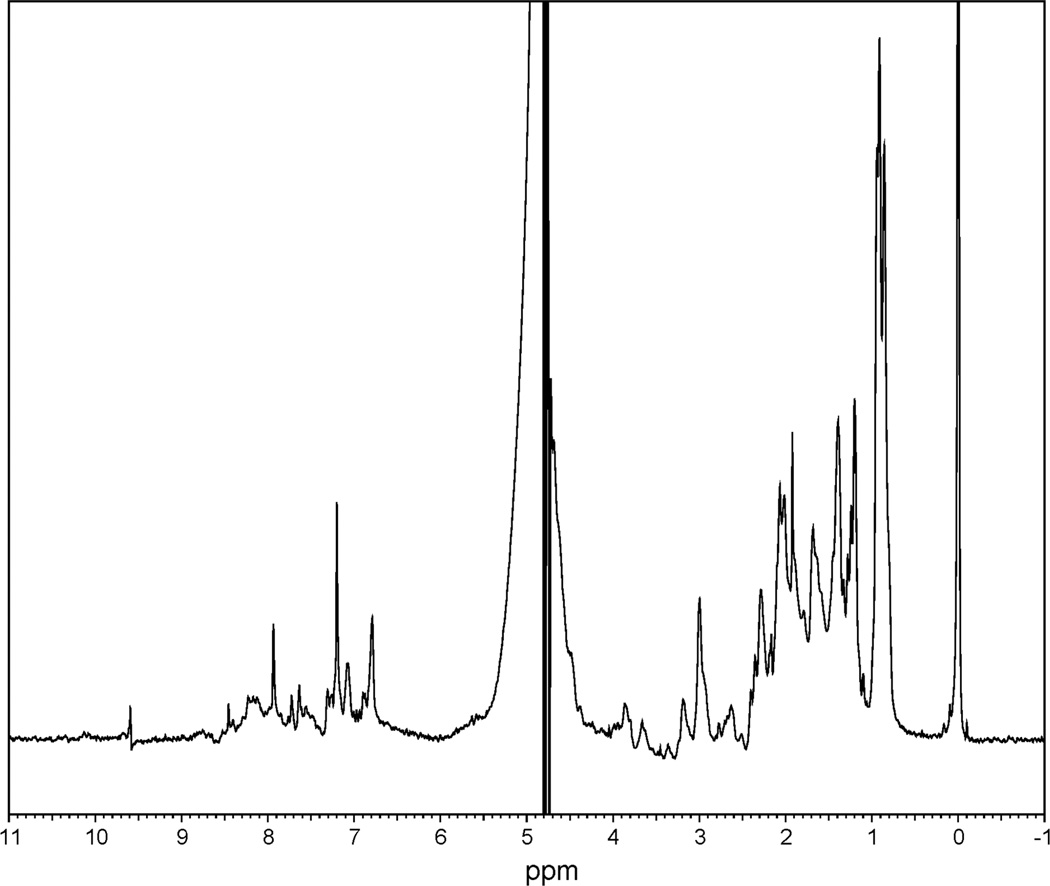
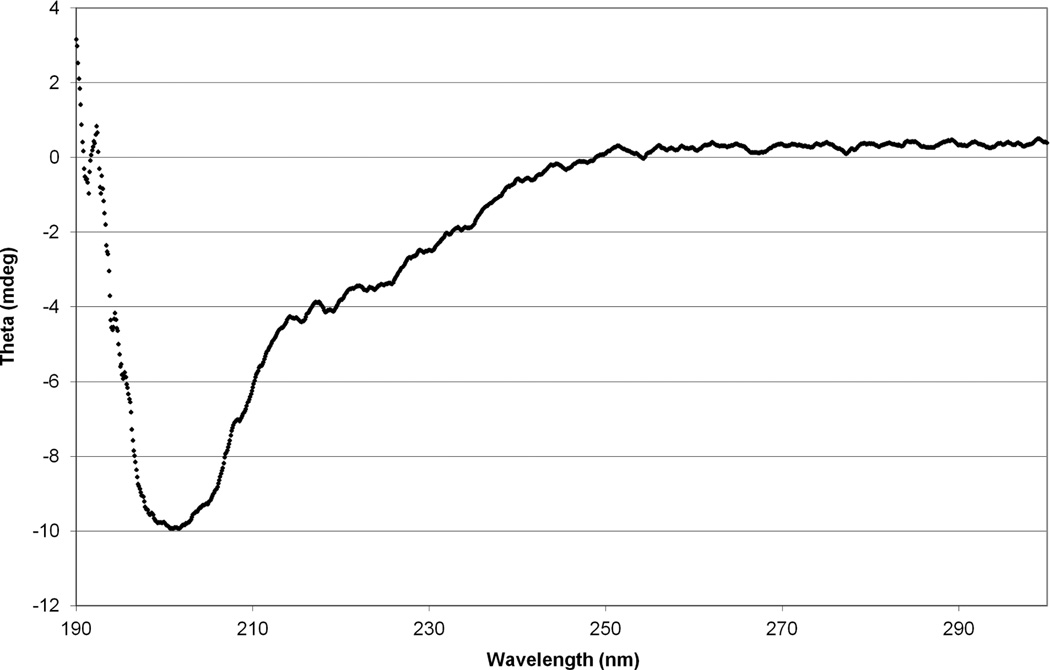
Since the plug seems able to enter and occlude the barrel, we sought to determine the secondary structure characteristics of the plug in solution. The NMR spectrum (Fig. 7) of the plug showed no peaks below 0.5 ppm or above 8.5 ppm, suggesting there is no hydrophobic protein core, but not ruling out secondary structure completely. To check for secondary structure, we performed circular dichroism on the plug (Fig. 8), and analysis of the spectrum found a mostly unfolded polypeptide with some beta-sheet structure.
Figure 7.
FhuA plug has no protein core. This NMR spectrum shows most of the peaks in the range of 0.5–3 ppm and 7–8.5 ppm, suggesting that the methylene groups and amine groups of the protein, respectively, are not in a hydrophobic core.
Figure 8.
FhuA plug has little secondary structure. This CD spectrum shows no obvious minima at the canonical secondary structure locations, but analysis by the K2D2 program suggests a ~26% beta sheet structure and ~8% alpha helix structure, with the rest being unstructured.
Discussion
We have shown for FhuA reconstituted in planar lipid bilayers that trypsin can, in the presence of 4 M urea, prevent the loss of conductance normally induced by the removal of urea (Fig. 2A). Combined with the gel shift assay (Fig. 3), we interpret these data to mean that in the presence of urea, at least some part of the plug comes out of the barrel and is exposed to the action of trypsin. The fact that this response was much less in the absence of urea (Fig. 2B) suggests that trypsin was acting on something that it has more access to in the presence of urea, i.e., the plug, and not the loops of the protein. Mass spectrum analysis indicates that the first 67 residues of FhuA are accessible to trypsin (Fig. 4), in agreement with data previously obtained by the Coulton laboratory(11). It should be noted, however, that while in that case complete conversion of FhuA to the faster-migrating species on SDS-PAGE occurred in the absence of urea, that conversion took 20 hours of incubation at 37 °C. This is not inconsistent with our result of very little whole FhuA conversion (in the absence of urea) to a faster migrating band, since our exposure was only 20 minutes at room temperature. A mutant FhuA lacking residues 21–128 was also exposed to trypsin(20), and K154 was found to be a trypsin-accessible site. However, we believe that the different results (trypsin action at K154 versus K67) observed are due to the differing constructs used in the trypsin digest experiments; hence the data obtained with this FhuA V21–128 are not necessarily comparable to our studies.
The fact, however, that trypsin action on whole FhuA was constrained to the intial 67 residues of the polypeptide, either in the presence (this work) or absence (this work and(11)) of urea, suggests that the plug domain of FhuA may normally be transiently exposed to solution, and urea may be accelerating the rate of plug movement into the solution and/or decelerating reentry of the plug domain back into the lumen of the barrel. In fact, molecular dynamics simulations of the action of TonB on BtuB(21) indicated that unwinding of the plug domain of BtuB by TonB was “a relatively low-force event”. This unwinding included the “unzipping” of the first beta strand from the rest of the four-stranded beta sheet of the plug domain of BtuB. In FhuA, the first 67 residues of the plug domain also encompass this first beta strand of the beta sheet present in the plug domain (Fig. 4), lending support to the notion that the action of urea on FhuA mimics to some extent the in vivo behavior of FhuA (that is, the action of TonB upon FhuA).
Since we were led to believe that the plug of whole FhuA came at least partly out of the barrel in the presence of urea and then reinserted into the barrel with the removal of urea, we thought to separately express and purify the plug and barrel of FhuA and attempt to observe interactions on planar lipid bilayers. It should be noted that this has been previously observed in vivo for the proteins FhuA(22), FepA(8) and FpvA(23). Likewise, we also observed interactions of the plug and barrel on the planar lipid bilayer. As seen in Figure 5, the addition of the plug to either the cis or the trans compartment was able to cause a loss of conductance, suggesting that i) the plug is plugging the barrel of FhuA and ii) like their full-length brethren, the FhuA barrels also insert bidirectionally into the membrane(17). Experiments with colicin M, however, suggest an alternative possibility. Colcin M, when added to the trans, but not cis, compartment after FhuA barrels had been inserted with a urea cycle, yielded an increase in conductance (Fig. S2), suggesting that the barrel channels may all be oriented extra-cellular side trans, and two plugs may be occluding the same barrel from different sides of the membrane.
We could not unplug the barrels that had been plugged by the exogenous plug, using 4 M urea (not shown). This may be attributed to a non-native fold of the plug into the barrel of FhuA. It is also possible that the lack of a linker between the plug and barrel is what is responsible for this inability to pull the plug out of the barrel with 4 M urea, since the whole polypeptide (barrel plus plug) had been essentially reconstituted save for the one covalent linkage between the plug and barrel.
We were able to reproduce previously published data showing conductance induced across the membrane with FhuA barrel (and no osmotic gradient) in the presence of symmetric 1 M salt solutions(24), a result also demonstrated for other FhuA mutants(25–27) and even wild type FhuA(28). However, we obtained much smaller conductances than we obtained in 0.1 M KCl with an osmotic gradient (Fig. S1). For this reason, we used 0.1 M KCl and an osmotic gradient in all our experiments with the FhuA barrel construct. The greater rate of barrel insertion into the membrane induced by 3 M glycerol (or 4 M urea) is explained by the negative staining images of the detergent solutions, showing the formation of vesicles in the presence of FhuA (Fig. 6). As demonstrated by Niles et al.(19), an osmotic gradient across the membrane greatly facilitates fusion of vesicles into the membrane. The phenomenon of glycerol inducing robust conductance across the membrane in conjunction with the FhuA barrel, as shown in Figure 4B (glycerol with whole FhuA does not have this effect), suggests that the barrels are open channels, and once inserted into the membrane, allow ions to flow freely through them.
The Braun(24) and Movileanu(28) laboratories demonstrated frequent flickering of the conductance of the FhuA barrel channels in symmetric 1 M KCl solutions, a result we cannot directly confirm due to the lack of temporal resolution in our system. They also demonstrated that the deletions of both the plug and loop 4 converts FhuA into a large open pore, different from the pores formed by the single deletions of plug and loop 4. It is therefore possible that the extracellular loops and plug can each modify the other’s behavior. Our results and their interpretation, however, are independent of possible changes in the local orientation and flexibility of the large extracellular loops that may accompany changes in plug conformation. The presence of channel currents across the membrane with wild-type FhuA observed by Mohammad et al,(28) is something that we nor others (e. g.,(17, 24, 29)) have observed. However, given the amount of protein in the system, and our hypothesis that the plug may transiently exit the barrel, it is not unreasonable that one may observe stochastically opened channels with the wild-type protein.
Isolated FepA plug was reported to be a mostly unstructured polypeptide(20), and because we can get the FhuA plug to occlude the FhuA barrel, we examined the secondary structure characteristics of the isolated FhuA plug, to see if the plug entered the barrel as a folded or unfolded structure. A one-dimensional proton NMR spectrum suggested a lack of a hydrophobic core within the polypeptide, but did not rule out isolated secondary structure without a tertiary fold (Fig. 7). Further analysis by circular dichroism (Fig. 8) revealed that approximately 25% of the polypeptide had assumed a beta-strand configuration. It is possible that these beta strands are free beta strands that are supposed to form the four-stranded beta sheet of the plug domain observed in the many crystal structures of TonB-dependent transporters solved to date.
Supplementary Material
Acknowledgements
We thank Dr. James Coulton and Dr. Karron James for teaching us the FhuA purification protocols, Dr. Ariel Lewis for assistance with the circular dichroism experiments, Dr. Richard Harris for obtaining the NMR spectrum, Mr. Edward Nieves for performing the Mass Spectrometry, and Mr. Frank P. Macaluso for performing the negative staining electron microscopy.
This work was supported by the National Institutes of Health grants GM-29210 (A.F.), and GM008572 (E.U.).
Abbreviations
- EDTA
ethylenediaminetetraacetic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- LDAO
lauryldimethylamine-oxide
- TBDT
TonB-dependent transporter
- BMCC
1-biotinamido-4-(4'-(maleimidoethyl-cyclohexane)-carboxamido) butane
- SBTI
soybean trypsin inhibitor
- HRV 3C
Human Rhino Virus 3C protease
- TEV
Tobacco Etch Virus protease
- DTT
Dithiothreitol
Footnotes
Supporting Information Available:
The supporting information describes our own experiences in using the barrel-only construct of FhuA in 1 M KCl, and our results with colicin M. This material is available free of charge via the Internet at http://pubs.acs.org.
References Cited
- 1.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postle K, Kadner RJ. Touch and go: tying TonB to transport. Mol Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 3.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Structure of TonB in complex with FhuA E. coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 4.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 5.Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 6.Eisenhauer HA, Shames S, Pawelek PD, Coulton JW. Siderophore transport through Escherichia coli outer membrane receptor FhuA with disulfide-tethered cork and barrel domains. J Biol Chem. 2005;280:30574–30580. doi: 10.1074/jbc.M506708200. [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty R, Storey E, van der Helm D. Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli. Biometals. 2007;20:263–274. doi: 10.1007/s10534-006-9060-9. [DOI] [PubMed] [Google Scholar]
- 8.Ma L, Kaserer W, Annamalai R, Scott DC, Jin B, Jiang X, Xiao Q, Maymani H, Massis LM, Ferreira LC, Newton SM, Klebba PE. Evidence of ball-and-chain transport of ferric enterobactin through FepA. J Biol Chem. 2007;282:397–406. doi: 10.1074/jbc.M605333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devanathan S, Postle K. Studies on colicin B translocation: FepA is gated by TonB. Mol Microbiol. 2007;65:441–453. doi: 10.1111/j.1365-2958.2007.05808.x. [DOI] [PubMed] [Google Scholar]
- 10.Ingham C, Buechner M, Adler J. Effect of outer membrane permeability on chemotaxis in Escherichia coli. J Bacteriol. 1990;172:3577–3583. doi: 10.1128/jb.172.7.3577-3583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moeck GS, Tawa P, Xiang H, Ismail AA, Turnbull JL, Coulton JW. Ligand-induced conformational change in the ferrichrome-iron receptor of Escherichia coli K-12. Mol Microbiol. 1996;22:459–471. doi: 10.1046/j.1365-2958.1996.00112.x. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 13.Danilevskaya ON, Gragerov AI. Curing of Escherichia coli K12 plasmids by coumermycin. Mol Gen Genet. 1980;178:233–235. doi: 10.1007/BF00267235. [DOI] [PubMed] [Google Scholar]
- 14.James KJ, Hancock MA, Moreau V, Molina F, Coulton JW. TonB induces conformational changes in surface-exposed loops of FhuA, outer membrane receptor of Escherichia coli. Protein Sci. 2008;17:1679–1688. doi: 10.1110/ps.036244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller P, Rudin DO. Induced excitability in reconstituted cell membrane structure. J Theor Biol. 1963;4:268–280. doi: 10.1016/0022-5193(63)90006-7. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Iratxeta C, Andrade-Navarro MA. K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct Biol. 2008;8:25. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udho E, Jakes KS, Buchanan SK, James KJ, Jiang X, Klebba PE, Finkelstein A. Reconstitution of bacterial outer membrane TonB-dependent transporters in planar lipid bilayer membranes. Proc Natl Acad Sci U S A. 2009;106:21990–21995. doi: 10.1073/pnas.0910023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viswanatha T, Liener IE. The inhibition of trypsin. III. Influence of urea. J Biol Chem. 1955;215:777–785. [PubMed] [Google Scholar]
- 19.Niles WD, Cohen FS, Finkelstein A. Hydrostatic pressures developed by osmotically swelling vesicles bound to planar membranes. J Gen Physiol. 1989;93:211–244. doi: 10.1085/jgp.93.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klug CS, Su W, Liu J, Klebba PE, Feix JB. Denaturant unfolding of the ferric enterobactin receptor and ligand-induced stabilization studied by site-directed spin labeling. Biochemistry. 1995;34:14230–14236. doi: 10.1021/bi00043a030. [DOI] [PubMed] [Google Scholar]
- 21.Gumbart J, Wiener MC, Tajkhorshid E. Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys J. 2007;93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braun M, Endriss F, Killmann H, Braun V. In vivo reconstitution of the FhuA transport protein of Escherichia coli K-12. J Bacteriol. 2003;185:5508–5518. doi: 10.1128/JB.185.18.5508-5518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nader M, Journet L, Meksem A, Guillon L, Schalk IJ. Mechanism of ferripyoverdine uptake by Pseudomonas aeruginosa outer membrane transporter FpvA: no diffusion channel formed at any time during ferrisiderophore uptake. Biochemistry. 2011;50:2530–2540. doi: 10.1021/bi101821n. [DOI] [PubMed] [Google Scholar]
- 24.Braun M, Killmann H, Maier E, Benz R, Braun V. Diffusion through channel derivatives of the Escherichia coli FhuA transport protein. Eur J Biochem. 2002;269:4948–4959. doi: 10.1046/j.1432-1033.2002.03195.x. [DOI] [PubMed] [Google Scholar]
- 25.Braun V, Killmann H, Benz R. Energy-coupled transport through the outer membrane of Escherichia coli small deletions in the gating loop convert the FhuA transport protein into a diffusion channel. FEBS Lett. 1994;346:59–64. doi: 10.1016/0014-5793(94)00431-5. [DOI] [PubMed] [Google Scholar]
- 26.Killmann H, Benz R, Braun V. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 1993;12:3007–3016. doi: 10.1002/j.1460-2075.1993.tb05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Killmann H, Benz R, Braun V. Properties of the FhuA channel in the Escherichia coli outer membrane after deletion of FhuA portions within and outside the predicted gating loop. J Bacteriol. 1996;178:6913–6920. doi: 10.1128/jb.178.23.6913-6920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammad MM, Howard KR, Movileanu L. Redesign of a plugged beta-barrel membrane protein. J Biol Chem. 2011;286:8000–8013. doi: 10.1074/jbc.M110.197723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonhivers M, Ghazi A, Boulanger P, Letellier L. FhuA, a transporter of the Escherichia coli outer membrane, is converted into a channel upon binding of bacteriophage T5. EMBO J. 1996;15:1850–1856. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.