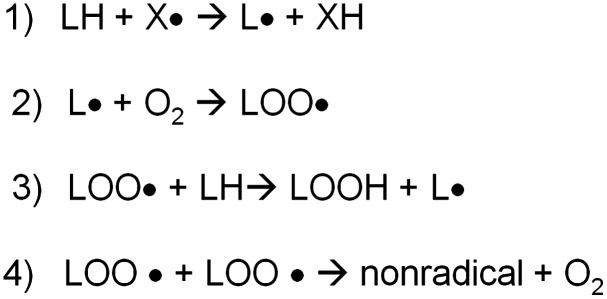Abstract
Several studies demonstrated that oxidative damage is a characteristic feature of many neurodegenerative diseases. The accumulation of oxidatively modified proteins may disrupt cellular functions by affecting protein expression, protein turnover, cell signaling, and induction of apoptosis and necrosis, suggesting that protein oxidation could have both physiological and pathological significance. For nearly two decades, our laboratory focused particular attention on studying oxidative damage of proteins and how their chemical modifications induced by reactive oxygen species/reactive nitrogen species correlate with pathology, biochemical alterations, and clinical presentations of Alzheimer's disease. This comprehensive article outlines basic knowledge of oxidative modification of proteins and lipids, followed by the principles of redox proteomics analysis, which also involve recent advances of mass spectrometry technology, and its application to selected age-related neurodegenerative diseases. Redox proteomics results obtained in different diseases and animal models thereof may provide new insights into the main mechanisms involved in the pathogenesis and progression of oxidative-stress-related neurodegenerative disorders. Redox proteomics can be considered a multifaceted approach that has the potential to provide insights into the molecular mechanisms of a disease, to find disease markers, as well as to identify potential targets for drug therapy. Considering the importance of a better understanding of the cause/effect of protein dysfunction in the pathogenesis and progression of neurodegenerative disorders, this article provides an overview of the intrinsic power of the redox proteomics approach together with the most significant results obtained by our laboratory and others during almost 10 years of research on neurodegenerative disorders since we initiated the field of redox proteomics. Antioxid. Redox Signal. 17, 1610–1655.
-
II. Protein (/Lipid) Oxidation and Protein Dysfunction
-
IV. Application of Redox Proteomics to Selected Neurodegenerative Disorders
I. Introduction
Redox proteomics is the subset of proteomics in which oxidatively or nitrosatively modified proteins are identified (115). Our laboratory was among the first that used redox proteomics to identify oxidatively modified brain proteins (91, 92, 233). Others first used redox proteomics to identify oxidized thiols (34, 88, 157, 250). Redox proteomics has been applied to numerous disorders known to be associated with oxidative stress (OS) (115). This comprehensive article focuses on applications and results of redox proteomics that provide insights into selected neurodegenerative disorders.
II. Protein (/Lipid) Oxidation and Protein Dysfunction
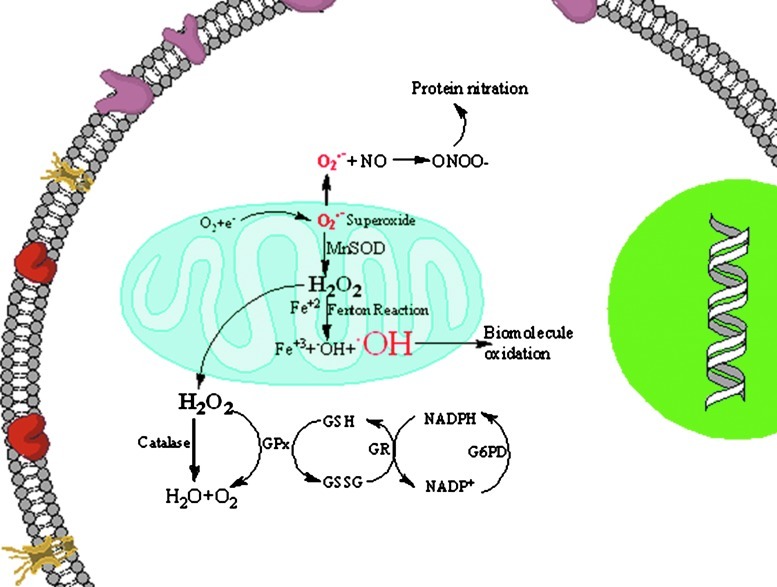
OS induced by free radicals plays an important role in the pathophysiology of a wide variety of diseases including neurodegenerative disorders (63, 180). Free radicals are generated in vivo from various sources, one of the major sources being the leakage of superoxide radical from the mitochondria (Fig. 1). Under physiological conditions, levels of superoxide anion radicals (O2.−) are maintained in the cell by the antioxidant enzyme, superoxide dismutase (SOD), which disproportionates O2.− to hydrogen peroxide (H2O2) and oxygen (Fig. 1). Further, the H2O2 formed is converted to water and oxygen by the enzymes catalase, peroxidase, or glutathione peroxidase (GPx). GPx uses reduced glutathione (GSH) to carry out its functions, and the levels of reduced GSH are maintained by the enzyme glutathione reductase (GR), which converts oxidized glutathione (GSSG) to GSH using NADPH for reducing equivalents. In the brain, the levels of catalase are greater than those for GPx. The importance of these enzymes in relation to neurodegeneration will be discussed in further detail next. During neurodegeneration, the balance just described for the regulation of free radical levels is lost, leading to increased production of free radicals, and also the generation of other types of reactive oxygen species (ROS) and reactive nitrogen species (RNS). When the levels of hydrogen peroxide increase in the cells and if redox transition metal ions such as Fe+2 or Cu+ are available nearby, Fenton reactions will occur, resulting in the formation of hydroxyl radicals, which are highly reactive and can damage biomolecules, including protein, lipids, carbohydrates, and nucleic acids (79). In neurodegenerative disorders, this imbalance in metal ion homeostasis can induce OS. If the levels of superoxide radicals are high and if there is an increased availability of nitric oxide, radical-radical recombination results in the formation of peroxynitrite, a highly reactive product with a half life of <1 s that can lead to nitration of biomolecules, proteins, and lipids (38). Hence, markers of OS, levels of antioxidant enzymes, and elevation of cellular stress response proteins reflect the level of oxidative damage in, and fate of, the cell.
FIG. 1.
Free radicals are generated by various mechanisms. One way by which free radicals are generated is via release of superoxide anion from the mitochondria, leading to increased formation of reactive oxygen and reactive nitrogen species and, consequently, damaging the biomolecules. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
Proteins constitute one of the major targets of ROS/RNS, which can elicit a variety of modifications in amino-acid residues, including cysteine (Fig. 2), methionine, tryptophan, arginine, lysine, proline, and histidine (63, 79, 384) among others. Among various types of modifications by ROS/RNS are the formation of protein carbonyls (PCO), 3-nitrotyrosine (3-NT) and protein-bound 4-hydroxy-2-trans-nonenal (HNE), the latter being a reactive product of lipid peroxidation.
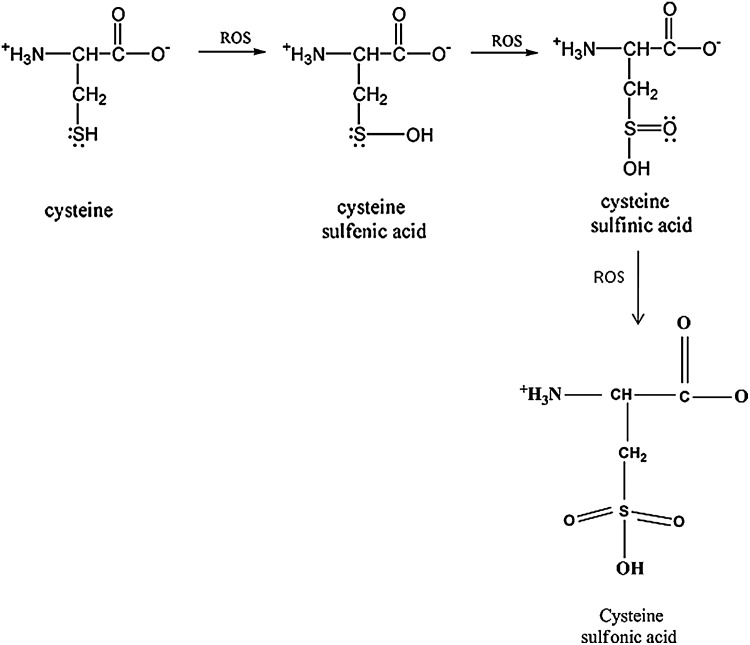
FIG. 2.
Cysteine oxidation at neutral pH. Cysteine plays an important role in the regulation of protein function. Cysteine is vulnerable to attack by reactive oxygen species, which can lead to the formation of cysteine sufinic acid and eventually to the cysteine sulfonic acid. Measurement of the sulfonic acid on a protein is another maker for the detection of oxidative stress.
A. Protein carbonyls
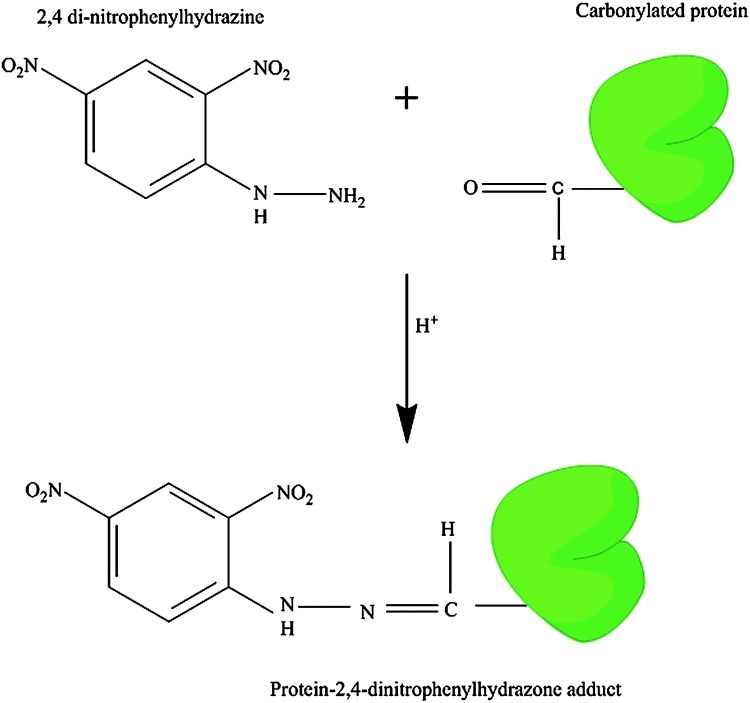
PCO result from several sources, among which are peptide backbone fragmentation, hydrogen atom abstraction at peptide alpha carbons, attack on several amino-acid side chains (see above), and by the formation of Michael adducts between Lys, His, or Cys residues and α- and β-unsaturated aldehydes formed during the peroxidation of polyunsaturated fatty acids (384). PCO are also formed by the secondary reactions of amino groups of lysine residues with reducing sugars or their oxidation production (glycation/glycoxidation reactions) (114, 352). Hence, protein carbonylation leads to oxidation of side-chains, backbone fragmentation, formation of new reactive species (peroxides, DOPA), release of further radicals, and occurrence of chain reactions. Most oxidative protein damage is irreversible; however, there are certain enzymes in vivo that can either repair or clear the damaged proteins (see below). PCO are stable products of protein oxidation compared with the other products of OS, for example, F2 isoprostanes, which are readily generated during sample storage, processing, and analysis. Consequently, PCO are a general and widely used index to determine the extent of oxidative modification in both in vivo and in vitro conditions (40, 79, 114, 352, 360, 401). Several sensitive assays were developed for detection of oxidatively modified proteins (114, 247), the most often used of which is the detection of the protein hydrazone derivative of the carbonyl group with 2,4-dinitrophenylhydrazine (DNPH). These protein hydrazones can be detected spectrophotometrically at 375 nm, but in solution samples, homogeneity or uniformity is one of the potentially confounding issues. Another means for detection of protein hydrazones is immunochemical detection using an anti-DNP-protein antibody that can give a clear indication of the amount of total PCO in a given sample. The latter method has been widely employed to detect PCO in biological samples (Fig. 3). Other methods for PCO analysis include use of biotin hydrazide coupled to fluorescein isothiocyanate (FITC)-labeled streptavidin (374).
FIG. 3.
Derivatization of protein carbonyl using 2,4-dinitrophenylhydrazine (DNPH). The carbonyl group reacts with the DHPH to form a protein-DNPH hydrazone at acidic pH. This product is stable at neutral pH. The DNPH-protein hydrazone measures are used for the determination of the amount of oxidative damage to the protein in biological samples. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
B. Protein nitration
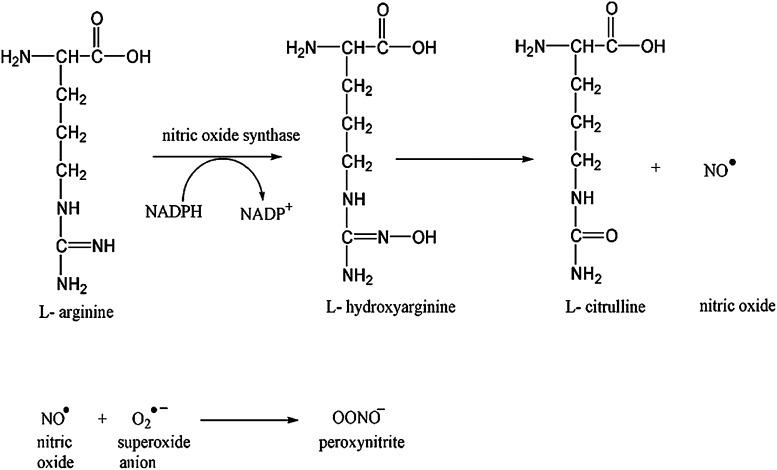
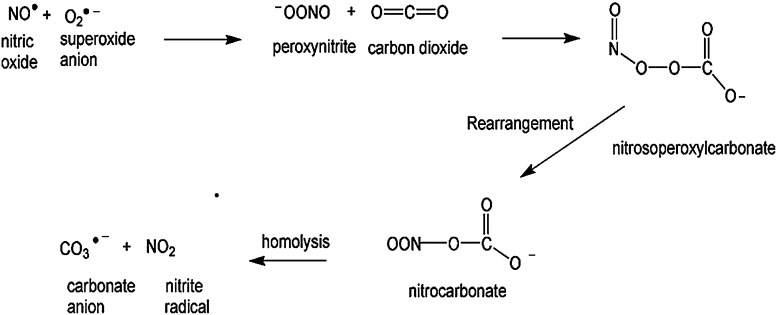
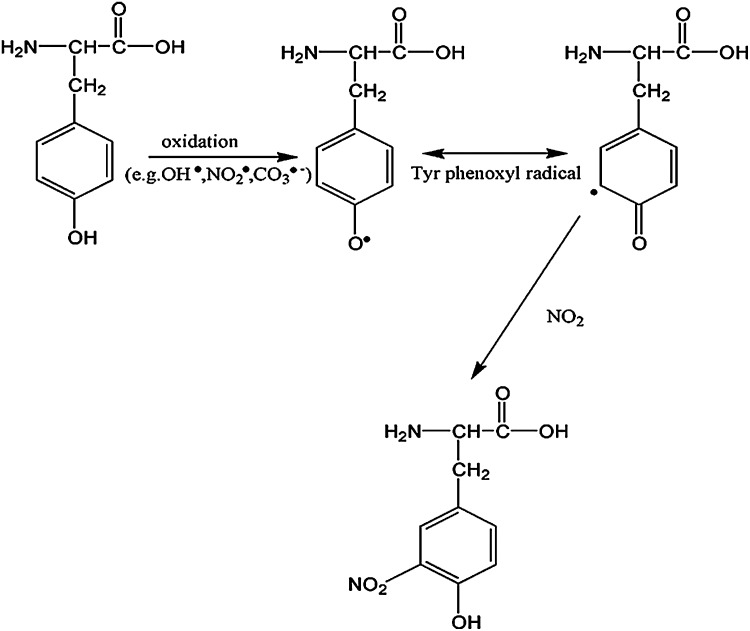
Protein nitration is a formal protein oxidation, resulting from an RNS reaction. In conjunction with the enzyme nitric oxide synthase (NOS), arginine produces nitric oxide (NO.) and L-citrulline. Nitric oxide can react with superoxide to form the strong oxidant, peroxynitrite (Fig. 4). Peroxynitrite has been shown to affect mictrotubule assembly and ATPases (237) via specific amino-acid residue oxidation. Peroxynitrite can also modify protein thiols as observed in cysteine and methionine oxidation (11) as well as tyrosine and tryptophan to promote protein nitration. Peroxynitrite can exist as an anion (ONOO−) or, rarely, the protonated peroxynitrous acid (ONOOH). Peroxynitrous acid undergoes homolysis to produce damaging hydroxyl radicals (OH•) and nitrogen dioxide radical. Formation of the acid form of peroxynitrite is CO2 dependent. A nitrosoperoxyl intermediate is formed from the combination of peroxynitrite and carbon dioxide, which rearranges to form nitrocarbonate. This species can be cleaved homolytically to form carbonate and NO2 radicals (Fig. 5), which react with a tyrosyl free radical to form 3-NT (Fig. 6).
FIG. 4.
Formation of peroxynitrite. During the conversion of L-arginine to L-Citrulline, nitric oxide is formed as one of the products. Nitric oxide can react with the superoxide anion, resulting in the formation of a highly reactive product, peroxynitrite.
FIG. 5.
The reaction of peroxynitrite and carbon dioxide results in the formation of nitrosoperoxylcarbonate, which undergoes rearrangement to form nitrocarbonate. Nitrocarbonate can undergo homolysis, resulting in the formation of nitrite radical and carbonate anion.
FIG. 6.
Formation of 3-NT from a tyrosine. The nitrite radical produced by the reaction of nitric oxide with carbon dioxide reacts with the tyrosine reside at meta-position, resulting in the formation of 3-NT. 3-NT, 3-nitrotyrosine.
Nitric oxide is multifunctional, as it is involved in signal transduction by activating guanylate cyclase and increasing intracellular cGMP. NO also plays a role in vasodilation, neurotransmission, cardiac function, and inflammation (82). Nitric oxide is constitutively produced by endothelial and neuronal NOS (eNOS, nNOS, respectively) and induced by inducible NOS (iNOS). NO has been associated with neurodegenerative diseases by acting as a neurotoxin when excessively produced; however, recent studies suggest that NO may have neuroprotective properties as well, that is, NO acts as a Janus molecule (82). As noted, there are three forms of NOS: neuronal (nNOS or Type I), inducible (iNOS or Type II), and endothelial (eNOS or Type III). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) acts as an NO sensor (71, 183). Nitric oxide is permeable to the plasma membrane and can bind to guanyl cyclase (204). This modification affects the synthesis of cyclic GMP, which alters several key GMP-related proteins, including cGMP phosphodiesterases (4), cGMP ion gated channels, and cGMP protein kinases. Type I NOS is a calcium-dependent enzyme, as it is stimulated by an increase in Ca2+ leading to excitoxicity and mitochondrial dysfunction. nNOS regulates cerebral blood flow, skeletal muscle contraction, and athleroscleorsis. nNOS also regulates iNOS expression through NF kappa B regulation. iNOS binds to calmodulin, a calcium-binding regulatory protein(138, 385). Although the primary function of Type III NOS is vasodilation, this enzyme still plays a role in mitochondrial dysfunction and smooth muscle contraction. This specific isoform has been found to interact with β-actin, vascular endothelial growth factor, and caveolin 1 (231, 323), which bolsters the role of eNOS in muscle contraction, cell proliferation, and apoptosis. Most recently, eNOS has been associated with heat shock proteins (HSPs) 70 and 90, which act as molecular chaperones by guiding damaged proteins to the proteasome for protein degradation. Nitric oxide can also bind to glutamate channels and indirectly to calcium and potassium channels (141). Glutamate, an excitatory neurotransmitter in neurons, binds to the N-methyl D-aspartic acid (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors and leads to Ca2+ entrance to neurons, which if excessive, causes a disruption of calcium homeostasis. This disruption can eventually lead to cell death, thereby providing additional support for the role of nitric oxide in apoptosis.
1. Peroxynitrite (ONOO−)
Peroxynitrite can react with tau (389), cytochrome c, (90), manganese superoxide dismutase (MnSOD) (261), Cu/ZnSOD (402), creatine kinase (218), and GAPDH (375) among many other proteins of importance to neuronal functions. Tau acts as a stabilizing protein for microtubules. Elevated oxidative and nitrosative stress are associated with hyperphosphorylation of tau. Once hyperphosphorylated, tau can no longer sustain microtubule assembly, causing its disintegration and eventual neuronal apoptosis. Cytochrome c is a mitochondrial protein that plays a pivotal role in cell death. As a mobile electron carrier in the electron transport chain (ETC) of mitochondria, cytochrome c transfers one electron from Complex III to Complex IV. Cytochrome c is highly soluble and can be released into the cytoplasm if the mitochondrial outer membrane is opened. Once released, cytochrome c stimulates cellular apoptosis by binding to apoptotic protease activating factor 1, which, in turn, binds to other apoptotic effectors to form the apoptosome. The apoptosome can then activate several caspases that subsequently trigger apoptosis. The inactivation of SOD results in an excess of superoxide and an overall increase in ROS production and OS. Inactivation of ONOO− targets, creatine kinase, and GAPDH results in lowered ATP production, inefficient energy metabolism, and dysfunction of other key cellular processes (71).
2. Nitrogen dioxide (NO2)
Nitrogen dioxide can increase protein nitration, which results in protein dysfunction. NO2 serves as an oxidant in inflammation mediated by the peroxidases, eosinophil peroxidase and myeloperoxidase (142). Nitrogen dioxide exposure increases the levels of nitrosative stress that can lower antioxidant levels. Lipoic acid, an endogenous mitochondrial complex cofactor and antioxidant, undergoes oxidation by nitrogen dioxide and can lead to increased tyrosine dimerization (382). This gas can oxidize the antioxidant, GSH, and increase activity of GR and GPx (354). The depletion of GSH shifts the cellular redox balance to oxidative and nitrosative stress. Nitrogen dioxide radicals can also be formed by the oxidation of peroxynitrite. As discussed next in Section 4, the levels of protein nitration are elevated in Alzheimer's disease (AD)/Parkinson disease (PD)/amyotrophic lateral sclerosis (ALS) and Huntington disease (HD) consistent with a role of NO2 in neurodegeneration.
C. HNE adduction to proteins
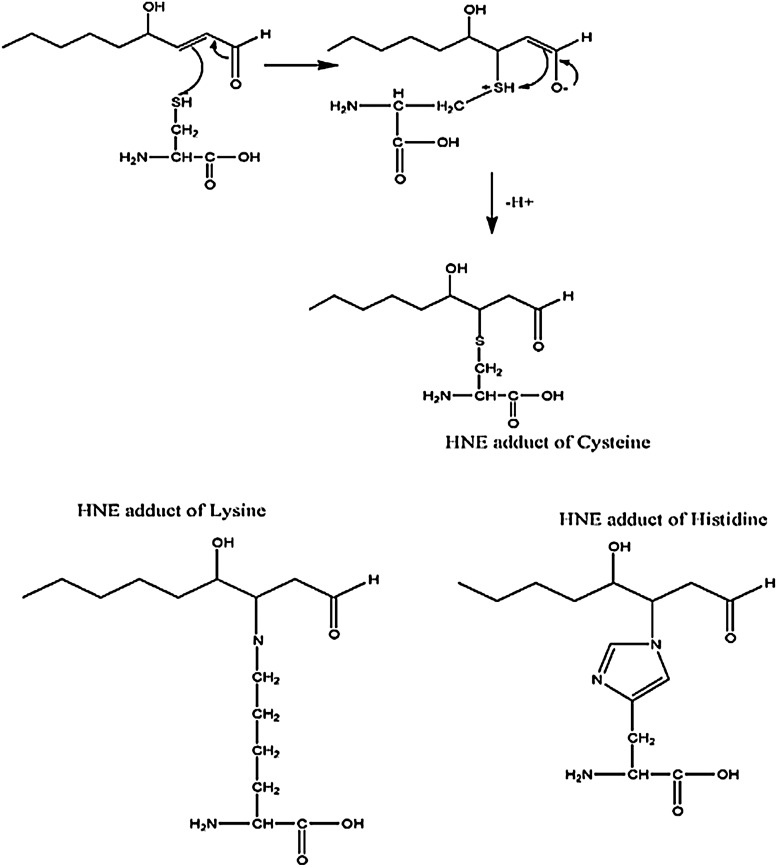
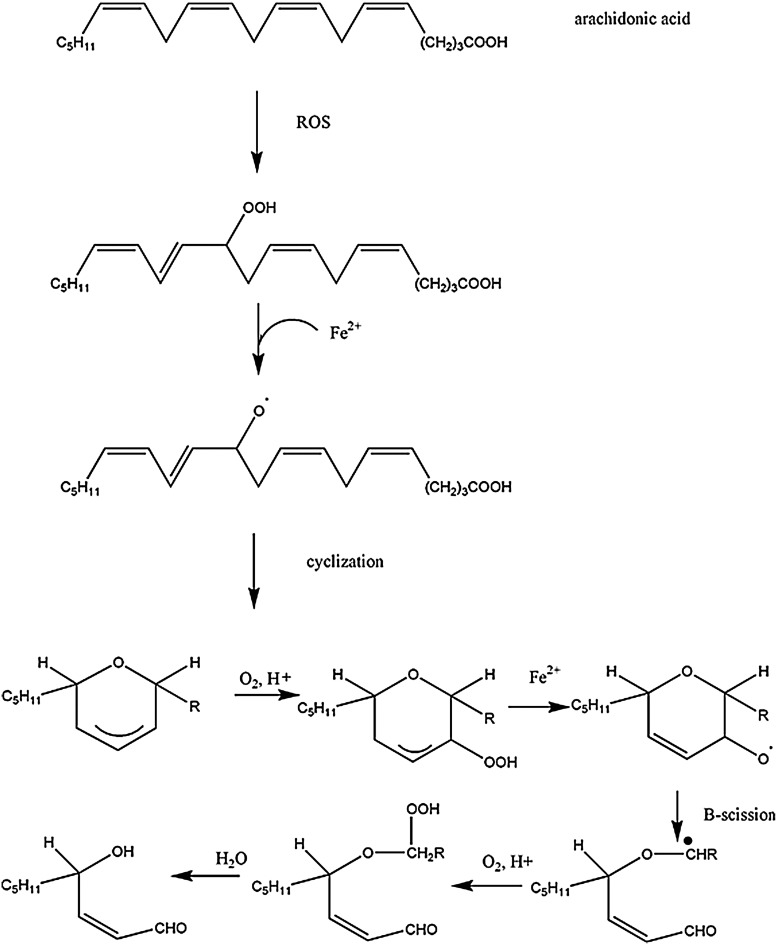
Lipids within the central nervous system (CNS) are exceptionally susceptible to oxidation due to the fact that polyunsaturated fatty acids are rich in the brain, and the concentration of oxygen in the lipid bilayer is high, whereas the antioxidant levels are relatively low. Lipid peroxidation, leading to numerous products, including α,β-unsaturated aldehydydes, is highly evident in neurodegenerative diseases (5). As a whole, lipid-peroxidation-derived reactive electrophilic aldehydes are capable of facile covalent attachment to proteins by forming stable adducts with cysteine, lysine, and histidine (Fig. 7) through Michael addition (66, 139). Lipid peroxidation occurs through continuous free radical chain reactions until termination occurs (Fig. 8). Lipid-resident free radicals attack an allylic hydrogen atom on acyl chains of lipids to form a carbon centered radical (step 1). This radical reacts with paramagnetic oxygen (O2) to produce peroxyl radicals (step 2). These peroxyl radicals can react with adjacent allylic H atoms on acyl chains of lipids forming a lipid hydroperoxide and a C-centered radical, thus propagating the chain reactions (step 3). Depending on a number of factors, including acyl chain length and degree of unsaturation, the lipid hydroperoxide can decompose to produce multiple reactive products such as acrolein, iso- and neuroprostanes, malondialdehyde, and HNE, all of which are significantly elevated in several neurodegenerative diseases, including AD, PD, ALS, and HD or models thereof (17, 56, 66, 144). Lipid peroxidation can be terminated by two radicals reacting and forming a nonradical and oxygen (step 4). α-tocopherol (vitamin E) is a “chain breaking” antioxidant and can terminate the propagation steps of lipid peroxidation. When the phenoxyl H of vitamin E is abstracted by radicals, an α-tocopherol radical forms that can be reverted back to vitamin E by vitamin C or GSH. An example of the steps I–IV in lipid peroxidation is shown in Figure 8.
FIG. 7.
One of the products of lipid peroxidation is HNE that can react with cysteine, lysine, and histine via Michael addition. Protein-bound HNE levels are used as an index of lipid peroxidation. HNE, 4-hydroxy-2-trans-nonenal.
FIG. 8.
Lipid peroxidation reaction summary. The process of lipid peroxidation involves an initiation process that begins with the hydrogen atom abstraction from an unsaturated fatty acid, resulting in the formation of lipid radical, which can then react with molecular oxygen, resulting in the formation of lipid peroxyl radicals. The lipid peroxyl radical can then abstract a H-atom from the other unsaturated fatty acid; this is referred to as a chain propagation reaction. When two lipid peroxyl radicals react, this will result in the termination of the lipid peroxidation process.
HNE is an α, β-unsaturated alkenal product of omega-6 fatty acid oxidation (Fig. 9). Increased levels of HNE cause disruption of Ca2+ homeostasis, glutamate transport impairment, and membrane damage, leading to cell death (66, 139). GSH prevents HNE damage in cells (79, 312). Similar to Figure 9, polyunsaturated fatty acids such as arachidonic acid and linoleic acid (337) undergo free radical mediated mechanisms by which a lipid peroxyl radical is formed. Ultimately, the resulting peroxyl radical is converted to an allylic carbocation via β-scission. The peroxyl radical is further oxidized to a lipid peroxide. Through hydration, the C-O bond breaks, resulting in 4-hydroxy-2-nonenal. As noted, once formed, HNE can covalently attach to proteins by Michael addition, which alters protein structure (361) and causes a loss of protein function and activity (139).
FIG. 9.
Formation of HNE from arachidonic acid. Oxidation of unsaturated fatty acids results in the formation of HNE.
D. Importance of clearance and detoxification systems
1. The proteasome, parkin, ubiquitin carboxy-terminal hydrolase-L1, and HSPs
The function of the proteasome is to degrade damaged, aggregated proteins. The 26S proteasome is a structure composed of two major subunits, the regulatory 19S cap and 20S catalytic core. These components combine through ATP binding to form the complete 26S proteasome. Oxidized proteins are degraded by the 20S proteasome in an ubiquitin-independent manner (118).
Both parkin and ubiquitin carboxy-terminal hydrolase-L1 (UCH-L1) are essential to the proper function of the proteasome. Parkin acts as an E3 ligase whose sole responsibility is to attach ubiquitin molecules to damaged proteins. Genetic mutations in parkin have been shown to be associated with familial PD (400). Although the role of parkin is still under investigation, it has been recently studied as a therapeutic for Parkinson's disease, as it is reportedly neuroprotective (1, 102). UCH-L1 removes ubiquitin molecule from the C-terminal end of the poly ubiquitin polymer after attachment of the protein to the proteasome. If ubiquitin units are not removed, the protein cannot be properly degraded, and ubiquitin molecules will not be recycled for future use. Levels of damaged proteins thereby increase, causing possible proteasomal overload. Oxidation modification of UCH-L1 has been observed in AD hippocampus (91, 103) and PD (179). Mutations in this protein support the concept of impaired protein degradation, mitochondrial dysfunction, and proteasomal overload associated with many neurodegenerative disorders (181).
HSPs act as chaperone proteins that aid in restoring misfolded or aggregated proteins, or in directing misfolded proteins to the proteasome. HSPs are involved in combating stress by protecting proteins from denaturation (83). HSPs 70 and 90 interact with eNOS, which is possibly a compensatory regulatory mechanism used to repair oxidative damage characteristic of neurodegenerative disease.
2. Superoxide dismutase
Maintenance of SOD is critical to achieving oxidative balance; otherwise, the cell would be in a constant state of OS. There are four different forms of SOD, including Cu/ZnSOD (SOD1), MnSOD (SOD2), NiSOD, and FeSOD. Mutations in SOD1 have been shown to cause familial ALS, and overexpression of SOD1 has been known to be associated with Down syndrome (DS) (177). These data are interconnected, because the SOD1 gene resides on chromosome 21, the locus of the trisomy for DS, the same chromosomal location for amyloid precursor protein (APP), the precursor of the toxic, AD-relevant peptide, amyloid beta-peptide (Aβ) (1–42). This is equally important, because SOD1 knockout mice have a normal lifespan and do not develop motor neuron disease (319), but SOD2 knockout mice die shortly after birth due to increased OS. This observation demonstrates the importance of mitochondrial resident MnSOD. Based on its location, modification of this protein can lead to greatly impaired proteasome function, causing an oxidized protein “overload” with the inability to correctly degrade oxidized proteins. This notion is further supported by research showing that specific nitration of Tyr9, Tyr11, and Tyr34 by peroxynitrite in MnSOD inactivates the enzyme (373).
3. Catalase
Catalase is reported to decrease lipid peroxidation products (87) Under OS conditions, as demonstrated in AD, PD, and ALS (25), catalase activity is lowered significantly, thereby reducing antioxidant potential (104).
4. Peroxiredoxins
Peroxiredoxins (Prxs) are functionally similar to catalase in that they detoxify free radicals in the cell by reducing H2O2. There are six forms of peroxiredoxin: Prx1, Prx2, Prx3, Prx4, Prx5, and Prx6. Prx1–Prx5 use thioredoxin (Trx) as an electron donor, while Prx6 uses GSH. There are two classes of Prxs: 1-Cys and 2-Cys. Peroxiredoxin VI (PRX VI) is the only 1-Cys Prx, while the other five isoforms are 2-Cys Prxs. The two classes differ by the number of active cysteine residues involved in catalysis (324). In AD brain, the levels of Prx-1 and Prx-2 were found to be increased, while the level of Prx-3 was significantly decreased, suggesting a role of ROS especially from mitochondria as a key player in the pathogenesis of AD (227). Redox proteomics studies from our laboratory led to the identification of Prx2 as a nitrated protein in early AD (EAD) brain, suggesting impaired regulation of RNS such as peroxynitrite lead to increase nitration of selective target proteins (321). In PD brain, the level of Prx2 was found to be significantly increased compared with age-matched controls (31).
5. Trx and Trx reductase
The Trx are a family of proteins that act as oxidoreductases. The dithiol center contributes to the protein's catalytic activity. Trx can reversibly reduce disulfide bonds. By removing hydrogen peroxide, Trx helps to reduce levels of cellular OS, thereby enhancing antioxidant ability. Although its antioxidant properties are important, especially its role in recycling oxidized Prxs (see above), Trx is a multifunctional protein involved in DNA synthesis, protein folding, bacterial and viral infections, transcriptional regulation, immune response, and cellular communication (249). Trx reductase, a selenoenzyme, reduces Trx by the cofactor, NADPH. There are two forms of Trx reductase, cytsolic (Trx reductase-1) and mitochondrial (Trx reductase-2). Another function of Trx reductase is its function in proper brain development. Alterations and loss of function of Trx reductase have been well documented in both AD and PD (237, 257). The well-established relationship of Trx and Trx reductase in maintaining redox balance attests to their importance in neurodegenerative disorders, specifically AD and PD.
The activity of Trx is regulated by a protein called Trx-binding protein-2 (TBP-2), also known as vitamin D3 upregulated protein 1 (VDUP1) or Trx-interacting protein. TBP-2/VDUP1/Txip interact directly with the redox-active domain of Trx via two cysteine residues (287). The interaction of TBP-2 with Trx will prevent the interaction of Trx with other molecules such as apoptosis signal-regulating kinase 1 and proliferation associated gene (292), thereby making cells more susceptible to oxidative damage and apoptotic cell death. Further, increasing evidence showed that TBP-2 also regulates important biological functions, such as the regulation of glucose and lipid metabolism (338). Upregulation of TBP-2/VDUP1 enhances paraquat-induced OS (212). Hence, an increase in the levels of TBP-2/VDUP1 might lead to an increase in OS, by suppression of Trx activity. In cancerous cells, the level of TBP-2 expression has been reported to be decreased, suggesting that this protein plays a role in cancer. Recent studies are focusing on silencing of TBP-2 to prevent cancer growth (411). Moreover, TBP-2 deficiency induces lipid dysfunction, and this might be critical in the aging process. Thus, Trx and TBP-2 play important roles in the pathophysiology of cancer and metabolic syndrome by direct interaction or by independent mechanisms.
6. Glutathione reductase
GSH, a tripeptide composed of glutamate, cysteine, and glycine, is synthesized by two enzymes: glutamate-cysteine ligase and GSH synthase. Cysteine is the limiting amino acid in GSH biosynthesis, (micromolar levels in the brain, while glutamate and glycine are in millimolar concentrations) (220). Free GSH is used to maintain the reduction potential of many cell types. Since pro-oxidants are readily available in the brain, two GSH molecules can form a disulfide bridge and be converted to GSSG via GPx. GR is an antioxidant enzyme that catalyzes the reduction of GSSG to GSH using NADPH, thus maintaining free GSH levels and increasing overall antioxidant ability. GR activity is decreased in AD (23, 87).
7. Vitamins in neurodegeneration
Plasma and cerebrospinal fluid (CSF) from AD patients show reduced levels of ascorbate compared with the control (62, 326), which might hinder the reduction of α-tocopherol radical back to α-tocopherol (155), thereby leading to increased oxidative damage. In addition, dietary vitamin E intake significantly reduced risk of PD (410), but similar studies with vitamin C are lacking. In ALS, the use of vitamin E did not show any significant protection. The studies conducted so far using vitamins suggest that more clinical trials are needed with vitamins C and E in patients with AD/PD/HD and ALS to explain preclinical promise of these antioxidants (216). The lack of protective effects in clinical trials of vitamins could be explained based on the fact that the reducing agents required for recycling of oxidants to its active forms were not included in the studies nor were the basal redox states of subjects determined.
8. Involvement of iron in neurodegeneration
Living organisms require iron to correctly function and perform their most essential metabolic processes. Iron is required to support the brain's high respiratory rate as well as for correct myelination, neurotransmitter synthesis, and gene/protein expression.
Fe homeostasis is frequently altered in neurodegenerative disorders (39), and iron progressively accumulates in the brain with age. In addition, during brain aging, iron is partially converted from its stable and soluble form (ferritin) into hemosiderin and other derivatives that contain iron at higher reactivity (109). Thus, the pathogenic role of iron in brain aging results not only from its accumulation but also from its increased reactivity. Under certain conditions, iron is a powerful pro-oxidant due to its high availability; the fascile electron chemistry that is fundamental for its functions may also be a source of OS-induced toxicity. Iron metabolism in humans is conservative: 1–2 mg of iron is absorbed per day, and the same amount is excreted. When an excess of iron is not efficiently removed by detoxification systems, it may, especially in the ferrous state (Fe+2), promote the conversion of H2O2 to·OH via the Fenton reaction and, in turn, lead to a greater turnover in the Haber-Weiss cycle. In addition, OS itself may increase the levels of free iron. This effect occurs through the release of iron from ferritin by superoxide anion, from heme proteins such as hemoglobin and cytochrome c by peroxides, and from iron-sulfur proteins by ONOO·−. All these phenomena lead to amplification of OS, and the excessive production of ROS is responsible for damage to proteins, DNA, and phospholipids leading to structural and functional alterations of neuronal cells.
Interestingly, the brain is endowed with a peculiar iron metabolism compared with other organs. First, the blood–brain barrier limits brain access to plasma iron. There is a highly specific transport mechanism that moves iron across the endothelial cells of the BBB into brain. However, little is known about the mechanism of iron release into the brain or the regulation of the transport mechanism. Insights into this transport mechanism could be crucial for understanding how excess iron can accumulate in the brain observed in many neurodegenerative diseases. Second, the concentration of iron varies widely in different brain regions. For example, those brain areas associated with motor functions (e.g., extrapyramidal regions) tend to have more iron than nonmotor-related regions (230), which might contribute to the observation that movement disorders are commonly associated with iron imbalance.
E. Role of iron in neurodegeneration
1. Fe homeostasis in AD
Several studies showed alteration of iron hemostasis in AD brain. T1 and T2 magnetic resonance relaxation times analysis in transgenic mice model of AD showed the presence of iron in amyloid deposits, and the T1 results were negatively correlated with age. Further, T2 in the subiculum of adult APP/PS1 animals was lower than in PS1 mice, suggesting a relationship between amyloid and iron loads in this region (137). AD patients who were carriers of the HFE mutation showed higher levels of iron, lower levels of transferrin (TF) and ceruloplasmin (CP), and higher CP/TF ratios, suggesting a link between HFE mutations and iron abnormalities, and OS in AD (167). Further, the iron transport protein TF functions were also reported to be disrupted in AD (108). AD hippocampus showed a moderate positive correlation with mini-mental state examination (MMSE) scores, and a negative correlation with the duration of the disease for iron using phase imaging (127). A recent study showed that APP protein possesses ferroxidase activity, can catalytically oxidize Fe(2+), and has a major interaction with ferroportin. Alterations in APP have been shown to lead to iron retention, and increased OS in HEK293T cells, primary neurons, and APP mice model of AD. The regulation of iron levels in AD has been linked to zinc based on the fact that zinc is a component of senile plaque (SP) and regulates ferroxidase activity (133). Further detailed investigation of the excessive accumulation of iron in the AD affected regions may lead to better understanding of AD.
2. Fe homeostasis in PD
PD brain has increased deposition of iron in microglia, astrocytes, oligodendrocytes, and dopaminergic neurons of the substantia nigra pars compacta (325). In addition, the levels of iron also were reportedly increased in the SN in both subchronic 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)- and 6-OHDA-induced PD animal models (211, 295). Studies with human BE-M17 neuroblastoma cells overexpressing wild-type, A53T, or A30P α-synuclein showed that iron together with dopamine or H2O2 stimulates the production of intracellular aggregates and induce toxicity. Further, the ratio of Fe(II) and Fe(III) (224) is altered in PD. For the synthesis of dopamine, Fe(II) is used as a co-factor by the enzyme tyrosine hydroxylase. Further, Fe(III) is capable of inducing OS by its interaction with neuromelanin (96) and may also be involved in the formation of α-synuclein oligomers (192, 396). In a recent study, Davies et al. (119) showed that α-synucluein acts as a cellular ferrireductase, and thereby helps in reducing iron (III) to bio-available iron (II). Further, PD patients also showed an increased level of divalent metal transporter-1 (DMT-1) in the same area where PD pathology and iron deposition accumulate, suggesting that increased levels of iron and DMT-1 might be involved in PD pathogenesis. The iron storage protein ferritin was found to be increased in postmortem PD brain (325), which might be a response to the increased iron content reported in PD. In contrast some studies showed a significant decrease of SN ferritin levels in PD (143).
3. Fe homeostasis in ALS
Lower and upper motor neurons degenerate in ALS, resulting in progressive paralysis and death. Increased levels of iron were reported in the spinal cord from ALS subjects (206) and may possibly correlate with increased levels of oxidative damage through the induction of Fenton chemistry. Fe accumulation may be due to its increased uptake by its specific transporter, lactoferrin, which is reportedly increased in ALS-affected motor neurons (246). Increased Fe deposition conceivably could be due to increased levels of ferritin, as this iron-binding protein was found in SOD1-G93A mice just before end-stage disease. Moreover, in ALS patients, CSF iron reducing ability is decreased, while the content of oxidized proteins is increased in both CSF and plasma (347). In order to better understand the role of iron in ALS, the expression of proteins associated with iron homeostasis (DMT, TF receptor, the iron exporter Fpn, and CP) has been studied in a transgenic mice model of ALS. mRNA levels of these proteins were higher in rostral compared with caudal spinal cord regions, and this finding correlates with the caudal-to-rostral progression of the disease in SOD1-G37R transgenic mice (210).
Other evidence supporting the involvement of Fe in this disorder is the prevalence of a HFE (hemochromatosis gene) mutation in ALS patients as the second most frequent mutation in this disease (397). HFE interacts with the TF receptor, and HFE mutations are associated with decreased expression of SOD1, α-tubulin, and β-actin. Thus, HFE polymorphisms in ALS could contribute to altered Fe homeostasis and, consequently, to increased oxidative damage in this disease (380).
4. Fe homeostasis in HD
Iron and ferritin accumulation has been detected in the putamen, caudate nucleus, and globus pallidus by MRI and postmortem investigations in HD brain, the same regions in which extensive pathological damage is observed (122).
Iron levels have been found to be higher early in the disease process and have therefore been considered as a putative risk factor. Indeed, iron-rich areas, like the caudate nucleus and the putamen, that receive major excitatory input from the cortex, are particularly affected in HD compared with other iron-rich regions with less excitatory transmission or areas with dense NMDA receptors but lower iron concentrations. This finding has led to the hypothesis of an enhancing adverse effect of iron and excitatory transmission. However, the cause of the increased iron levels in HD still remains unknown. Mutation of huntingtin (Htt), with a CAG trinucleotide expansion (>38 repeats), is the genetic cause of HD; however, Htt is essential both for proper regulation of the iron pathway and iron response protein. Mutant Htt also is involved in the stimulation of autophagy and proteosome systems that, under normal conditions, degrade ferritin after its Fe-mediated oxidation. Ferritin plays an important role in Fe homeostasis by sequestering this metal; in turn, Fe levels regulate ferritin expression, which increases with Fe accumulation. Simmons et al. (349) analyzed the specific localization of ferritin in the brain from transgenic R6/2 mice and HD patients and found that ferritin was predominantly increased in microglia. Those cells appeared dystrophic, suggesting that they may be dysfunctional and contribute to HD progression. Moreover, low serum ferritin level and slightly elevated CP levels in the HD brain indicate a more generalized dysregulation of iron metabolism. Further studies are needed to determine the exact interactions and role of iron in the pathogenesis of HD. Iron metabolism has been shown to be altered in the animal models of HD; in addition, in vitro studies showed that the oxidation by mutant htt is dependent on iron. Alterations in Fe signaling and increased expression of the TfR protein were reported in a model of HD (STHdhQ111/Q111) compared with wild-type cells (STHdhQ7/Q7) (381). Further, the expression of HTT was found to be elevated in response to increasing Fe levels (191).
F. Some known consequences of protein oxidation
Oxidation of proteins often makes the protein dysfunctional or nonfunctional; therefore, protein oxidation has both physiological and pathological consequences (72, 159). Further, the oxidation of proteins could lead to the alteration in the secondary and tertiary structure of proteins. For example, during secondary structure formation of proteins the hydrophobic amino acid domains are usually buried inside the proteins; oxidation of the proteins induces a conformation change of the proteins, thereby leading to exposure of the hydrophobic amino-acid residues to an aqueous environment, promoting protein aggregation and accumulation of the oxidized proteins as cytoplasmic inclusions (79), as observed in AD (40).
Oxidation of proteins may also prevent the subunit association of proteins contributing to the loss of tertiary structure of a protein and consequently affecting its function. Hence, it is recommended that identification of oxidatively modified proteins by redox proteomics should be followed by functional assessment of the identified proteins. These functional studies may identify metabolic or structural consequences caused by oxidative modification (302, 376). A number of previous studies showed that oxidation of proteins could lead to alterations in protein expression and gene regulation, protein turnover, modulation of cell signaling, induction of apoptosis, necrosis, etc., eventually leading to loss of cells and function (72). Further, oxidation of proteins increases the susceptibility of a protein to degradation by the 20S proteasomes and, consequently, decreased levels of the proteins in general. However, in certain diseases, oxidation of proteasome components renders the proteasome inactive, consequently leading to accumulation of damaged proteins within the cells.
Redox proteomics (see next) analyses were used to identify specific oxidatively modified brain proteins in neurodegenerative diseases.
III. Overview of Redox Proteomics
Two-dimensional (2D) proteomics was first introduced by O'Farrell (289) and by Klose (228), enabling the greater separation of proteins based on isoelectric point and relative mobility. Redox proteomics approaches makes use of this method that is discussed next to identify oxidatively modified proteins in various biological samples. While immunochemical techniques just discussed are useful for identifying overall oxidative modification levels, they do not provide specific information regarding individual proteins that have been modified. Additionally, it becomes important to identify the specific amino-acid sites of oxidative modification in order to better understand effects on protein structure and function. These topics are the field of redox proteomics. Note that in the context of this article, we use the term redox proteomics to refer to proteomics techniques that are used to identify oxidatively modified proteins, specifically PCO, −3-NT-, and HNE-modified proteins. Traditional, and still the most often used, redox proteomics approaches that identify oxidatively modified proteins rely on 2D polyacrylamide gel electrophoresis (2D-PAGE), Western blot analyses, and mass spectrometry (MS) (Fig. 10). Redox proteomics can be applied to the identification of several oxidative modifications such as PCO, 3-NT, HNE, and glutathionylation (157). Other nongel-based approaches that utilize liquid chromatography (LC) or affinity chromatography in combination with MS also have been developed for identification of oxidized proteins. This section of the article is intended to familiarize the reader with some of the current proteomics techniques available for the identification, quantification, and enrichment of PCO, 3-NT-, and HNE-modified proteins that are applicable to neurodegenerative diseases such as AD, PD, HD, and ALS, as well as other disorders (68, 115). Data obtained from any of the redox proteomics approaches next require further analyses (e.g., enzymatic activity studies, computational simulations, etc.) in order to completely understand the effects of oxidative modification on protein structure and function and the relevance to neurodegenerative diseases or redox biology. Insights into pathology, biochemistry, and consequent clinical presentation of neurodegenerative disease resulting from redox proteomics are discussed in Section 4 next for each disorder.
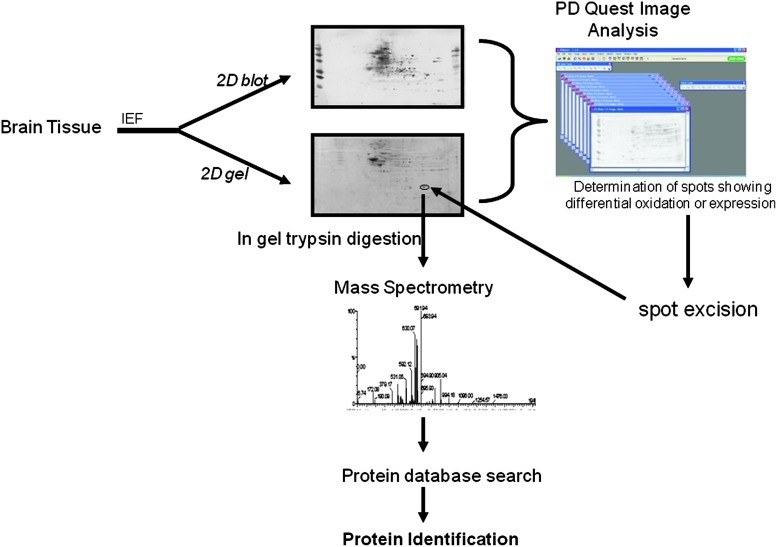
FIG. 10.
Outline of redox proteomics approach. The identification of oxidatively modified protein involves first the separation of proteins by isoelectric point (IEF) followed by the separation of proteins based on relative mobility (Mr). The separation of the proteins is followed by transferring the proteins onto nitrocellulose or polyvinylidene fluoride membrane, probing with the antibody of interest, and determination of oxidatively modified protein by image analysis. Once a protein is identified as oxidatively modified, the protein spot will be excised from the gel, digested with trypsin, and subjected to mass spectrometry for correct identification of the proteins. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
A. Global, gel-based approaches
Success in redox proteomics with 2D-PAGE has been possible due to the availability of primary antibodies that are specific for particular oxidative modifications. For instance, as just noted for the detection of PCO-modified proteins, proteins are commonly derivatized with a reagent such as DNPH using Schiff base chemistry. The resulting protein with a DNP hydrazone adduct is separated using 2D-PAGE, which separates proteins based on isoelectric point (pI) and migration rate. The proteins on the 2D gel are transferred onto a nitrocellulose or polyvinylidene fluoride membrane and probed with an anti-DNPH antibody. A number of secondary antibodies linked to enzymes (e.g., alkaline phosphatase, horseradish peroxidase) can be used to visualize PCO-modified protein spots based on chemiluminescence, fluorescence, or colorimeteric assays. Other modifications such as HNE, 3-NT, and glutathionylation can be identified using this overall approach; however, no sample derivatization is required, and the antibodies are accordingly adjusted (366).
Software algorithms such as PDQuest or Dymension Delta2D etc., which compare spot-to-spot pixel density between or among samples, can be used to determine specific protein spots that change in an oxidative level after normalization of spots on the 2D blot to the corresponding spot on a 2D gel (concurrently run on a separate aliquot of the same sample). Individual spots of interest are excised from the 2D gel and undergo in-gel digestion with trypsin. Tryptic peptides are either analyzed with matrix-assisted lazer desorption ionization (MALDI)-MS or electrospray ionization (ESI)-tandem MS (MS/MS). MALDI-MS analyses rely on peptide mass fingerprinting (PMF) that identify the oxidized protein. Masses of tryptic peptides are measured in the MS and are searched against the appropriate species database using MASCOT, a probability-based scoring algorithm (68). In some cases, modified peptides may be identified, as the precursor masses are shifted by the mass of the modification (e.g., 3-NT is a 45 Da shift).
While earlier proteomics studies often employed MALDI and PMF that identify proteins, newer and more precise ESI-MS/MS methods are now commonly employed. In ESI-MS/MS experiments, intact peptide masses are measured, and several precursor peptide ions are isolated in the MS and fragmented using collision-induced dissociation (CID). The energy introduced to ions during CID causes fragmentation along the peptide backbone, such that b- and y-type fragment ions are generated. The intact precursor mass and list of b- and y-fragment masses are used to determine the amino-acid sequence of the peptide and subsequent identification of the protein with MASCOT or SEQUEST database searching algorithms. When setting up the database searching criteria, users can include dynamic modifications on specific residues (e.g., oxidation of methionine, etc.). In this manner, it is possible to identify specific sites of modification; however, identifications can be limited by the low abundance and ionization efficiency of oxidatively modified peptides.
Other 2D-gel-based approaches rely on derivatization strategies that introduce fluorescent tags into the modified protein. For example, Yoo and Regnier utilized biotin hydrazide to derivatize PCO groups in yeast cells exposed to hydrogen peroxide (406). Biotin-tagged samples were separated with 2D-PAGE, and an avidin-FITC probe was used to visualize PCO-modified proteins in the gel. This approach was recently applied for the identification of PCO-modified proteins that vary with age in serum of neonatal and fetal pigs (86). The 2D-PAGE-based approach using FITC detected as little as 0.64 pmol of PCO-modified proteins (406). One of the limitations with this and the aforementioned 2D-PAGE approaches is that they require additional starting material, as a second 2D gel has to be run in parallel for protein identification. Additionally, other limitations of 2D-PAGE include poor resolution and sensitivity to highly acidic/basic proteins, hydrophobic proteins, very small/very large proteins, and limited dynamic range.
B. Targeted, gel-free approach
1. Enrichment of PCO modified proteins
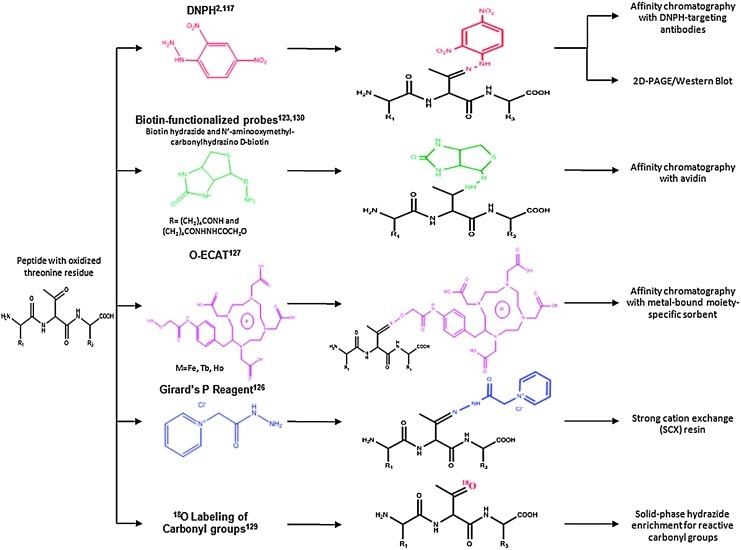
Nongel based strategies for the enrichment of PCO-modified proteins have been developed. A summary of these methods is provided in Figure 11 and has been recently reviewed (263). Most of these methods are based on the Schiff base chemistry that is possible with the carbonyl group. The most traditional Schiff base method relied on the 2D Western analysis of DNPH-derivatized carbonyl proteins (283) and had early applications in the identification of oxidized proteins in AD brain (74). Using shotgun proteomics methods, tryptic peptides are separated with strong cation exchange (SCX) and/or reverse-phase (RP) LC and detected by MS. Through the incorporation of a heavy isotope (13C6) version of DNPH, this method also can be applied to the quantitation of PCO-modified proteins between two samples (383). To date, this has only been tested on simple protein mixtures.
FIG. 11.
Summary of methods for the derivatization and enrichment of carbonylated proteins are shown using an example tripeptide that contains an oxidized threonine residue. We note that other commonly carbonylated residues include Pro, Arg, Lys, His, and Trp, among others. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
As just noted, investigators have made use of biotin functionalized probes, such as biotin hydrazide and biocytin hydrazide, to isolate PCO-modified proteins. Biotin probes are useful due to the strong binding affinity of biotin with avidin (125). In this approach, as shown in Figure 11, for an illustrative tripeptide containing an oxidized threonine residue, carbonyl groups on modified proteins react with biotin through formation of hydrazone or oxime groups. Biotin-tagged proteins are isolated using immobilized avidin or streptavidin columns, often after the Schiff base has been reduced with sodium cyanoborohydride. This affinity chromatography approach has been applied to the analysis of metal-catalyzed oxidized human albumin (377), and complex mixtures from yeast (259), rats exposed to 2-nitropropane (275), rat plasma (274), human plasma (263), and cardiac mitochondrial proteins (97). In all these studies, amino-acid sites of modification can be determined in the MS due to the mass shift associated with the biotin tag. An advantage of this approach is that it allows the identification of carbonylated proteins that arise due to direct oxidation of side chains (e.g., threonine, arginine, lysine, and proline), or modification by lipid peroxidation products such as HNE and advanced glycation endproducts (AGEs) (263). It is often the case that other oxidative modifications such as oxidation of histidine, methionine, and tryptophan residues are also identified with these approaches. An MS study of HNE-modified creatine kinase has taken advantage of newer instrumentation (138).
Due to the limitations in ESI ionization efficiency of biotinylated peptides, the Girard's P reagent, which contains a quaternary amine group, has also been applied to enrich and detect PCO-modified proteins (276). The quaternary amine group is used to isolate PCO-modified tryptic peptides with an SCX resin at pH 6.0. Peptides are separated and detected with RP LC-MS/MS. The quaternary amine group helps increase ionization efficiency of PCO-modified peptides during ESI. In order to better facilitate database searching and improve confidence in peptide identification specific to PCO-modified peptides, a heavy isotope (2H5) version of Girard's P reagent was developed and successfully applied to oxidized TF protein spiked into a matrix of yeast lysate (276).
A different type of reagent, oxidation-dependent element coded affinity tags (O-ECAT), has been used to enrich for PCO-modified proteins by employing rare earth metals such as Tb or Ho (241). The structure of the O-ECAT tag is shown in Figure 11, whereby the aminooxy group forms an oxime with aldehydes or ketones and the 1,4,7,10-tetraazacyclododecane, N, N′, N′′, N′′′-tetraacetic acid (DOTA) serves as the metal chelator group. Two samples can be coded with the different metals and mixed before tryptic digestion and affinity purification based on an antibody against the DOTA moiety. Due to the different mass shift caused by the metals [i.e., Tb (158.92 Da), Ho (164.93 Da)] in the reagent tags, the relative heights of the doublet pairs that arise in the MS spectrum can be used for assessing relative quantitation levels of oxidized proteins in the different samples. This method was demonstrated in recombinant human serum albumin, whereby a number of oxidation sites were mapped (241). O-ECAT pairs co-elute in the RP separation, making data analysis more straightforward. An exciting advantage of this approach is that the multiplexing capabilities can be increased with the introduction of a wide range of metals.
The last approach shown in Figure 11 for the redox proteomics identification of PCO-modified proteins is a label-free method that relies on 18O labeling of carbonyl groups (242, 327). The amount of 18O that is incorporated into carbonyl groups is titrated in a controlled manner such that the prepared ratio of 18O: 16O introduces an isotopic signature specific to carbonylated peptides. The isotopic cluster associated with carbonylated peptides shows an 18O:16O ratio that matches with the prepared experimental conditions. Using a software algorithm, peaks displaying a specific isotopic cluster pattern can be readily sorted from other noncarbonylated peaks. While this approach does not require multiple steps of chromatographic separation that can result in sample loss, there are limitations in the number of modification sites identified.
2. Enrichment of HNE-modified proteins
Several of the approaches for the chemical enrichment of PCO-modified proteins can be applied for the identification of HNE modifications, such as biotin functionalized probes (98, 275, 392). For example, the biotin functionalized probe N′aminooxymethylcarbonylhydrazino-D-biotin, also known as aldehyde-reactive probe, has been applied to identify HNE-modified proteins in human monocyte cell lines before and after ascorbic acid treatment (97). Solid-phase hydrazide approaches have been used to enrich HNE-modified peptides (242, 318). This enrichment strategy has been coupled with electron capture dissociation (ECD) of peptides whereby neutral loss of HNE (158 Da) from CID MS/MS triggers MS3 analysis to identify the sequence of the HNE-modified peptide (318). The use of ECD in combination with CID increases the number of identified HNE-modified peptides in comparison to CID alone.
3. Enrichment of 3-NT modified proteins
Helman and Givol developed an anti-3-NT antibody immobilized on a sepharose affinity column to capture nitrated peptides from lysozyme (188). The recovery rate of 3-NT-containing peptides was ∼55% as measured with UV-Vis detection. Recovery rate was increased by varying incubation time on the column and incorporating more washes (35). Immunopurification strategies have been recently employed for the characterization of 3-NT-modified proteins in CSF of human patients to better understand HIV-associated neurocognitive disorders (37).
MacMillan-Crow and Cruthirds were the first who successfully developed a method for the immunoprecipitation of 3-NT-proteins (262) followed by 2D Western analysis. The 1A6 monoclonal antibody is widely used and has been employed to understand OS during renal ischemia/reperfusion (110), effects of in vivo nitroglycerin treatment on 3-NT-modifications of prostacyclin synthase in rats (193), the inhibition of MnSOD with 3-NT modification (262), effects of nitration on Ca+2-ATPase activity in aged adult hearts (229), and other applications (393). This antibody has fewer issues with nonspecific binding relative to polyclonal antibodies (22) and anti-3-NT agarose conjugates (124). A common problem with immunoprecipitation, in general, is the difficulty in identification of low-abundance proteins, and this limitation is not specific to 3-NT-modified proteins (171, 217).
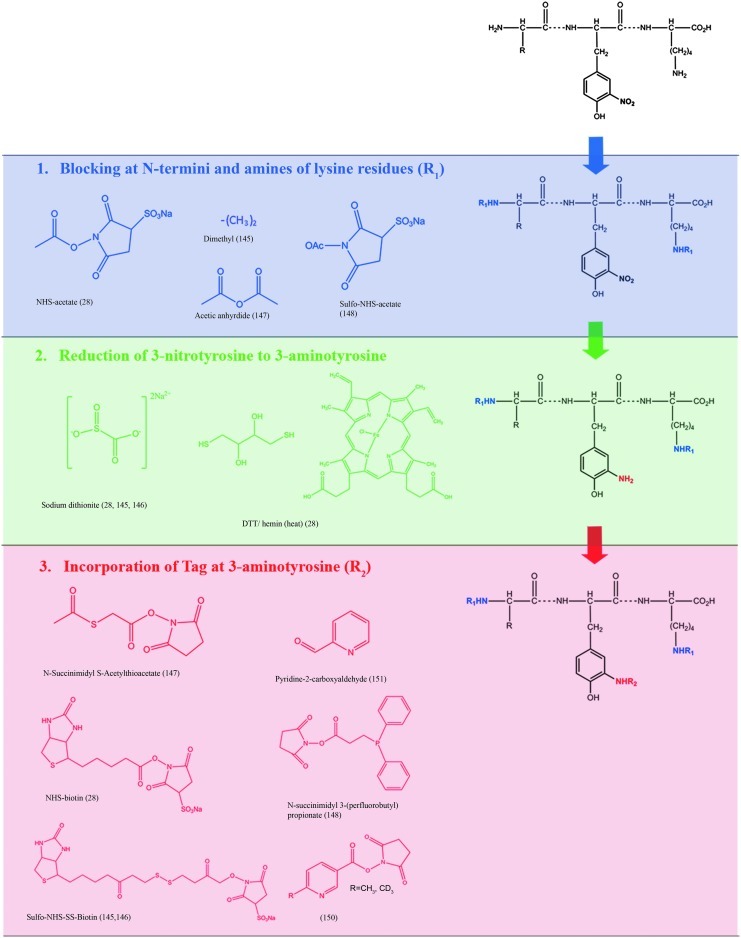
The number of redox proteomic strategies that have been developed for the isolation and detection of 3-NT-modified proteins is steadily rising. Initial enrichment approaches suffered due to the limited reactivity of the nitro group of 3-NT. To this end, chemical modification steps that convert 3-NT to the more reactive 3-aminotyrosine (3AT) have been employed. Figure 12 provides a summary of various strategies for the enrichment and detection of 3-NT-modified proteins. Next, we provide a brief description of these methods.
FIG. 12.
Summary of strategies for the enrichment of 3-NT-modified proteins. In nongel based methods, 3-NT modified proteins were detected by blocking N-termini and amines of lysine residues. See text for references. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
Similar to PCO-modified proteins, affinity chromatography methods based on biotin/avidin interactions are employed for 3-NT-modified proteins (4, 199, 286, 332). The incorporation of the biotin tag has been shown to be more effective after blocking of N-termini (ɛ-amines) and ɛ-lysine (ɛ-amines) residues followed by reduction of 3-NT to 3AT (286). Blocking strategies, as shown in Figure 12, have employed various chemical reagents that introduce acetyl groups to primary ɛ-amines (4) or dimethyl groups (199). After blocking N-termini and lysine residues, 3-NT is reduced to 3AT with either sodium dithionite (199, 286, 332) or dithiol threitol (DTT) and hemin (4). The next step is the incorporation of a biotin tag to 3AT. Several biotin tags have been shown to be effective such as NHS-biotin (4), sulfo-NHS-SS-biotin (282, 286, 332), or biotin (199). The enrichment of the biotin-tagged peptides then occurs using an avidin (4) or a streptavidin (286) column. Overall, this particular chemical tagging strategy has been useful for identifying 3-NT-sites in in vitro nitrated bovine serum albumin (BSA) (286) and other small protein mixtures (199). However, its use for complex protein practices, such as the brain, may be problematic, which presently limits the utility of this approach.
Recently, the isobaric tag for relative and absolute quantitation (iTRAQ) reagent has been used to tag 3AT groups as opposed to biotin (101, 195). Due to the multiplexing capabilities of iTRAQ, up to eight different samples can be pooled together to search for 3-NT modifications in 3-NT-modified proteins. The iTRAQ tag relies on gas-phase fragmentation chemistry that generates reporter ions that show up at low mass-to-charge (m/z) values (i.e., 113, 114, 115, 116, etc.). Using software programming, “enrichment” of 3-NT-modified peptides can take place postanalysis. This approach has been demonstrated in simple mixtures from 3-NT-Angiotensin II and 3-NT-bovine BSA (101, 195). As just noted, translation of this powerful method for simple systems may be more difficult in complex samples such as brain homogenates.
Another approach uses a different chemical approach for isolating 3-NT-modified proteins based on Ni2+-nitrotyrosine affinity (NTA) column magnetic agarose beads (112). Similar to the strategies just mentioned, shown in Figure 12a, b, N-termini, and lysine residues are initially blocked through reaction with sulfo-NHS-acetate, and 3-NT are converted to 3AT. A Schiff base is formed by reacting 3AT with pyridine-2-carboxyaldehyde, resulting in a metal-chelating motif that can be captured using an NTA column with a magnetic separator to sort non-nitrated and 3-NT-peptides (112). An enrichment procedure based on the solvophobic properties of fluorinated carbonated groups and their preference to be localized in a fluorine-rich environment has been manipulated for the identification of 3-NT-Angiotensin II and 3-NT-bovine serum albumin spiked into HeLa cell lysate digests (23). Fluorinated carbon-tagged peptides are captured by solid phase extraction with fluorinated carbon-linked silica beads, a chemistry based on fluorine-fluorine interactions. Using this new approach, 28 nitrated peptides from human hepatoma cell line, Huh7, have been identified (23).
IV. Application of Redox Proteomics to Selected Neurodegenerative Disorders
A. Alzheimer's disease
AD is the most common form of dementia in the elderly population. In the United States, more than five million people are diagnosed with AD, which is clinically characterized by progressive memory loss, cognitive impairment, loss of language and motor skills, and changes in behavior not due to any other cause. The definitive diagnosis of AD is obtained at autopsy by the presence of three characteristic hallmarks of AD, that is, synapse loss, extracellular SPs, and intracellular neurofibrillary tangles (NFTs). The major component of SP is Aβ, a 40–42 amino acid peptide that is derived from proteolytic cleavage of an integral membrane protein (APP) by the action of beta- and gamma-secretases. NFT are largely composed of hyperphosphorylated tau protein (135, 176).
Aβ(1–42) has been considered to play a causal role in the development and progression of AD (340). The putative role of Aβ(1–42) in AD pathogenesis is further supported by a number of in vitro and in vivo studies. The Aβ peptide exists in different aggregation states, and a number of studies suggest that the small oligomers of Aβ are the actual toxic species of this peptide rather than Aβ fibrils (131, 170). In addition, studies of familial AD and individuals with DS, who develop AD-like dementia at late ages, further provided a strong association of the role of Aβ in AD pathogenesis and progression (255). The trisomy of DS is on chromosome 21, the chromosome that is also the locus for APP.
A number of studies from our laboratory and others showed that the single methionine residue at position 35 in Aβ (1–42) play a critical role in inducing OS associated with this neurotoxic peptide (67, 70, 105). In other reactions involving oxidation of the Met35 formation of methionine sulfoxide (MetO) occurs, which can be reduced back to methionine by MetO reductase. The activity of MetO reductase is reduced in AD brain (161).
AD brain, CSF, and plasma demonstrate increased levels of OS in AD (27, 69, 75, 278). In AD brain, increased OS has been well documented with markers for protein, DNA, and RNA oxidation as well as lipid peroxidation (7, 72, 74). Protein oxidation is indexed in the AD brain by an increase in carbonylated, protein-bound HNE, and 3-NT-modified proteins (74, 368). However, the initiating event leading to AD pathogenesis has not been determined, though it has become evident that OS is implicated in the development of AD (7, 69, 75).
1. PCO in AD
PCO levels were reported to be elevated in AD brain (69, 74, 114, 190, 310, 352). We showed (190) 42% and 37% increased PCO content in the Alzheimer's hippocampus and inferior parietal lobule (IPL), respectively, relative to these brain regions in control and to AD cerebellum (CB), whereas carbonyl content in controls was comparable in these three brain regions. Others found that brain carbonyl levels were increased with age (352). Smith and collaborators (353) observed a strong PCO signal in NFTs, neuronal cell bodies, and apical dendrites as well as neuronal and glial nuclei in hippocampal sections of AD brains. In the frontal cortex of subjects with the Swedish APP670/671 FAD mutation, increased levels of PCO, diene conjugates, and lipid peroxides compared with sporadic AD were found (43). Further, the levels of carbonyl reductase (CR) protein are increased in brain of AD and DS subjects (26), suggesting enzyme induction due to increased levels of PCO. The authors suggested a possible role of Aβ in this induction. However, this group did neither measure the activity of CR nor identify the mechanism by which the increased CR levels occur. We speculate that, in addition to induction of CR, oxidative modification of this protein or oxidative dysfunction of the 20S proteasome might lead to increased accumulation of protein oxidation. The levels of PCO were found to be significantly increased in synaptic and nonsynaptic mitochondria in the frontal cortex of AD (18). A recent study from our laboratory showed increased levels of PCO in the mitochondria isolated from AD lymphocytes (367).
A number of oxidatively modified proteins have been detected in AD brain and plasma. By using redox proteomics, our laboratory first identified the specific targets of carbonylation in AD IPL (91, 92). After this study, a number of other targets of oxidation have been reported from our laboratory in different brain regions, and these studies also showed that oxidatively modified proteins are prone to inactivation (80, 368, 369). Brain from subjects with amnestic mild cognitive impairment (MCI) showed increased levels of PCO compared with the age-matched controls (10, 65, 76, 222). Further, global OS measurements revealed significantly higher levels of PCO in the MCI IPL relative to preclinical AD (PCAD) (and controls), despite equal levels of neuropathology (10).
2. Identification of carbonylated proteins in brain of subjects with AD
a. Sample: the brain
Human postmortem brain tissue is of high importance for the study of human diseases of the CNS. However, several factors may interfere with tissue and molecular preservation of brain samples obtained from brain banks. Some factors are related with premortem events such as prolonged agonal state, hypoxia, acidosis, fever, and seizures. Others are related with long postmortem delay between death and sample processing for storage or fixation, temperature, characteristics of the fixative solutions, and processing of frozen material. Finally, a third group of factors seems unpredictable and concerns unexpected variations from case to case or from region to region from the same brain, despite similar premortem and postmortem conditions.
All these factors are of major concern, because they may interfere with molecular studies and lead to erroneous conclusions and special care should be taken to consider these circumstances when dealing with human postmortem brain tissue for research. In particular, brain protein preservation largely depends on the postmortem interval (PMI) and the postmortem temperature of storage.
The Alzheimer's Disease Center Brain Bank at the University of Kentucky has prolonged and robust experience and operates under detailed guidelines that conform to the National Institute on Aging/National Institutes of Health “Biospecimen Best Practice Guidelines for Alzheimer's Disease Centers.” All brain samples used for most of the studies from our laboratory reported in this article were obtained with low PMI (<4 h), thus ensuring proper protein preservation for redox proteomics studies. Due to loss of structural and biochemical integrity and increased likelihood of oxidation, any time longer than 4 h PMI almost surely will cause confounds in interpretation of redox proteomics results.
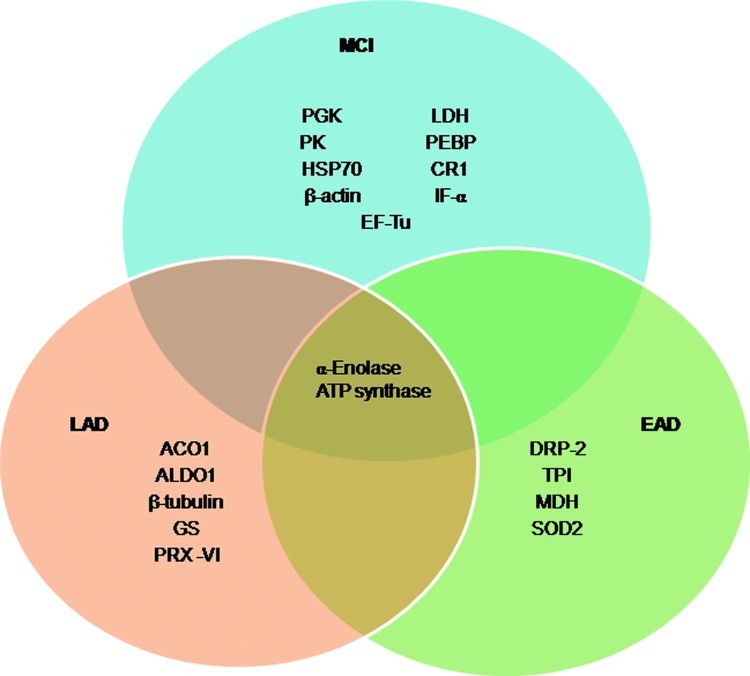
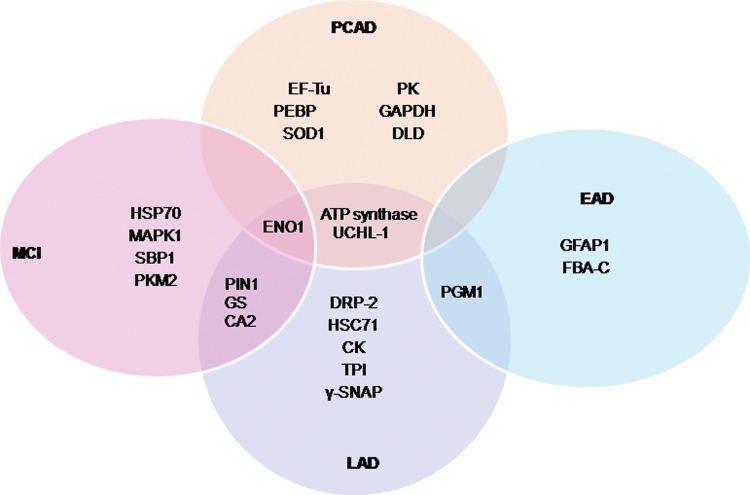
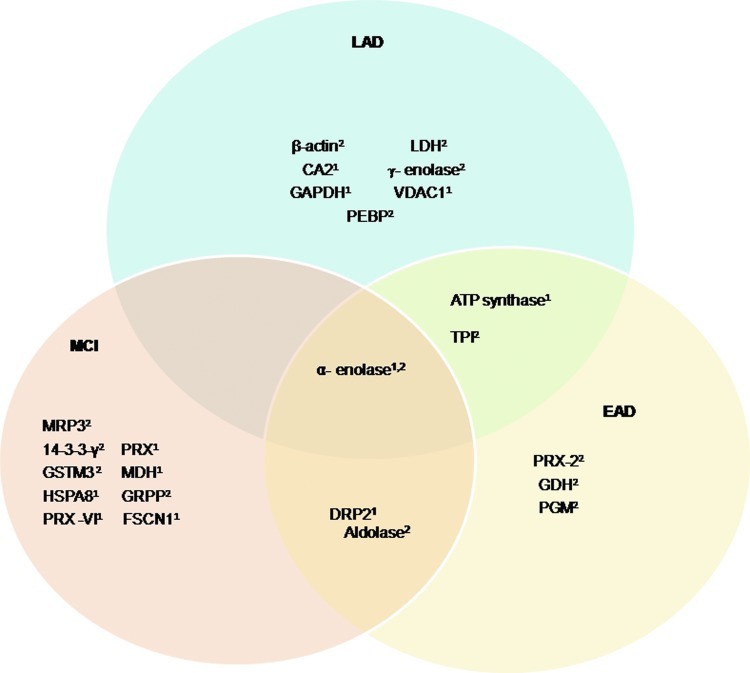
Using a redox proteomics approach, we reported specific carbonylation of the following brain proteins in subjects with AD: alpha-enolase (Eno1), UCH-L1, dihydropyrimidinase-related protein 2 (DRP2, also designated collapsin response mediator protein 2 [CRMP2]), heat shock cognate 71, creatine kinase BB (CK), peptidyl prolyl cis-trans isomerase 1 (Pin1), phosphoglycerate mutase 1 (PGM1), glutamine synthetase (GS), triosephosphate isomerase (TPI), ATP synthase alpha chain (α-ATP synthase), gamma-soluble N-ethylmaleimide sensitive factor (NSF) attachment proteins (γ-SNAP), and carbonic anhydrase 2 (68, 74, 77). These data support the notion that protein carbonylation perturbs energy metabolism, pH regulation, Aβ production, tau hyperphosphorylation, and mitochondrial functions. These proteins and their functions are discussed in detail next.
b. Energy dysfunction
Decreased ATP production could eventually lead to cellular impairment. Using a redox proteomics approach, we identified, compared with control brain, CK, Eno1, TPI, PGM1, and α-ATP synthase as carbonylated energy-related proteins. All these proteins are involved directly or indirectly in the production of ATP in brains (74), and the oxidative modification of glycolytic enzymes likely leads to their inactivation. For example, CK, enolase, PGM1, GAPDH, and ATPase activities are reportedly diminished in AD brain (198, 270, 363). Glucose is the primary source of energy for the brain, which, though having a relatively small mass as a percentage of body mass, accounts for 20% of glucose metabolism and more than 30% of oxygen consumption. Glucose metabolism is essential for proper brain function; a minimum interruption of glucose metabolism causes brain dysfunction and memory loss (270). PET scanning shows a consistent pattern of reduced cerebral glucose utilization in AD brain (198).
Eno1 catalyzes the penultimate step of glycolysis by converting 2-phosphoglycerate to phosphoenolpyruvate. This glycolytic enzymes demonstrates increased oxidation in AD and models of AD (50, 74, 77). Lowered enzymatic activity of enolase has been previously established in the brain of subjects with MCI (76) and subjects with AD (271, 363). Carbonylation of this protein supports the hypothesis of altered energy metabolism as a common theme in neurodegenerative disease. ATP, the cell's energy currency, is extremely important at nerve terminals for normal neurotransmission. Decreased levels of cellular ATP at nerve terminals may lead to loss of synapses and synaptic function, both of which can affect propagation of action potentials and contribute to memory loss in exhibited by AD and MCI patients.
Although the main function of enolase is its role in glycolysis, it has also been shown to play a role in plasminogen regulation and activation of the MEK/extracellular regulated kinases (ERK) pro-survival pathways (73, 358). Plasminogen undergoes proteolysis by tissue-type plasminogen activator (TPA) and converted to its active form, plasmin. TPA is brain specific, and plasmin enhances the degradation of Aβ; however, if TPA is not regulated by oxidatively inhibited enolase, Aβ degradation is lessened (73, 240). Aβ aggregation is observed primarily in SPs; therefore, plasminogen can influence Aβ degradation and enolase regulation. Protein modification of Eno1 may disrupt neuronal energy metabolism and ion homeostasis, thereby impairing ion-motive ATPases, signal transduction, membrane asymmetry (23), and glucose and glutamate transporters (238). Such metabolic and oxidative compromise, known to exist in AD (219, 238, 269), may thereby render neurons susceptible to excitotoxicity and apoptosis. The oxidative modification of energy-related proteins correlates with the altered energy metabolism reported in brain in MCI and AD, which can contribute to neurodegeneration (165, 272). These results support the notion that energy metabolism is a key feature in the progression of AD pathogenesis. Since glycolysis is the main source of ATP production in brain, impairment of glycolysis may lead to shortage of ATP in brains, thus to cellular dysfunction (74, 271). Moreover, decreased ATP shortage can also induce hypothermia, causing abnormal tau phosphorylation through differential inhibition of kinases and phosphatases (311).
CK catalyzes the conversion of creatine to phosphocreatine at the expense of ATP, which is later used in the production of high-energy phosphate used for generation of ATP. Immunochemical approaches in Alzheimer's superior and medial temporal gyri have previously identified CK as a carbonylated protein. Further, using a redox proteomics approach, this protein was also found to be carbonylated in the inferior parietal region of AD brain compared with that of the age-matched control (91). Moreover, CK activity is reported to be diminished in AD brain (8). Loss of its activity in AD (117), resulting from its oxidation (91), suggests decrease energetics in neurons and synaptic elements, consequently in impaired brain function in AD.
ATP synthase goes through a sequence of coordinated conformational changes of its α and β subunits to produce ATP. ATP synthase δ subunit is located on the exterior column of the enzyme. It is one component of the F0 subunit of ATP synthase. With a sufficient proton gradient, the rotor of this mitochondrial complex moves so that ADP and Pi bind in a tight conformation and produce ATP. The rotor then moves 120° counterclockwise to the open position, thereby releasing ATP into the cell. Reduced levels of ATP strengthen the rationale that energy metabolism is altered in AD (160). ATP synthase has been previously shown to be oxidatively modified in late-stage AD (304). The oxidation of ATP synthase leads to the inactivation of this mitochondrial complex. Failure of ATP synthase could contribute to a decrease in the activity of the entire ETC and impaired ATP production, resulting in possible electron leakage and increased ROS production, suggesting an alternate rationale for the OS seen in AD (10, 74). Altered expression of mitochondrial proteins, functional deficits, and lowered activity in different complexes of the ETC are observed in AD (184, 281). These changes, coupled with the changes in complex I, III, and IV, may cause electron leakage from the mitochondria to produce ROS. This action may also affect the proton gradient and overall mitochondrial function, which suggests a complementary mechanism for the acknowledged existence of OS in AD (365).
TPI isomerizes dihydroxyacetonephosphate to glyceraldehyde-3-phosphate (G3P) in glycolysis. This reaction is imperative for the continuation of glycolysis and the overall production of ATP. As just noted, ATP is essential in maintaining ATPases, ion-motive pumps, and potential gradients. In the AD brain, TPI is oxidatively modified as shown by our group in late-stage AD (363), but there is no significant reduction in its activity in AD (271).
PGM1 is a glycolytic enzyme that catalyzes the interconversion of 3-phosphoglycerate to 2-phosphoglycerate. Due to its involvement in glycolytic pathway, carbonylation, and reported decreased expression and activity of PGM1 in the AD brain compared with the age-matched controls are consistent with the loss of total cellular energetics in AD (362). Taken together, oxidative inactivation of these ATP-related enzymes may be related to known metabolic defects in AD detected by PET scanning (309, 317).
c. Excitotoxicity
Oxidative modification of GS led to structural alteration of GS and a reduced activity (72). Since GS catalyzes the rapid amination of glutamate to form glutamine, oxidative modification of GS could lead to impairment of the glutamate-glutamine cycle in AD brains, thereby leading to elevated extracellular levels of glutamate (72). Impairment of this important cycle may contribute to the glutamate dysregulation in AD brains (44) followed by an influx of Ca2+ and activation of NMDA and AMPA receptors that cause neuronal excitotoxic death (266). Moreover, alteration in GS activity have consequences on neuronal pH due to the potential accumulation of ammonia, providing another possible mechanism for neuronal degeneration. Lastly, since GS is essential for amino acid and nucleotide synthesis, oxidative dysfunction of GS can lead to important negative sequelae for brain metabolism.
d. Proteosomal dysfunction
UCH-L1 belongs to a family of UCHL that play important roles in the ubiquitin–proteolytic pathway involved in protein degradation of altered proteins and has been implicated in many neurodegenerative diseases (103, 186). UCH-L1 was found to be carbonylated protein in AD brain or by Aβ(1–42) (50, 91, 103). Loss of activity of UCH-L1 in the AD brain is consistent with the observed increased protein ubiquitinylation, decreased proteasome activity, and accumulation of damaged proteins in AD brains (69). Loss of UCH-L1 function causes neuroaxonal dystrophy (329), significant protein oxidation, and accumulation of synuclein in gracile axonal dystrophy mice (395). Thus, oxidative inactivation of UCH-L1 possibly contributes to both protein aggregation and OS observed in AD brain. Moreover, UCH-L1 oxidative dysfunction could affect activity of the 26S proteasome, which is known to be altered in AD (221). Thus, these pathophysiological observations in AD brain may be related to oxidized UCH-L1: brain protein with excess ubiquitinylation, decreased activity of the 26S proteaome, and consequent accumulation of aggregated, damaged proteins. Oxidative damage of UCH-L1 was also identified in familial AD by redox proteomics and accompanied by reduced enzyme activity (368), further confirming that oxidative modification generally impairs protein functionality.
e. Neuritic abnormalities
DRP2, also known as CRMP2, is critical to neuroplasticity for memory consolidation (236). DRP2 plays an important role in maintaining microtubule assembly, cellular migration, and cytoskeletal remodeling. DRP2 also interacts with collapsin and regulates dendritic length. DRP-2 has been reported to be associated with NFTs, which may lead to decreased levels of cytosolic DRP-2. This, in turn, would eventually lead to shortened neuritic and axonal growth, thus accelerating neuronal degeneration in AD (408), a classic hallmark of AD pathology. CRMP2 is a calmodulin binding protein, which on binding calmodulin, alters the function of CRMP2 and stimulates calpain mediated proteolysis. In addition to AD, decreased expression of CRMP2 protein also was observed in fetal and adult DS subjects (260, 399). Since memory and learning are associated with synaptic remodeling, oxidative modification and subsequent loss of function of this protein could conceivably be involved in the observed cognitive impairments in MCI and AD (74, 107, 260). Moreover, the decreased function of CRMP2 could be responsible for shortened dendritic length and synapse loss observed in AD (28, 107). Shortened dendritic length would likely lead to less neuronal communication with adjacent neurons that could contribute to memory loss and cognitive decline associated with AD.
f. APP regulation, tau hyperphosphorylation, and cell-cycle regulation
Pin1 is a regulatory protein that recognizes phosphorylated Ser-Pro or phosphorylated Thr-Pro motifs in target proteins. After binding to this motif on the target protein by the WW domain of Pin1, the PPIase active site domain of Pin1 alters the sterochemistry of the Pro residue of the target protein from cis to trans and vice versa, thereby regulating the activity of the target protein (65, 339). Pin1 plays an important role in the cell growth and is required for proper progression through the cell cycle in dividing cells (252, 268). In addition, Pin1 plays an important role in regulating the phosphorylation-dephosphorylation of tau protein, APP, and other proteins such as cyclin dependent kinase-5, etc. In AD brain, Pin1 is found to be colocalized with phosphorylated tau and also shows an inverse relationship to the expression of tau in AD brains (196, 316). Further, Pin1 was also identified by redox proteomics as oxidatively modified protein in AD hippocampus (65, 363). Oxidation and decreased levels and activity of this protein could favor the formation of NFTs, SP, and subsequent synapse or cell loss due to cell arrest (20).
Decreased activity of Pin1 is consistent with increased phosphorylation of Tau protein, which could destabilize the microtubule assembly (59, 116), eventually leading to disruption of the axonal cytoskeleton. Consistent with this notion, Lu and co-workers showed that Pin1 overexpression could restore the function of Tau protein in an AD model (412), suggesting oxidative alteration of Pin 1 could be one of the initial events that trigger tangle formation and oxidative damage in AD brains. Moreover, given that Pin1 regulates APP, it is conceivable that oxidative dysfunction of Pin1 could be associated with two major pathological hallmarks of AD: plaques and NFT.
As just mentioned, Pin1 also regulates the activity of CDK5, a protein that is important in keeping neurons from entering the cell cycle. In postmitotic neurons, entrance into the cell cycle leads to neurons becoming trapped, resulting in apoptosis (65, 412). Hence, oxidative modification of Pin1, identified by redox proteomics (65, 363), could conceivably be related to the observation of elevated cell-cycle protein in the AD brain (282).
g. Synaptic abnormalities and LTP
The synaptosomal protein, γ-SNAP, is a member of SNAPs that play an important role in SNARE complexes for vesicular neurotransmitter release, hormone secretion, and mitochondrial integrity. γ-SNAP's important role in vesicle docking is key for the release of neurotransmitters, which is necessary for proper neuronal communication. The oxidation of γ-SNAP could contribute to the impaired learning and memory observed in AD, as well as to the alteration of synaptic circuitry and AD pathogenesis (267, 334). Loss of synaptic connections is found in many regions of AD brain (334). The strongest correlation with cognitive decline in AD is with the synaptic density (335). Consequently, oxidative dysfuntion of γ-SNAP would be consistent with clinical, pathological, and biochemical changes in AD.
h. pH maintenance
CAII is a Zn2+ metallo-enzyme that catalyzes reversible hydration of carbon dioxide to bicarbonate. CAII shares high (68%) similarity to the mitochondrial counterpart carbonic anhydrase 5a (CA-5a) and 5b (CA-5b), implicating the potential coupling or interaction with each other to function in metabolic processes, cellular transport, gluconeogenesis, and mitochondrial metabolism. CAII regulates cellular pH, CO2, and HCO3− transport, and maintains H2O and electrolyte balance by reversible hydration of CO2. CAII affects synaptic remodeling, consistent with the notion that a deficiency of CAII leads to cognitive defects varying from disabilities to severe mental retardation, suggesting the importance of CAII in cognitive function (351). Though levels of CAII are elevated, CAII activity is diminished in the AD brain (271), likely caused by the oxidative modification of the enzyme. Aβ(1–42) leads to oxidative dysfunction of CAII, which might lead to diminution of the major cellular buffering system in brain, thereby promoting protein aggregation, aggregation of Aβ peptide, and subsequent neurodegeneration (50, 77). A recent study reported elevated levels of CAII in AD plasma (208).
i. Mitochondrial abnormalities
Dysfunction of mitochondria has been reported to alter APP metabolism, enhancing the intraneuronal accumulation of amyloid β-peptide and enhancing the neuronal vulnerability (61). Several other studies indicate that Aβ decreases the activity of mitochondrial respiratory chain complexes (258, 277), and the activity of many of the different mitochondrial enzymes appears to be reduced in AD brain (48, 194). Thus, increasing evidence suggests an important role of mitochondrial dysfunction in the pathogenesis of AD. These altered enzymes could play an important role in mitochondrial dysfunction and cell death. Further, as just noted, the cytosolic accumulation of ATP synthase α-chain with NFTs in AD has been reported (341). Moreover, the identification of ATP synthase alpha as an excessively nitrated protein suggests impaired function and also interactions among the subunits. This, in turn, could lead to reduced activity of F1F0-ATPase (ATP synthase, complex V) that could compromise brain ATP synthesis and induce damaging ROS production, and, if severe, could lead to neuronal death (197).
3. Carbonylated proteins in brain of subjects with amnestic MCI
In brain from subjects with amnestic MCI compared with age-matched controls, CA II, Hsp70, mitogen-activated protein kinase I (MAPKI), syntaxin binding protein I (SBP1), Eno1, GS, pyruvate kinase M2, and Pin1 showed significant increased carbonylation. CA II, Eno1, GS, and Pin1were discussed just now in the context to AD pathology or clinical presentation, and similar considerations apply to amnestic MCI.
Hsp70 is neuroprotective against intracellular Aβ; however, this protein is carbonylated in AD, thereby reducing its cellular protection (264). Several other HSPs have been found to be oxidatively modified in AD (74), including Hsp90 and Hsp60 (123), while Hsp 27 and Hsp 32 levels are elevated in amnestic MCI (123). Impairment of these proteins could contribute to proteasomal overload and dysfunction, observed in AD (221). Aβ-treated synaptosomes show that HSPs are oxidatively modified (50), further illustrating the vulnerability of HSPs to Aβ-induced OS.
Pyruvate kinase catalyzes the final step in glycolysis, the conversion of phosphoenolpyruvate to pyruvate with the concurrent transfer of the phosphate group from phosphoenolpyruvate to ADP, thereby generating ATP. Under aerobic conditions, pyruvate can be transported to the mitochondria, where it is converted to acetyl coenzyme A, the latter entering the tricarboxylic acid (TCA) cycle and further metabolic processes that produce considerably more ATP through oxidative phosphorylation. Anaerobically, pyruvate can be reduced to lactate. Additionally, enzymatic activity is reduced, thus suggesting that oxidative modification leads to loss of protein function. Considerations just given for loss of ATP and altered PET scans in AD also apply to MCI subjects.
SBP1 is a neuron-specific protein that binds strongly to syntaxin 1 and is important for synaptic vesicle exocytosis and neurotransmitter release, a key process for neurotransmission. As just discussed, oxidation leads to loss of function of SBP1, which could impair neurotransmission and subsequently might contribute to loss of neuronal function, eventually leading to loss of memory and cognition and neurodegenerative processes involved in progression of MCI to AD.
Recent studies suggested mitogen-activated protein kinases (MAPKs) as key regulators of the formation of plaques and tau hyperphosphorylation in AD (147). MAPKs pathways transduce intracellular signaling to increase expression of different proteins; dysregulation of MAPK-dependent pathways suggests a systematic disorder of protein translation regulation in MCI brains. ERK activation is present in EAD astroglia, while in more advanced AD, it is associated with neuronal cell bodies and dystrophic neurites around plaques, suggesting that ERK activation in astroglia may be an important early response to the onset of AD pathology (169). More recently, abnormal phosphorylation of tau was reported to correlate with increased activity of ERK1/2 in postmortem AD brains (300). Oxidative modification of MAPKs might make them more prone to phosphorylation or may be an alternative mechanism of their activation, thus initiating signaling cascades, ultimately leading to hyperphosphorylation of tau. Based on the existing literature, we hypothesize that amyloid-induced oxidation of MAPK might contribute to increased phosphorylation of tau in AD, leading to cell death.
4. EAD carbonylated proteins
In EAD, a transitional stage between MCI and AD, three proteins, that is, PGM1, glial fibrillary acidic protein (GFAP), and fructose bisphosphate aldolase C (FBA-C), were identified by redox proteomics as carbonylated brain proteins compared with control (370).
GFAP is an intermediate filament protein that is highly expressed in reactive astrocytes. Increased production of GFAP is a hallmark of astrogliosis in neurodegenerative diseases. GFAP is exclusively found in astrocytes and has been shown to undergo activation in AD (280). The oxidation of proteins in EAD is consistent with the idea that OS-associated inflammation is a key mediator in the progression of AD.
FBA-C is a glycolytic enzyme that catalyzes the conversion of fructose 1,6-bisphosphate into dihydroxyacetone phosphate (DHAP) and G3P. Previous studies showed that the levels and activities of PGM1 and FBA-C are decreased in AD brain (363) as just discussed, and could contribute to decreased brain energetics.
5. PCAD vs. amnestic MCI protein carbonylation in brain
PCAD was just discussed. Eno1 and HSP90 were identified by redox proteomics with increased carbonylation in MCI IPL relative to that in PCAD (10). As just discussed, Eno1 oxidative dysfunction contributes to loss of cellular energetics, loss of activation of pro-survival pathways, and loss of Aβ degradation (73). Given the oxidation of enolase in amnestic MCI and late-stage AD, enolase conceivably might be involved in the progression and pathogenesis of AD. Further, redox proteomic analysis of MCI IPL relative to PCAD IPL identified HSP90 with increased carbonylation. As just discussed, HSPs serve as molecular chaperones and help guide damaged proteins to the proteasome. HSP 90 is critical to suppressing inflammation through degradation of hypoxic-inducing factor 1 alpha (285); oxidation of this protein may contribute to the widespread inflammation in AD/MCI. Lastly, since amnestic MCI patients have memory loss, while PCAD patients have normal cognition, these two redox proteomics-identified oxidatively modified proteins conceivably could be involved in memory loss in amnestic MCI.
6. Protein-bound HNE in brain and progression of Alzheimer's disease
As just discussed, HNE is a reactive product of lipid peroxidation, and this α,β-unsaturated alkenal binds to Cys, His, or Lys residues of proteins, thereby changing the conformation and function of proteins (66, 79, 139, 187, 361). In AD, HNE has been found to be significantly elevated in AD brain, plasma, and CSF (187, 238, 297, 348). PCAD subjects have clinically normal antemortem psychometric scores but brain pathology that meets the neuropathological criteria for AD and exhibit no significant brain cell loss or neuronal atrophy (202). Although no alteration of protein-bound HNE was found in PCAD IPL, increased levels of total HNE and acrolein in hippocampus were reported (51). This section of this comprehensive article deals specifically with the HNE modifications observed in the other three progressive stages of AD: MCI, EAD, and LAD.
In amnestic MCI, several proteins have been identified by redox proteomics as HNE-conjugated in the hippocampus and IPL brain regions. These proteins include Eno1, phosphoglycerate kinase, lactate dehydrogenase B, pyruvate kinase, ATP synthase, neuropolypeptide h3, HSP70, CR1, β-actin, initiation factor alpha, and elongation factor Tu (EF-Tu) (320). Since altered energy metabolism and reduced cholinergic activity are two well-documented hypotheses of AD, the HNE modification of several cholinergic, glycolytic, and ATP generating proteins support the notion of involvement of these pathways in AD.
ATP synthase, Eno1, and pyruvate kinase have been just discussed and also have been found to be oxidatively modified in AD brain. Another glycolytic enzyme that is found to be HNE modified in MCI brain is phosphoglycerate kinase, which catalyzes the dephosphorylation of 1,3-bisphosphoglycerate to 3-phosphoglycerate. This reaction undergoes substrate-level phosphorylation by phosphoryl transfer from 1,3-bisphosphoglycerate to ADP to produce one molecule of ATP. Impairment of this glycolytic enzyme results in decreased energy production and irreversible downstream effects, such as multidrug resistance (132). This result could conceivably be related to the identification of multidrug resistant protein 1 (MRP1) as a protein with elevated HNE binding in AD (364).
Lactate dehydrogenase B anaerobically reduces pyruvate to lactate through lactic acid fermentation using NADH as a cofactor. The NAD+ generated in this process is used in glycolysis to oxidize G3P to 1, 3-bisphosphoglycerate, an important reason for this reaction. Lactate is a substrate for gluconeogenesis and given that glucose is the major supplier of energy to the brain, proper lactate production is crucial (223). Lactate dehydrogenase enzymatic activity is significantly reduced in MCI hippocampus (320), which provides supplemental evidence for the correlation between protein dysfunction and enzyme activity impairment. Dysfunction of this enzyme could yield excess pyruvate and a reduction in the production of glucose.
Actin is a principal protein that plays a central role in maintaining structural integrity, cell morphology, and structure of the plasma membrane. Actin microfilaments play a role in the neuronal membrane cytoskeleton by maintaining the distribution of membrane proteins, and segregating axonal and dendritic proteins (33). In the CNS, actin is distributed widely in neurons, astrocytes, and blood vessels. It is particularly concentrated in growth cones, dendritic spines, and presynaptic terminals. HNE conjugation of actin can lead to loss of membrane cytoskeletal structure, decreased membrane fluidity, and trafficking of synaptic proteins and mitochondria. Moreover, actin is involved in the elongation of the growth cone, and loss of function of actin could play a role in the loss of synapse and neuronal communication documented in AD (267).
CR is an enzyme that reduces carbonyl-containing compounds to their resultant alcohols, thereby reducing the level of PCO. Subsequent malfunction or downregulation of this enzyme would be consistent with increased PCO, which, because of the polarity of the carbonyl moiety, could expose normally buried hydrophobic amino acids to the protein surface, resulting in a disruption of conformation. CR has been shown to reduce the lipid peroxidation product, HNE (129). CR expression is altered in DS and AD patients (26). This enzyme was found to be modified in persons with corticobasal degeneration, a neurological disorder whose symptoms closely mirror that of PD (100). The gene for CR is located in close proximity to the gene for the antioxidant enzyme, Cu/ZnSOD (244). Interestingly, the genes for SOD1, CR, and APP are located on chromosome 21, which is a trisomy in DS patients (154, 232). A potential link between DS and AD by irregular meiotic recombination in chromosome 21 (308) has been postulated. Current research supports a possible relationship among CR, DS, and Aβ in neurodegeneration.
EF-Tu and eukaryotic initiation factor α (eIF-α) are intimately involved in protein synthesis machinery. Human mitochondrial EF-Tu is a nuclear-encoded protein and functions in the translational apparatus of mitochondria. Mammalian EF-Tu acts as a GTPase by hydrolyzing one molecule of GTP for each A site amino-acylated tRNA of the ribosome. As just discussed, mitochondria play pivotal roles in eukaryotic cells in producing cellular energy and essential metabolites as well as in controlling apoptosis by integrating various death signals (294). Mitochondrial protein synthesis inhibition, either by deleting mtDNA or by blocking translation in the organelle, is associated with the impairment of differentiation in different cell types, including neurons (390). Loss of neuronal differentiation can lead to an incomplete development of the neuron, which would result in reduced neurotransmission.
eIF-α, which binds aminoacyl-tRNA to acceptor sites of ribosomes in a GTP-dependent manner (306), is involved in cytoskeletal organization by bundling and binding actin filaments and microtubules. The expression level of eIF-α is regulated in aging, transformation, and growth arrest. Due to eIF-α regulation in differing states of cell life and its key position in protein synthesis and cytoskeletal organization, this protein is an important determinant of cell proliferation and senescence (379). Inhibition of eIF-α promotes apoptosis (306), indicating that eIF-α activity is critical to normal cell function.
Taken together, increased levels of HNE-bound eIF-α and EF-Tu suggest an impairment of protein synthesis machinery, either in cytosol or mitochondria, associated with an impairment of the rate and specificity of ribosome functions. Numerous studies have provided indirect evidence that suggests alterations in protein synthesis may occur in AD (128, 146, 330). The dysfunction of the protein synthesis apparatus, mediated in part by redox proteomics identified oxidatively dysfunctional EF-Tu and eIF-α, could compromise the ability of brain cells to generate the countless factors needed to regulate cell homeostasis, thus contributing to impaired neuronal function and to the development of neuropathology in AD.
Neuropolypeptide h3 is critical for modulation of choline acetyltransferase, an enzyme essential in the synthesis of acetylcholine. The loss of choline acetyltransferase leads to reduced levels of acetylcholine, causing poor neurotransmission (291). NMDA receptors activate the production of this enzyme, and modulation of the NMDA receptor mediates cholinergic deficits (213). AD patients have considerable cholinergic deficits, consistent with dysregulation of acetylcholine levels and loss of cholinergic neurons (328). The oxidative modification of this protein further supports the involvement of cholinergic neurons in AD, an early hypothesis of this disorder (156). Neuropolypeptide h3 undergoes HNE modification in MCI hippocampus and nitration in late-stage AD (68). Neuropolypeptide h3 has several other names including phosphatidylethanolamine binding protein (PEBP), hippocampal cholinergic neurostimulating peptide, and Raf kinase inhibitor protein (RKIP). As a PEBP, PEBP may be important in phospholipid asymmetry. Apoptosis is initiated when phosphatidylserine resides on the outer leaflet of the membrane. Loss of function and changes in conformation of PEBP conceivably could lead to loss of phospholipid asymmetry, a signal for neuronal apoptosis, which further supports the role of PEBP as a parapoptosis inhibitor (359). Loss of PEBP may impact lipid asymmetry, as loss of activity is observed in AD and MCI and mouse models of familial AD (23, 24, 166) and can potentially disrupt cellular homeostasis. PEBP levels are decreased in AD, which promotes amyloid beta accumulation in the Tg2576 transgenic mouse model of AD (166). RAF kinases are serine/threonine protein kinases involved in cell signaling in the mitogen-activated protein cascade and NF-kappa B. RKIP disrupts this signaling pathway by interacting with RAF1-MEK 1/2 and NF-kappa B inducing kinase, causing the inhibition of NF-kappa B activation and regulating apoptosis. As demonstrated by the various functions through its numerous monikers, neuropolypeptide h3 is a highly important protein and oxidative modification is likely detrimental to neurons.
EAD, as just mentioned, is thought to be a transitional stage of AD in which patients exhibit progressive cognitive deficits and display mild dementia on clinical evaluation. Redox proteomics analysis identified two HNE-conjugated proteins in this stage of AD that overlap those in the preceding stage of AD, MCI. These proteins include Eno1 and ATP synthase, which were just discussed. Additionally, triose phosphate isomerase, malate dehydogenase, MnSOD, and DRP2 (CRMP2) undergo HNE conjugation in EAD brain as identified by redox proteomics (322).
Oxidative impairment of mitochondrial resident MnSOD is likely a contributing factor to the mitochondrial dysfunction associated with AD. Activity for MnSOD is significantly reduced in EAD brain and CSF compared with the age-matched control, which is consistent with the concept of mitochondrial dysfunction as a factor in the progression of AD. MnSOD was also found to be nitrated and subsequently inactivated in mice by peroxynitrite (121, 149). Overexpression of SOD2 increases Aβ degradation, while partial deficiency promotes Aβ deposition, thereby likely contributing to cognitive decline observed in a transgenic mouse model of AD (134).
Malate dehydrogenase (MDH) catalyzes the reversible oxidation of malate to oxaloacetate by NAD+ in the TCA cycle. MDH links glycolysis to the ETC by transferring NADH to NADH dehydrogenase (Complex I) through the malate-aspartate shuttle resulting in the production of ATP. In contrast to elevated HNE binding to MDH, MDH levels were increased in AD patients, but the level of protein oxidation of MDH was not significant, which probably highlights a compensatory mechanism in response to OS (234). Activity of MDH increases during aging (58, 293), which further supports the hypothesis of mitochondrial dysfunction in AD.
In late-stage AD, redox proteomics identified four HNE-modified proteins that overlap EAD (Eno1, ATP synthase, MnSOD, and CRMP2) (304, 322). Both Eno1 and ATP synthase are consistently HNE modified in all transitional stages of AD, providing evidence for the altered energy metabolism and mitochondrial dysfunction hypotheses associated in the progression of AD (66, 160, 175). Other HNE-modified brain proteins in AD were identified by redox proteomics (304): Aldolase (ALDO1) cleaves fructose 1,6-bisphosphate and produces the two glycolytic intermediates, G3P and DHAP. Fructose 1,6-bisphosphate is neuroprotective and preserves GSH in cortical neurons during OS conditions (391). ALDO1 catalyzes a critical step, as it generates two substrates that are used to eventually produce 2 molecules of ATP and more in TCA and ETC chain. Consequently, HNE modification results in decreased energy metabolism. Levels of ALDO1 are significantly decreased in AD hippocampus (41) and PD (173). Enzyme activity is reduced (41), and impairment can cause increased levels of fructose 1,6-bisphosphate, inhibition of complete glycolysis, and ATP depletion.
Aconitase catalyzes the isomerization of citrate to isocitrate in the TCA cycle. As an iron-sulfur protein, its Fe-S cluster participates in the hydration—dehydration reaction that occurs. The three cysteine residues in the Fe-S core can undergo Michael addition and form acrolein, HNE, and maldondialdehyde conjugated adducts, thereby increasing lipid peroxidation markers (248, 350). Enzymatic activity of this enzyme is significantly reduced in AD, thereby yielding in protein dysfunction (304). The TCA cycle takes place in the mitochondria; therefore, aconitase impairment results in mitochondrial dysfunction, a common theme of neurodegenerative diseases (405). As just noted, decreased ATP production can lead to voltage-gated channel and ion-motive pump disruption as well as synapse loss, an early event in Alzheimer's disease pathology (335).
α-Tubulin is an isoform of tubulin that alternates with β-tubulin to form a prominent cytoskeletal structure, the microtubule. Microtubules are used to transport cargo (i.e., vesicles and organelles) from the cell body to the periphery and vice versa. On HNE modification, α-tubulin is structurally altered and microtubules depolymerize (162). Therefore, cargo cannot reach their destination and the cytoskeleton is altered (284). This could contribute to the notion that synaptic domains are the first to be damaged in AD neurons (235).
Prxs are a family of antioxidant enzymes that are pivotal in antioxidant defense as just discussed. PRX VI is a 1-Cys Prx that plays a role as a second messenger for growth factors and cytokines. Prx VI, a GPx that exhibits Ca2+-independent phospholipase A2 activity (331), is cytosolic and is expressed in astrocytes and in neurons at low levels (94). In addition, the decrease in the activity of this enzyme may also lead to decreased phospholipase A2 activity. Phospholipase A2 is a target for regulation by Pin1, which as just discussed, has been reported to be downregulated and showed oxidative dysfunction in the AD brain (65, 363). PRX VI has been found to be protective against mitochondrial dysfunction, a feature that pinpoints its effectiveness as an antioxidant (136). PRX VI also plays important roles in cell differentiation and apoptosis, and HNE modification may lead to tau hyperphosphorylation and NFT formation in addition to development of OS.
7. Protein-bound 3-NT in brain and progression of Alzheimer's disease
In the AD brain compared with age-matched controls, enolase, GAPDH, α-ATP synthase, beta actin, CA II, voltage-dependent anion channel protein, TPI, lactate dehydrogenase (LDH), and neuropolypeptide h3 were identified by redox proteomics as nitrated proteins (93, 371). Most of these proteins were also found to be targets of protein carbonylation and HNE-modification (see above), and, hence, not discussed in detail in this section.
GAPDH is known for its functional involvement in glycolysis and, consequently, in energy production; therefore, this nitrated protein has altered activity with consequent decreased glucose metabolism (71). In addition, inhibition of GADPH can lead to accumulation of trioses, with subsequent nonenzymatic conversion to methyl glyoxal (MG), a highly reactive alpha-ketoaldehyde that readily oxidizes proteins, lipids, and other cellular components, leading to further cytotoxicity (172). Further, MG binds to Cys, Lys, and His residues by Michael addition at a faster kinetic rate than does HNE. In addition, GAPDH is known to affect APP and Tau (71). Consequently, this multifunctional protein could have important biochemical, clinical, and pathological sequelae of relevance to AD (71).
8. Nitrated brain proteins in MCI
Eno1, glucose-regulated protein precursor, ALDO1, glutathione-S-transferases (GST) Mu, multidrug resistant protein 3 (MRP3), 14-3-3 protein gamma, MDH, PR VI, DRP-2 (CRMP2), Fascin 1 (FSCN1), and HSPA8 protein were identified as nitrated proteins in MCI by redox proteomics (372). Most of these proteins also were found to be oxidatively modified in AD, and have been discussed pertaining to AD.
The brain proteins that are found nitrated in MCI but not in AD includeGST Mu, MRP3, 14-3-3 protein gamma, and FSCN1. One of the mechanisms for the removal of toxic metabolites from cells is accomplished via GST and MRP proteins. GST conjugates HNE to GSH, resulting in the formation of GS-HNE adducts that are effluxed out of cells via MRP efflux pumps. Hence, oxidation and functional impairment of these proteins would lead to increased accumulation of HNE in the cell and, consequently, in cell death. In the MCI brain, increased levels of protein-bound HNE have been found (78). In the AD brain, GST protein levels and activity were reported to be decreased; in addition, GST was found to be oxidatively modified by HNE (364).
14-3-3-protein gamma is a member of the 14-3-3 protein family. These proteins are involved in a number of cellular functions including signal transduction, protein trafficking and metabolism. In the AD brain (239), CSF (60), and ICV animal model of AD (158), the levels of 14-3-3 proteins are increased, which conceivably could lead to altered binding to two of its normal binding partners, glycogen synthase kinase 3 and tau, and may promote tau phosphorylation and polymerization, conceivably contributing to the formation of tangles and subsequent neurodegeneration in AD.
FSCN1, also known as p55, is a structural protein involved in cell adhesion and cell motility (403). P55 protects cells from OS and is used as a marker for dendritic functionality. FSCN1 was also shown to interact with protein kinase C (16), thereby playing important rules in post-translational protein modification. Impairment of this protein is conceivably related to faulty neurotransmission from the affected dendritic projections, to altered intracellular signaling, and may contribute to the progression of AD.
9. Nitrated proteins in EAD
Protein nitration is increased in EAD subjects compared with age-matched controls (321). In the EAD brain, redox proteomics analysis identified the increased nitration of Prx2, TPI, glutamate dehydrogenase, neuropolypeptide h3, PGM1, H-transporting ATPase, Eno1, and ALDO1 (321). All these proteins were identified as either the target of protein carbonylation or HNE modification and have been just discussed.
In summary, redox proteomics analyses of brain proteins throughout the spectrum of AD have identified proteins whose oxidative dysfunction is consistent with the clinical presentation, pathology, and/or biochemistry of this disorder, demonstrating the power and utility of this technique. Oxidative dysfunction of proteins involved in ATP production, excitotoxicity, detoxification, protein degradation, neuritic abnormalities, and mitochondrial abnormalities are likely involved in neurodegeneration at various stages of this dementing disorder. Taken together, the redox proteomics studies in amnestic MCI, EAD, and late-stage AD identified Eno1 as the common target of protein carbonylation, HNE modification, and nitration between AD, EAD, and MCI, consistent with the notion that this protein should be critical to AD progression and pathogenesis. Figures 13, 14, and 15 show the common targets of protein carbonylation, HNE, modification and nitration, respectively, among AD, MCI, and EAD. The identification of these common targets of protein oxidative modification among different stages of disease is consistent with the concept that losses of function of these proteins are key in the progression and pathogenesis of AD. Continued studies are in progress in our laboratory to understand the role of oxidatively modified proteins in AD pathogenesis.
FIG. 13.
Venn diagram of HNE-modified proteins identified during the progression of AD. Alpha-enolase and ATP synthase are the common targets of oxidative modification between AD, MCI, and EAD, and oxidative modification of these proteins might be key in the progression and pathogenesis of AD. AD, Alzheimer disease; EAD, early AD; MCI, mild cognitive impairment. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
FIG. 14.
Venn diagram of excessively carbonylated proteins throughout the pathogenesis of AD. Enolase, ATP synthase alpha, and UCH-L1 are the common targets of oxidation between AD, MCI, EAD, and PCAD. PCAD, preclinical Alzheimer disease; UCH-L1, ubiquitin carboxy-terminal hydrolase-L1. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
FIG. 15.
Nitrated proteins identified during the progression of AD in the hippocampus1 and IPL2 regions. DRP2 and aldolase are common targets of nitration between MCI and EAD. The identification of α-enolase as the only common target of protein nitration in AD, MCI, and EAD suggest that nitration of enolase might be critical to the progression and pathogenesis of AD. DRP2, dihydropyrimidinase-related protein 2; IPL, inferior parietal lobule. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
B. Parkinson disease
PD, the second most common age-related neurodegenerative disorder and in its late stages, a significant contribution to persons with dementia, is characterized by a decline in motor function in the form of resting tremors, muscle rigidity, akinesia, and bradykinesia. PD has two forms: familial and sporadic, the latter being the more common form. Mutations in α-synuclein, parkin, DJ-1, LRRK2, UCH-L1, and PINK1 contribute to early onset familial PD, while sporadic cases of PD have been demonstrated in both familial and sporadic cases (245). Parkin acts as an E3 ligase that attaches ubiquitin to damaged proteins. The polyubiquitination of these proteins cause them to be tagged for degradation by the proteosome. A component of the E3 ligase-ubiquitin complex, SKP1-A has been found to have substantially lowered levels in the substantia nigra in PD patients (150). SKP1-A deficiency prevents proper cell-cycle propagation and triggers the development of aggregates that can cause cellular apoptosis. Cells with sufficient SKP1-A activity have increased cell survival and dopamine function, thereby establishing a new model to study sporadic PD (265). This neurodegenerative disorder is associated with protein aggregates of α-synuclein, a protein whose main functions involve mitochondria and synaptic vesicle formation (46). These aggregates are the major component of Lewy bodies located primarily in the putamen and substantia nigra, brain regions closely associated with motor movement. Recently, aggregates of oxidatively modified α-synuclein have been exhibited in the substantia nigra of patients before the appearance of PD (148). Consistent with increased aggregation of oxidized proteins, recent studies suggest α-synuclein exists as a tetramer in PD rather than as a monomer or smaller aggregates in the normal state (30). The chief neurotransmitter involved in motor function, dopamine, is dramatically lost in neurons, causing substantial neuronal death. HNE was also found to alter dopamine transport (279), contributing to dopamine loss, which is paramount to disease pathogenesis. In addition, to the presence of Lewy bodies, genetic mutations in the proteasome-relevant genes, parkin and UCH-L1, may be associated with the disease. Oxidative damage is well known in PD brains (9, 153, 407) and has been associated with the overexpression of wild-type or mutant (-synuclein (295). Increased oxidative damage in several metabolic enzymes including glyceraldehyde 3-phosphate dehydrogenase, ALDO1, Eno1, and SOD have been observed in sporadic cases of PD, contributing to the hypothesis of reduced glucose metabolism in neurodegeneration(148). Similarly, iron imbalance is exhibited in PD and may be a consequence of the elevation of OS and changes in iron binding proteins as just discussed (12). Moreover, OS in PD has been linked to cell death in PD brains by mitochondrial dysfunction, excitotoxicity, and the toxic effects of nitric oxide (209).
1. Redox proteomics in PD
Although OS has been well documented in PD, a few redox proteomics experiments involving human subjects have been completed. However, significant research using animal models of PD has been completed and will be discussed in this section. By shifting focus to the brain region most severely affected in this disease, the substantia nigra, two classic proteins, α-enolase and β-actin, were identified as being oxidatively modified in a hemiparkinsonian animal model (120). Different animal models of PD are known; however, redox poteomics studies so far were conducted only in A30P (-synuclein transgenic mice. These mice develop an age-dependent accumulation of (-synuclein in neurons of the brain stem (168, 215), suggesting (-synuclein aggregation-associated OS is involved in the pathology in A30P (-synuclein transgenic mice.
Using redox proteomics, several significantly oxidatively modified brain stem proteins were identified in symptomatic mice with overexpression of a A30P mutation in (-synuclein compared with the brain proteins from the nontransgenic mice (313). These proteins were identified as carbonic anhydrase (CA-II), Eno1, and lactate dehydrogenase 2 (LDH2) (313). The activities of these enzymes were also significantly decreased in the A30P (-synuclein transgenic mice brains when compared with the brain proteins from nontransgenic control. This observation is consistent with the notion that oxidative modification of proteins leads to loss of their activities (72, 238).
Each of these proteins has been just discussed in the section on AD. Oxidative inactivation of LDH may contribute to mitochondrial dysfunction in PD patients. γ-Enolase, one of the subunits of functional enolase and neuronal-specific, was identified in an intermembrane space/outer mitochondrial membrane fraction. Hence, oxidative inactivation of enolase may alter normal glycolysis and mitochondrial function in brains, and may contribute to the alteration of energy metabolism in PD. Consistent with this notion, LDH2, enolase, and CA II are associated with mitochondrial function. Increasing data implicate mitochondrial dysfunction and oxidation in PD (209, 333, 343). Furthermore, MPTP and rotenone lead to complex I dysfunction with increased oxidative modification of proteins and (-synuclein aggregation (343). 6-hydroxydopamine, a neurotoxin, deletes GSH, a potent antioxidant, in brain striatum (305), causing striatal neurodegeneration in vivo (273). GAPDH, a critical enzyme in glycolysis and a protein with many other important functions (71), is also oxidized in sporadic PD (185). Moreover, DJ-1, PINK1, and parkin all appear to modulate mitochondrial function (85, 298, 386). The observation that each of the redox proteomics identified oxidatively modified brain proteins in A30P mutant synuclein mice is associated with mitochondria provides strong evidence of mitochondrial dysfunction and aggregated synuclein as a key player in PD pathogenesis. This implication suggests that OS-mediated mitochondrial dysfunction may be responsible, at least partially, for neurodegeneration in the brains of A30P (-synuclein transgenic mice. However, a proteomic analysis showed that dopamine quinone, an oxidized and damaging form of dopamine, could alter brain mitochondria, with implications for PD (388).
C. Amyotrophic lateral sclerosis
ALS is a progressive neurodegenerative disorder that affects motor neurons of the cerebral cortex, brainstem, and the anterior horn of the spinal cord (106). The majority of ALS clinical presentations are sporadic (sALS), with fewer than 10% of ALS cases inherited in an autosomal dominant manner, that is, familial ALS (fALS). Both sALS and fALS are clinically indistinguishable and show similar features. The molecular mechanisms responsible for disease pathogenesis and progression are still unknown; however, studies have shown that patients with fALS have mutations in copper/zinc (Cu/Zn) superoxide dismutase (SOD1), a relevant component of the antioxidant defense system (254). More than 100 SOD1 mutations have been identified in fALS patients (13), most of which result from substitution of one single amino acid, such as SOD1G85R, SOD1G37R, and SOD1G93A. It is now well established that SOD1-mediated toxicity in ALS is due to a “gain” of toxic properties that are independent of SOD1 activity (145, 178). To explain the toxicity of ALS mutant SOD1 (mSOD1) proteins, two hypotheses have been proposed. The first hypothesis states that mSOD1 proteins are or become misfolded and consequently oligomerize to form intracellular aggregates (106, 387), which also include other essential proteins that are, therefore, no longer available to perform their correct function. The second—the oxidative damage hypothesis—proposes that toxicity is caused by aberrant chemistry of the active Cu/Zn sites of the misfolded enzyme (38), which contributes to further exacerbate OS conditions by increasing the levels of ROS within the cell (38, 251). This latter mechanism can lead to misfolding of Cu, Zn SOD (315). Elevated levels of ROS and the formation of insoluble protein complexes of mSOD1 protein have been shown in spinal cords of the G93A transgenic mice and preceed motor neuron degeneration (253). It is reasonable to assume that these two phenomena (protein aggregation and OS) are linked, as oxidative damage to the SOD1 was demonstrated in the G93A-SOD1 transgenic mice. Interestingly, recent studies (47) showed that misfolded/oxidized wild-type SOD-1 (wtSOD1) gained toxic functions similar to mSOD1. After mild treatment with hydrogen peroxide or another oxidizing reagent, wtSOD1 becomes more susceptible to misfolding (47). The aberrant chemistry after oxidant insult induces alteration of wtSOD1 dimer conformation, which, in turn, may dispel the Cu/Zn ions and dissociate into monomeric units. Further, misfolded wtSOD1 showed many properties that were thought to be characteristic of the mutant protein such as ubiquitination, association with chaperones, insolubility, and formation of protein aggregates. In addition, misfolded/oxidized wtSOD1 may be released outside the cell where it can induce other molecular pathways that lead to motor neurons degeneration, as it does mSOD1 (47). Iron dysregulation has been observed in both forms of ALS. Although modifications in proteins associated with iron transport and cellular iron sensing were demonstrated in a human SOD1 expressing mouse, the mechanisms for the pathogenesis of ALS by iron dysregulation remain unclear (210). All these data contribute to further validate the hypothesis that oxidative damage, including mSOD itself, plays a central role in the pathogenesis and progression of ALS.
Multiple pathological studies have reported evidences of increased OS in ALS postmortem tissue compared with control samples (29). Elevated PCO levels have been shown in the spinal cord (342) and motor cortex (144) from sALS cases, and increased 3-NT levels were found within spinal cord motor neurons in both SOD1–fALS and fALS patients (2). Immunoreactivity to the brain and endothelial forms of NOS, but not the inducible form, was also elevated in ALS motor neurons relative to controls (3), suggesting alterations in RNS as well as ROS. Markers for lipid peroxidation were detected in spinal cord from sALS patients, but were absent in control spinal cords (344), and levels of 8-hydroxy-2′-deoxyguanosine (8-OHdG), a marker of oxidized DNA, were elevated in whole cervical spinal cord from ALS subjects (151) and were most prominent within the ventral horn (144).
Other studies were performed in CSF from ALS patients to measure OS markers. Elevated levels of 8-OHdG (205) and HNE (355) were observed in CSF samples from ALS patients. Transgenic mouse and cell culture models of ALS showed a similar pattern of oxidation that was confirmed by the findings of increased levels of oxidative damage to protein, lipid, and DNA observed in the human disease (15, 145). An increase in central nervous tissue PCO content have also been reported in both fALS (49) and sALS (342) subjects.
1. Redox proteomics studies in ALS transgenic mice
Studies from our laboratory and others applied redox proteomics approaches to identify selective protein modifications in the spinal cord of G93A-SOD1 transgenic mice in comparison with wild-type mice (302). Perluigi et al. (302) identified proteins that were significantly modified by HNE in the spinal cord tissue of a model of fALS, G93A-SOD1 transgenic mice, including DRP-2 (CRMP2), Hsp70, and, possibly, Eno1. As previously noted in this article, CRMP2 is a member of the DRP gene family involved in axonal outgrowth and pathfinding through the transmission and modulation of extracellular signals (174). Immunoreactivity of human CRMP2 was shown in the NFTs of AD human brain, suggesting that CRMP2 plays a role in neuritic degeneration characteristic of AD (408). Dysfunction of the CRMP2-repairing activity in brain indicates that depletion of CRMP2 may result in neuronal abnormalities, thus accelerating the neuritic degeneration in many neurodegenerative disorders. The finding of increased oxidative modification of CRMP2 in G93A-SOD1 transgenic mice provided a potential link between oxidation-mediated loss in protein function and neuritic regeneration and plasticity known to be altered in ALS (182).
mSOD1 is aggregated with Hsp70, Hsp40, and α-crystallin in transfected cells (345). As just noted, Hsp70 is a chaperone protein that helps newly synthesized proteins to be folded and transported across the membrane (84). Pathologically, human SOD1-immunoreactive inclusions in the spinal cord of ALS patients and of transgenic mice are frequently stained with antibody against heat-shock cognate Hsc70 (398). Moreover, overexpression of Hsp70 leads to reduction of protein aggregates and enhanced viability of G93A-SOD1 overexpressed motor neurons (57). A recent study proposed a potential anti-ALS drug candidate, which had the ability to induce the heat shock response (225), suggesting that Hsp70 could play a role in the folding of SOD1 and prevent aggregate formation. We suggested that diminished degradation of mSOD1 is possibly due to inactivation of Hsp70, which is impaired by covalent binding by HNE. These data suggest that mechanisms regulating Hsp70 chaperone activity could play a crucial role in the pathophysiology of motor neurons disease, in particular in the context of mutations of SOD1.
By redox proteomics, our laboratory also identified the proteins that showed increased carbonyl levels in the spinal cord of G93A-SOD1 transgenic mice compared with those of wild-type mice: SOD1, translationally controlled tumor protein (TCTP), UCH-L1, and αB-crystallin (314).
Researchers (15) previously identified immunochemically SOD1 as one of the oxidatively modified proteins in G93A-SOD1 transgenic mouse spinal cord. Although G93A-SOD1 showed dismutation activity identical to that of wtSOD1, the activity of SOD1 in fALS patients with mutations reportedly is decreased 50% in motor cortex, parietal cortex, and CB (54). Moreover, free radical production in the G93A-SOD1 transgenic animals is induced by SOD1 mutation (15), alteration of tumor necrosis factor α (TNF-α), and TNF-α-modulating cytokines (189). Another oxidatively modified protein in transgenic mice identified by redox proteomics was TCTP. TCTP possesses calcium-binding activity (44) and has a tubulin binding region (226). TCTP levels are highly regulated in response to various stress conditions and extracellular signals, and similar to chaperones and other antiapoptotic protein (378), may exert a cytoprotective function for cells. Once oxidatively modified, the putative cytoprotective function and the calcium binding affinity of TCTP possibly are impaired in G93A-SOD1 mice. Consistent with this notion, free cytosolic calcium was increased in lymphocytes from ALS patients (111).
UCH-L1 has been just discussed. This protein belongs to a family of UCHL that play important roles in the ubiquitin–proteolytic pathway involved in protein degradation (395). Thus, oxidative inactivation of UCH-L1 possibly contributes to both protein aggregation and OS observed in G93A-SOD1 transgenic mice and ALS patients. Oxidative damage of UCH-L1 was also accompanied by reduced enzyme activity, thus further confirming that oxidative modification impairs protein functionality.
αB-Crystallin is a member of the small heat shock protein (sHSP) family hat are synthesized under stress conditions as well as normal conditions. The major function of sHSP is to stabilize other proteins under stress conditions, whereas the high-molecular-weight HSPs usually play roles in protein folding during biosynthesis (163). Moreover, α-crystallins were recruited to aggregates when cells were treated with a proteasome inhibitor, and the degradation of αB-crystallin, along with ubiquitin conjugation, was decreased in bovine lens epithelial cells when αB-crystallin was oxidized (200). Consistent with this notion, inclusions in ALS patients contain αB-crystallin, metallothionein, GS, and tubulin immunoreactivities (3).
Casoni et al. (89) showed increased nitration of α- and γ-enolase, ATP synthase beta chain, and heat shock cognate 71-kDa protein and actin in presymptomatic FALS mice using a redox proteomics approach. Further, these researchers showed that the nitration occurs at 16 sites in proteins oxidized. One of the sites of Eno1 nitration at Tyr(43) is also a target of phosphorylation, suggesting the nitration of the protein may affect the function of the proteins as discussed earlier. In addition, a study by Basso et al. (32) showed that nitration of the proteins play an important role in the aggregation of proteins, suggesting that oxidation/nitration has a key role in aggregation. All these data together may indicate that multiple oxidative modifications extensively and simultaneously affect certain proteins, inducing misfolding and finally aggregation. It appears that oxidation and protein aggregation should not be considered separate events, but they are both involved in the formation of insoluble, toxic proteinaceous inclusions, which represent a characteristic feature of ALS and many age-related neurodegenerative diseases.
The oxidatively modified proteins identified by redox proteomics from our laboratory (314) play a significant role in protein aggregation processes in the spinal cords of G93A-SOD1 transgenic mice. Indeed, ubiquitin protein epitopes and αB-crystallin were found in fibrillar neuronal inclusions in the cortex of sporadic ALS patients (21, 243). This article from our laboratory led to several commentary papers, all of which supported our hypothesis that aggregation and OS in ALS should be viewed as a continuum and not as separate processes (81, 113, 140, 314).
D. Huntington disease
HD is a progressive autosomal dominant disorder caused by expansion of CAG trinucleotide repeats in exon 1 of the huntingtin gene on chromosome 4 that encodes a stretch of polyglutamines (poly(Q)) in the N-terminus of the Htt protein (36, 356). Clinical symptoms of HD that typically manifest in midlife generally include psychiatric abnormalities, most commonly depression and mood disturbances, and involuntary choreiform movements and dementia that develop over a period of 15–20 years. Neuropathologically, the disease is characterized by bilateral striatal atrophy with marked neuronal loss and astrogliosis within the caudate and putamen. Htt is a widely expressed 350 kDa protein, and its specific role is not completely elucidated; Htt is implicated in vesicle trafficking in the endosome/lysosome pathway (42) and in the regulation in cortical cells of the production of brain-derived neurotrophic factor, a pro-survival factor for striatal neurons (130, 307). Importantly, wild-type Htt is believed to have a pro-survival role in the cell. The antiapoptotic function of wtHtt has been demonstrated in several in vitro studies (104, 409). These results demonstrated that expression of the full-length protein protected conditionally immortalized striatal-derived cells from a variety of apoptotic stimuli. WtHtt appeared to act downstream of mitochondrial cytochrome c release, preventing the formation of a functional apoptosome complex and the consequent activation of caspase-9.
Conversely, the N-terminal fragments of mutant Htt accumulate in the nuclei of affected neurons and form intranuclear aggregates (126). Indeed, the formation of mHtt aggregates is regarded as a hallmark of HD (336). Transgenic mice expressing the N-terminal fragment of Htt with 82 CAG repeats develop a progressive neurological disorder. In this mouse model, the presence of pathological poly(Q) results in the formation of both intranuclear and cytoplasmic ubiquitinated aggregates containing the protease-resistant mutated N-terminal Htt fragment in neurons of affected areas. mHtt is targeted for proteolysis but is resistant to removal.
Several hypothesis have been proposed to explain the pathogenesis of disease, including excitotoxicity (99), mitochondrial dysfunction (52), and impaired energy metabolism. Numerous evidences suggest a role of oxidative damage in HD brains (302, 342), and, consistent with this suggestion, antioxidant supplementation appeared to slow the progression of animal models of the disease (14). Among different mechanisms proposed, one of the most supported hypothesis of neuronal loss is linked to mitochondria dysfunction (53). mHtt-mediated mitochondrial alterations would be expected to affect oxidative damage in cells, and, indeed, there is evidence of elevated ROS generation and oxidative damage markers in HD postmortem brain tissue, animal models, and HD cell lines (53).
Emerging evidence points to the fact that mutant Htt may directly interact with neuronal mitochondria, consequently leading to their degeneration (189, 296). Functional changes in mitochondria caused by mHtt have been recently confirmed by the demonstration that poly(Q) can affect mitochondrial calcium handling. Mitochondrial Ca2+ homeostasis is compromised in HD due to altered opening of the permeability transition pore (299). Mitochondrial/energetic defects occur as a primary event in HD. Reduced ATP production can result in partial cell depolarization by making neurons more vulnerable to endogenous levels of glutamate (53). The concomitant increase of Ca+2 influx into neurons may trigger further free radical production, exacerbating oxidative damage to cellular components. Iron dysregulation is evident in HD brain. Ferritin, an iron binding protein, is significantly lowered in HD patients, causing a reduction in iron binding capacity and contributing to an imblance in iron homeostasis (45). Consistent with severe mitochondrial defects and impaired energy metabolism, biochemical studies in HD postmortem tissue have revealed alterations in the activity of several key enzymes of oxidative phosphorylation and the TCA cycle in brain regions targeted in HD. Activities of complexes II, III, and IV of the ETC are markedly and selectively reduced in caudate and putamen of advanced HD patients (55). One of the most profound defects detected in HD to date is the dramatic reduction in activity of aconitase in affected brain regions and muscle. The particular susceptibility of mitochondrial aconitase to oxidative damage may be related to the iron-sulfur cluster [4Fe-4S] in its active site (164). Studies in vitro established that aconitase is particularly sensitive to reaction with superoxide, which causes release of one iron atom from the cluster (152). Inactivation of aconitase may block normal electron flow to oxygen, leading to an accumulation of reduced metabolites such as NADH.
1. Redox proteomics-transgenic mouse model of HD
Our laboratory used proteomics to investigate the expression of proteins and their oxidative modification in the striatum from R6/2 transgenic mice, one of the most widely used models of HD (303). The protein expression levels of Eno1, dihydrolipoamide S-succinyltransferase, pyruvate dehydrogenase, and aspartate aminotransferase were significantly changed in 10-week-old transgenic mice compared either with their age-matched control or with 4-week-old transgenic mice. In addition, redox proteomics analysis showed that the specific PCO levels of α- and γ-enolase isoforms, aconitase, CK, HSP90, and VDAC-1 were significantly increased in the same sets of comparison. Our study indicated loss of activity caused by oxidative modification of the enzymes involved in glucose metabolism, such as α- and γ-enolase, pyruvate dehydrogenase, and aconitase, and of CK might lead to a reduced ATP production, consistent with the observations in the HD patients. In addition, loss of energy production by mitochondria leads to decreased production of ATP and, thus, disruption of cellular functions that depend on ATP, including metabolic intermediate synthesis and maintaining of ionic gradients as just noted. Taken together, these data point to the fact that bioenergetic defects are a profound feature of HD pathology. The finding of increased carbonyl levels and decreased activity of CK further determines which crucial enzymes are finely involved in energetic impairments in this disorder. The CK system, consisting of a cytosolic and a mitochondrial isoform (MtCK) together with their substrates creatine and phosphocreatine, is one of the most important immediate energy buffering and transport systems of the cell, especially in muscle and neuronal tissue (394). CKs are prime targets of oxidative damage (74), leading to inactivation of both isoforms. Consistent with this notion, we showed that oxidative modifications of CK decrease its activity during aging and neurodegenerative diseases (6). Recent reports have demonstrated that creatine therapy provides neuroprotection and delays motor symptoms in the transgenic animal model of HD (404), thus suggesting enhancement of cerebral energy metabolism as a protective mechanism against neurodegeneration.
Formation of neuronal inclusions with aggregated Htt is associated with the progressive neuropathology in HD. Accumulation of misfolded proteins is one of the major causes of neurodegenerative disorders like AD, ALS, and HD. Neuronal cells recognize the aggregated Htt protein as abnormally folded and, by recruiting molecular chaperones and proteasomal components, try to disaggregate and/or degrade the mutant protein. Conversely, the toxicity of mHtt may reduce the availability of HSPs, thereby disrupting their normal chaperone and anti-apoptotic functions and reducing their cytoprotective effects. Consistent with this view, we found increased carbonyl levels of HSP90 by redox proteomics in an ALS mouse model (314). We proposed that diminished degradation of mutant aggregated Htt is possibly related to inactivation of HSP90, which once oxidatively modified is not able to facilitate misfolded protein degradation by the proteasome. As just reported, others (225) reported the efficacy of a treatment with a co-inducer of HSPs to delay disease progression in a transgenic mouse model of ALS. Pharmacological activation of the HSP response conceivably could be a successful therapeutic approach for treating neurodegenerative disorders.
The slow, progressive nature of neuronal injury in chronic neurodegenerative disorders may be explained by cycling of free radicals and mitochondrial dysfunction. Thus, the identification of target proteins that are oxidatively modified may provide a crucial insight into the etiologic role of oxidative damage in mechanisms of neuronal death in HD and other neurodegenerative disorders. Loss of activity of these proteins by oxidative modification or by altered expression may contribute to abnormal metabolism and neurochemical changes ultimately leading to neuronal death.
2. Proteomics of HD brain
In agreement with the findings reported on transgenic mice, a proteomic analysis of human brain postmortem samples obtained from striatum and cortex of subjects with HD compared with samples of age- and sex-matched controls was performed (357). Antioxidant defense proteins that were strongly induced in striatum, but also detectable in cortex, were identified as Prxs I, II, and VI, as well as GPxs 1 and 6. The activities of other antioxidant enzymes such as MnSOD and catalase were also increased in HD. Aconitase showed decreased activities in striatum of HD patients. PCO were increased in HD, and GFAP, aconitase, γ-enolase, and creatine kinase B were identified as the main oxidative targets. Taken together, these results indicate that OS and damage to specific macromolecules would participate in the disease progression in human as well as animal models of the disease.
Based on the evidence that bioenergetic metabolism is implicated in the pathogenesis of HD, a previous multicenter, randomized, double-blind, placebo-controlled trial of coenzyme Q10 (CoQ10), given alone or in combination with remacemide hydrochloride, demonstrated beneficial effects (201). However, experimental results did not reach statistical significance, which might reflect choices of dosage. In fact, CoQ10 also has shown promise in other neurodegenerative conditions, such as PD, but at higher dosages than were used in HD trial (346). Therefore, current clinical trials to examine the safety and tolerability of higher dosages of CoQ10 in HD patients and healthy subjects are now in progress.
E. Down syndrome
DS, also called trisomy 21, is associated with neurodegeneration. After reaching 40–45 years of age DS patients develop a form of dementia that has almost identical clinical and neuropathologic characteristics of AD. There is considerable literature supporting a major role of OS in DS clinical phenotype (95, 203). Increased oxidative damage is demonstrated by oxidative DNA damage (urinary 8-OHdG), lipid peroxidation, and isoprostane 8,12-iso-iPF2alpha-VI levels (214), indicating a “pro-oxidant state,” which associated with overwhelmed antioxidant defenses—both enzymatic and nonenzymatic—constitute a clue to understanding the complexity of the DS phenotype. Indeed, an abnormal expression of genes located on chromosome 21, in association with responses to environmental stimuli, might alter the expression of disomic genes as well. OS is known to occur in DS from very early stages: already during embryonic development mitochondrial dysfunction has been reported as a marker of oxidative damage (19). Moreover, amniotic fluid from mothers carrying a DS fetus has oxidatively modified proteins as observed by redox proteomics (301).
The cause of DS is to be found in the genes expressed on chromosome 21, which when present with an extra copy lead to the overxpression of related protein products. Among these, the excess activity of SOD-1 is due to a third copy of its gene. This increased activity results in the accumulation of hydrogen peroxide that might reach toxic levels and might be related not only to the neuronal death observed in DS but also involved in the impairment of other functions. In addition, the overexpression of the APP gene expressed on chromosome 21 is likely related to the overproduction of Aβ(1–42) peptide, the major protein in the SPs, which is considered one of the important factors leading to the development of the AD pathology in DS subjects. Aβ(1–42) peptide has been found in the brain of children with DS as young as age 8 years, and the deposits increase with age. Interestingly, although are extensive deposits in the brain, there is no linear relationship with AD. There is a gap between the presence of abnormal brain pathology and the early signs of AD, suggesting that other factors (genetic or environmental) may play an important role in the development of AD.
Among these, accumulation of ROS causes abnormal lipid peroxidation metabolism, which leads to structural damage to membranes and the generation of more toxic products. ROS-related activity also leads to DNA damage. All these findings lead to the concept that OS might play an important role in the development of AD in persons with DS; however, OS alone does not explain the whole process. Elevated OS has been demonstrated in the brains of DS patients, as indexed by increased levels of TBARS, total PCO and AGEs in the cortex from DS fetal brain compared with controls (290) and a marked accumulation of 8-hydroxyguanosine (8OHG), oxidized protein, NT, in the cytoplasm of cerebral neurons in DS (288). Ishihara et al. (207) have recently demonstrated an increased level of ROS and mitochondrial dysfunction in primary cultured astrocytes and neurons from DS transgenic mice—Ts1Cje, suggesting that the gene-dosage hypothesis is sufficient to explain, at least the major part, the OS phenomena observed in this in vitro model of the disease.
1. Redox proteomics in DS transgenic mice
Ishihara et al. (207) identified by a redox proteomics approach the putative target proteins that were modified by lipid-peroxidation-derived products. ATP synthase mitochondrial F1 complex b subunit, Eno1, and TPI1 were identified as proteins modified by 3-hydroperoxy-9Z,11E-octadecadienoic acid. Neurofilament light polypeptide, internexin neuronal intermediate filamentα, neuron specific enolase, Prx6, phosphoglycerate kinase 1, and TPI were shown to be HNE-modified proteins. Dysfunction of these proteins impairs ATP generation, the neuronal cytoskeleton system, and anti-oxidant enzyme functionality. Some of these proteins have been previously identified as target proteins for HNE-modification (303, 320). Thus, these proteins appear to be common targets for lipid peroxidation-derived products in senescence and various OS-related disorders, and might play a central role in the degenerative process associated with oxidative damage in OS-related disorders, including DS.
V. Conclusions and Future Directions
Redox proteomic analysis of oxidatively modified proteins in AD, PD, HD, and ALS showed that the proteins involved in glucose metabolism, mitochondrial function, structural, and protein degradation are commonly affected in these neurodegenerative diseases (Fig. 16). These studies suggest that there might be a common mechanism by which neurodegeneration occurrs in different diseases. One of the common observations is the presence of OS and involvement of protein aggregates. Further, studies are needed to tease out detailed relationship of these observations.
FIG. 16.
Redox proteomics determined functional pathways in different neurodegenerative disorders. A comparative analysis of the functional pathways involved in the redox proteomics-identified brain proteins from AD, PD, HD, and ALS showed that the proteins involved in glucose metabolism, mitochondrial function, cellular structure, and protein degradation are affected in common in these neurodenerative diseases. HD, Huntington disease; PD, Parkinson disease. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars.)
With the increasing average life span of humans, age-related neurodegenerative disorders are expected to be a major health concern in our society. Many groups have demonstrated the role of OS in the pathogenesis and progression of neurodegenerative diseases including AD, PD, ALS, HD, and DS, among others. Proteins are one of the major targets of oxidative damage, and it has now become clear that chemical modifications induced by ROS affect both conformational and functional integrity of target proteins and lead, in most cases, to their dysfunction. In order to better understand the biological effects of such modifications, redox proteomics is an emerging tool that can provide powerful insights which can be used for further investigations. Data obtained from redox proteomics studies highlighted a number of common proteins and/or functional categories that are primarily affected in different neurodegenerative disorders. These alterations involve energy production, mitochondrial functions, neuritic abnormalities, proteasome, detoxification, excitotoxicity, and synapse function. Based on these findings, it is reasonable to assume that neurodegeneration is driven, at least in part, by the impairment of the pathways just mentioned. However other mechanisms, apart from OS, may play a role and explain, for example, the selective vulnerability of neural systems and diversity of clinical manifestations in the different neurodegenerative disease discussed.
New data need to be added to obtain a comprehensive OS signature of brain disorders. In fact, some diseases have not been fully investigated and as evidenced in the section related to disease, there is a lack of redox proteomics data on both human samples and animal models thereof. Further insight also can be obtained, for example, through the characterization of other oxidative modifications and advancement of proteomics techniques. A consideration that emerges from this comprehensive article of redox proteomics in some neurodegenerative disorders is the need to identify additional “specific” markers to the list currently available that will have the power to discriminate between different diseases, to better understand the specific neurodegenerative mechanisms involved, to identify therapeutic targets, and eventually be useful for early diagnosis of each disorder separately.
Consequently, the search for important information on biomarkers has centered on investigating CSF/blood composition in addition to what can be observed in the brain, because many proteins are specific to neuronal cells and cannot be found systemically. Currently, a definite diagnosis of AD can only be made by postmortem neuropathological examination, but brain tissue is inappropriate for early diagnosis of cognitive decline. Growing studies aim at identifying putative body fluid biomarkers for early diagnosis and stage progression of AD with particular attention to blood-derived markers. So far, CSF biomarkers reflecting the amyloid cascade hypothesis and cytoskeletal degeneration (Aβ, total tau, and phosphorylated-tau) have been found to be promising and reliable biomarkers for AD. CSF represents the most suitable biological fluid that studies neurodegenerative diseases, as it can reflect the biochemical changes occurring in the brain, but its analysis is not always easily feasible for large-scale screening, because of the costs involved, and because the invasive nature of lumbar puncture is uncomfortable and not without risk. Recently, neuroimaging through the use of MRI or PET is gaining high interest for the possibility to test several promising markers, as suggested by the funding of the Alzheimer's Disease Neuroimaging Initiative project. Concerns on the use of these diagnostic methods involve the cost and availability of the instruments of analysis, which impede the routine use of these techniques for the diagnosis of the asymptomatic early stages of AD.
The application of redox proteins to AD/PD/ALS/HD/DS revealed important targets of brain protein oxidation. The use of animal models together with redox proteomics approaches could provide potential insights into the mechanisms of neurodegeneration in and could also be of value for the development of therapeutic approaches to prevent or delay these neurodegenerative disorders.
In terms of future MS approaches for redox proteomics: due to the low-abundance nature of oxidative modifications, 2D-gel, LC, and MS-based approaches will continue to be developed for the enrichment and detection of these modifications. Many of the methods just described can be applied for the identification and quantitation of oxidatively modified proteins in neurodegenerative diseases. In the near future, it will become important to understand the nature of protein isoforms that arise from variable oxidative modifications to many residues and the contribution of a specific isoform to improper protein function. These questions will rely on developments in top-down proteomics and their coupling to bottom-up proteomics approaches as was recently demonstrated for identification of oxidized calmodulin isoforms in activated macrophages (256).
Very recently, new proteomics platforms have been developed that analyze body fluid and a number of medium/high molecular abundant proteins emerged as potential candidates. Assays are required for the validation of these candidates. Multiple reaction monitoring (MRM)-based approaches are an attractive alternative to ELISAs due to the sensitivity and selectivity of the technique, the capacity to multiplex, and the limited availability of antibodies. In addition, accuracy in the quantitation of analytes by MRM can be improved by combining with tandem mass tags, as this allows the incorporation of an internal reference into the analysis. This approach becomes of high importance for the validation of candidate biomarkers from discovery experiments. What is currently emerging in the field of biomarker research is the fact that only a combination of different markers could, most likely, offer a certain diagnosis and be able to capture all aspects of the disease.
The future of redox proteomics to gain insights into mechanisms, biomarkers, and therapeutic targets of neurodegenerative diseases is bright, and we believe redox proteomics will continue to make valuable contributions to eventual molecular-level understanding of neurodegenerative disorders.
Abbreviations Used
- α-ATP synthase
ATP synthase alpha chain
- γ-SNAP
soluble N-ethylmaleimide sensitive factor (NSF) attachment proteins
- 2D
two-dimensional
- 3AT
3-aminotyrosine
- 3-NT
3-nitrotyrosine
- 8-OHdG
8-hydroxy-2′-deoxyguanosine
- Aβ
amyloid beta-peptide
- AD
Alzheimer disease
- AGE
advanced glycation endproducts
- ALDO1
aldolase
- ALS
amyotrophic lateral sclerosis
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid
- APP
amyloid precursor protein
- BSA
bovine serum albumin
- CB
cerebellum
- CID
collision-induced dissociation
- CK
creatine kinase BB
- CNS
central nervous system
- CP
ceruloplasmin
- CR
carbonyl reductase
- CSF
cerebrospinal fluid
- Cu/ZnSOD
copper/zinc superoxide dismutase
- DHAP
dihydroxyacetone phosphate
- DMT-1
divalent metal transporter-1
- DNPH
2,4-dinitrophenylhydrazine
- DOTA
1,4,7,10-tetraazacyclododecane, N, N′, N′′, N′′′-tetraacetic acid
- DRP2/CRMP2
dihydropyrimidinase-related protein 2/collapsin response mediator protein 2
- DS
Down syndrome
- EAD
early AD
- ECD
electron capture dissociation
- EF-Tu
elongation factor Tu
- eIF-α
eukaryotic initiation factor α
- Eno1
alpha-enolase
- eNOS
endothelial nitric oxide synthase
- ERK
extracellular regulated kinases
- ESI
electrospray ionization
- ETC
electron transport chain
- fALS
familial ALS
- FBA-C
fructose bisphosphate aldolase C
- FITC
fluorescein isothiocyanate
- FSCN1
Fascin 1
- G3P
glyceraldehyde-3-phosphate
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- GPx
glutathione peroxidase
- GR
glutathione reductase
- GS
glutamine synthetase
- GSH
glutathione
- GSSG
oxidized glutathione
- GST
glutathione-S-transferase
- HD
Huntington disease
- HNE
4-hydroxy-2-trans-nonenal
- HSP
heat shock protein
- Htt
huntingtin
- IEF
isoelectric point
- iNOS
inducible nitric oxide synthase
- IPL
inferior parietal lobule
- iTRAQ
the isobaric tag for relative and absolute quantitation
- LC
liquid chromatography
- LDH
lactate dehydrogenase
- LDH2
lactate dehydrogenase 2
- MALDI
matrix assisted laser desorption ionization
- MAPK
mitogen-activated protein kinases
- MAPKI
mitogen-activated protein kinase I
- MCI
mild cognitive impairment
- MDH
malate dehydrogenase
- MetO
methionine sulfoxide
- MG
methyl glyoxal
- MnSOD
manganese superoxide dismutase
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- MRM
multiple reaction monitoring
- MRP1
multidrug resistant protein 1
- MRP3
multidrug resistant protein 3
- MS
mass spectrometry
- NFT
neurofibrillary tangles
- NMDA
N-methyl D-aspartic acid
- nNOS
neuronal nitric oxide synthase
- NOS
nitric oxide synthase
- NTA
Ni2+-nitrotyrosine affinity
- O-ECAT
oxidation-dependent element coded affinity tags
- OS
oxidative stress
- PAGE
polyacrylamide gel electrophoresis
- PCAD
preclinical Alzheimer disease
- PCO
protein carbonyl
- PD
Parkinson disease
- PEBP
phosphatidylethanolamine binding protein
- PGM1
phosphoglycerate mutase 1
- Pin1
peptidyl prolyl cis-trans isomerase 1
- PMI
postmortem interval
- Prx
peroxiredoxin
- PRX VI
peroxiredoxin VI
- RKIP
Raf kinase inhibitor protein
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RP
reverse-phase
- sALS
sporadic ALS
- SBP1
syntaxin binding protein I
- SCX
strong cation exchange
- sHSP
small heat shock protein
- SOD
superoxide dismutase
- SOD1
Cu/Zn superoxide dismutase
- SOD2
Mn superoxide dismutase
- SP
senile plaque
- TBP-2
Trx-binding protein-2
- TCA
tricarboxylic acid
- TCTP
translationally controlled tumor protein
- TF
transferrin
- TNF-α
tumor necrosis factor-α
- TPA
tissue plasminogen activator
- TPI
triosephosphate isomerase
- Trx
thioredoxin
- UCH-L1
ubiquitin carboxy-terminal hydrolase 1
- VDUP1
vitamin D3 upregulated protein 1
- wtSOD1
wild-type SOD-1
Acknowledgments
This work was supported, in part, by an NIH grant to D.A.B. [AG-05119]. The authors thank past and current postdoctoral scholars, graduate students, and visiting scientists in the Butterfield laboratory and their collaborators for their contributions to their redox proteomics studies in neurodegenerative disorders.
References
- 1.Abboud K. Bassila JC. Ghali-Ghoul R. Sabra R. Temporal changes in vascular reactivity in early diabetes mellitus in rats: role of changes in endothelial factors and in phosphodiesterase activity. Am J Physiol Heart Circ Physiol. 2009;297:H836–H845. doi: 10.1152/ajpheart.00102.2009. [DOI] [PubMed] [Google Scholar]
- 2.Abe K. Pan LH. Watanabe M. Kato T. Itoyama Y. Induction of nitrotyrosine-like immunoreactivity in the lower motor neuron of amyotrophic lateral sclerosis. Neurosci Lett. 1995;199:152–154. doi: 10.1016/0304-3940(95)12039-7. [DOI] [PubMed] [Google Scholar]
- 3.Abe K. Pan LH. Watanabe M. Konno H. Kato T. Itoyama Y. Upregulation of protein-tyrosine nitration in the anterior horn cells of amyotrophic lateral sclerosis. Neurol Res. 1997;19:124–128. doi: 10.1080/01616412.1997.11740784. [DOI] [PubMed] [Google Scholar]
- 4.Abello N. Barroso B. Kerstjens HA. Postma DS. Bischoff R. Chemical labeling and enrichment of nitrotyrosine-containing peptides. Talanta. 2010;80:1503–1512. doi: 10.1016/j.talanta.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Adibhatla RM. Hatcher JF. Lipid oxidation and peroxidation in CNS health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2010;12:125–169. doi: 10.1089/ars.2009.2668. [DOI] [PubMed] [Google Scholar]
- 6.Aksenov M. Aksenova M. Butterfield DA. Markesbery WR. Oxidative modification of creatine kinase BB in Alzheimer's disease brain. J Neurochem. 2000;74:2520–2527. doi: 10.1046/j.1471-4159.2000.0742520.x. [DOI] [PubMed] [Google Scholar]
- 7.Aksenov MY. Tucker HM. Nair P. Aksenova MV. Butterfield DA. Estus S. Markesbery WR. The expression of key oxidative stress-handling genes in different brain regions in Alzheimer's disease. J Mol Neurosci. 1998;11:151–164. doi: 10.1385/JMN:11:2:151. [DOI] [PubMed] [Google Scholar]
- 8.Aksenova MV. Burbaeva G. BB creatine kinase isoenzyme activity in the blood serum of patients with senile dementia, Alzheimer's disease and schizophrenia. Zh Nevropatol Psikhiatr Im S S Korsakova. 1989;89:113–116. [PubMed] [Google Scholar]
- 9.Alam ZI. Jenner A. Daniel SE. Lees AJ. Cairns N. Marsden CD. Jenner P. Halliwell B. Oxidative DNA damage in the parkinsonian brain: an apparent selective increase in 8-hydroxyguanine levels in substantia nigra. J Neurochem. 1997;69:1196–1203. doi: 10.1046/j.1471-4159.1997.69031196.x. [DOI] [PubMed] [Google Scholar]
- 10.Aluise CD. Robinson RA. Cai J. Pierce WM. Markesbery WR. Butterfield DA. Redox proteomics analysis of brains from subjects with amnestic mild cognitive impairment compared to brains from subjects with preclinical Alzheimer's disease: insights into memory loss in MCI. J Alzheimers Dis. 2011;23:257–269. doi: 10.3233/JAD-2010-101083. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez B. Radi R. Peroxynitrite reactivity with amino acids and proteins. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 12.Andersen JK. Iron dysregulation and Parkinson's disease. J Alzheimers Dis. 2004;6:S47–S52. doi: 10.3233/jad-2004-6s602. [DOI] [PubMed] [Google Scholar]
- 13.Andersen PM. Genetic factors in the early diagnosis of ALS. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(Suppl 1):S31–S42. doi: 10.1080/14660820052415899. [DOI] [PubMed] [Google Scholar]
- 14.Andreassen OA. Dedeoglu A. Ferrante RJ. Jenkins BG. Ferrante KL. Thomas M. Friedlich A. Browne SE. Schilling G. Borchelt DR. Hersch SM. Ross CA. Beal MF. Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington's disease. Neurobiol Dis. 2001;8:479–491. doi: 10.1006/nbdi.2001.0406. [DOI] [PubMed] [Google Scholar]
- 15.Andrus PK. Fleck TJ. Gurney ME. Hall ED. Protein oxidative damage in a transgenic mouse model of familial amyotrophic lateral sclerosis. J Neurochem. 1998;71:2041–2048. doi: 10.1046/j.1471-4159.1998.71052041.x. [DOI] [PubMed] [Google Scholar]
- 16.Anilkumar N. Parsons M. Monk R. Ng T. Adams JC. Interaction of fascin and protein kinase C alpha: a novel intersection in cell adhesion and motility. EMBO J. 2003;22:5390–5402. doi: 10.1093/emboj/cdg521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansari MA. Joshi G. Huang Q. Opii WO. Abdul HM. Sultana R. Butterfield DA. In vivo administration of D609 leads to protection of subsequently isolated gerbil brain mitochondria subjected to in vitro oxidative stress induced by amyloid beta-peptide and other oxidative stressors: relevance to Alzheimer's disease and other oxidative stress-related neurodegenerative disorders. Free Radic Biol Med. 2006;41:1694–1703. doi: 10.1016/j.freeradbiomed.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansari MA. Scheff SW. Oxidative stress in the progression of Alzheimer disease in the frontal cortex. J Neuropathol Exp Neurol. 2010;69:155–167. doi: 10.1097/NEN.0b013e3181cb5af4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arbuzova S. Hutchin T. Cuckle H. Mitochondrial dysfunction and Down's syndrome. Bioessays. 2002;24:681–684. doi: 10.1002/bies.10138. [DOI] [PubMed] [Google Scholar]
- 20.Arendt T. Synaptic plasticity and cell cycle activation in neurons are alternative effector pathways: the “Dr. Jekyll and Mr. Hyde concept” of Alzheimer's disease or the yin and yang of neuroplasticity. Prog Neurobiol. 2003;71:83–248. doi: 10.1016/j.pneurobio.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Arima K. Ogawa M. Sunohara N. Nishio T. Shimomura Y. Hirai S. Eto K. Immunohistochemical and ultrastructural characterization of ubiquitinated eosinophilic fibrillary neuronal inclusions in sporadic amyotrophic lateral sclerosis. Acta Neuropathol. 1998;96:75–85. doi: 10.1007/s004010050862. [DOI] [PubMed] [Google Scholar]
- 22.Aulak KS. Koeck T. Crabb JW. Stuehr DJ. Proteomic method for identification of tyrosine-nitrated proteins. Methods Mol Biol. 2004;279:151–165. doi: 10.1385/1-59259-807-2:151. [DOI] [PubMed] [Google Scholar]
- 23.Bader Lange ML. Cenini G. Piroddi M. Abdul HM. Sultana R. Galli F. Memo M. Butterfield DA. Loss of phospholipid asymmetry and elevated brain apoptotic protein levels in subjects with amnestic mild cognitive impairment and Alzheimer disease. Neurobiol Dis. 2008;29:456–464. doi: 10.1016/j.nbd.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bader Lange ML. St. Clair D. Markesbery WR. Studzinski CM. Murphy MP. Butterfield DA. Age-related loss of phospholipid asymmetry in APP(NLh)/APP(NLh) x PS-1(P264L)/PS-1(P264L) human double mutant knock-in mice: relevance to Alzheimer disease. Neurobiol Dis. 2010;38:104–115. doi: 10.1016/j.nbd.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baillet A. Chanteperdrix V. Trocme C. Casez P. Garrel C. Besson G. The role of oxidative stress in amyotrophic lateral sclerosis and Parkinson's disease. Neurochem Res. 2010;35:1530–1537. doi: 10.1007/s11064-010-0212-5. [DOI] [PubMed] [Google Scholar]
- 26.Balcz B. Kirchner L. Cairns N. Fountoulakis M. Lubec G. Increased brain protein levels of carbonyl reductase and alcohol dehydrogenase in Down syndrome and Alzheimer's disease. J Neural Transm Suppl. 2001;(61):193–201. doi: 10.1007/978-3-7091-6262-0_15. [DOI] [PubMed] [Google Scholar]
- 27.Baldeiras I. Santana I. Proenca MT. Garrucho MH. Pascoal R. Rodrigues A. Duro D. Oliveira CR. Peripheral oxidative damage in mild cognitive impairment and mild Alzheimer's disease. J Alzheimers Dis. 2008;15:117–128. doi: 10.3233/jad-2008-15110. [DOI] [PubMed] [Google Scholar]
- 28.Baloyannis SJ. Costa V. Mauroudis I. Psaroulis D. Manolides SL. Manolides LS. Dendritic and spinal pathology in the acoustic cortex in Alzheimer's disease: morphological and morphometric estimation by Golgi technique and electron microscopy. Acta Otolaryngol. 2007;127:351–354. doi: 10.1080/00016480601126986. [DOI] [PubMed] [Google Scholar]
- 29.Barber SC. Shaw PJ. Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. Free Radic Biol Med. 2010;48:629–641. doi: 10.1016/j.freeradbiomed.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Bartels T. Choi JG. Selkoe DJ. Alpha-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basso M. Giraudo S. Corpillo D. Bergamasco B. Lopiano L. Fasano M. Proteome analysis of human substantia nigra in Parkinson's disease. Proteomics. 2004;4:3943–3952. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- 32.Basso M. Samengo G. Nardo G. Massignan T. D'Alessandro G. Tartari S. Cantoni L. Marino M. Cheroni C. De Biasi S. Giordana MT. Strong MJ. Estevez AG. Salmona M. Bendotti C. Bonetto V. Characterization of detergent-insoluble proteins in ALS indicates a causal link between nitrative stress and aggregation in pathogenesis. PLoS One. 2009;4:e8130. doi: 10.1371/journal.pone.0008130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Battaini F. Pascale A. Lucchi L. Pasinetti GM. Govoni S. Protein kinase C anchoring deficit in postmortem brains of Alzheimer's disease patients. Exp Neurol. 1999;159:559–564. doi: 10.1006/exnr.1999.7151. [DOI] [PubMed] [Google Scholar]
- 34.Baty JW. Hampton MB. Winterbourn CC. Detection of oxidant sensitive thiol proteins by fluorescence labeling and two-dimensional electrophoresis. Proteomics. 2002;2:1261–1266. doi: 10.1002/1615-9861(200209)2:9<1261::AID-PROT1261>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 35.Bayir H. Kagan VE. Clark RS. Janesko-Feldman K. Rafikov R. Huang Z. Zhang X. Vagni V. Billiar TR. Kochanek PM. Neuronal NOS-mediated nitration and inactivation of manganese superoxide dismutase in brain after experimental and human brain injury. J Neurochem. 2007;101:168–181. doi: 10.1111/j.1471-4159.2006.04353.x. [DOI] [PubMed] [Google Scholar]
- 36.Beal MF. Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington's disease. Nat Rev Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- 37.Beasley A. Anderson C. McArthur J. Sacktor N. Nath A. Cotter JR. Characterization of nitrotyrosine-modified proteins in cerebrospinal fluid. Clin Proteomics. 2010;6:29–41. [Google Scholar]
- 38.Beckman JS. Chen J. Crow JP. Ye YZ. Reactions of nitric oxide, superoxide and peroxynitrite with superoxide dismutase in neurodegeneration. Prog Brain Res. 1994;103:371–380. doi: 10.1016/s0079-6123(08)61151-6. [DOI] [PubMed] [Google Scholar]
- 39.Berg D. Youdim MB. Role of iron in neurodegenerative disorders. Top Magn Reson Imaging. 2006;17:5–17. doi: 10.1097/01.rmr.0000245461.90406.ad. [DOI] [PubMed] [Google Scholar]
- 40.Berlett BS. Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 41.Bigl M. Bruckner MK. Arendt T. Bigl V. Eschrich K. Activities of key glycolytic enzymes in the brains of patients with Alzheimer's disease. J Neural Trans. 1999;106:499–511. doi: 10.1007/s007020050174. [DOI] [PubMed] [Google Scholar]
- 42.Bogdanov MB. Andreassen OA. Dedeoglu A. Ferrante RJ. Beal MF. Increased oxidative damage to DNA in a transgenic mouse model of Huntington's disease. J Neurochem. 2001;79:1246–1249. doi: 10.1046/j.1471-4159.2001.00689.x. [DOI] [PubMed] [Google Scholar]
- 43.Bogdanovic N. Zilmer M. Zilmer K. Rehema A. Karelson E. The Swedish APP670/671 Alzheimer's disease mutation: the first evidence for strikingly increased oxidative injury in the temporal inferior cortex. Dement Geriatr Cogn Disord. 2001;12:364–370. doi: 10.1159/000051282. [DOI] [PubMed] [Google Scholar]
- 44.Bommer UA. Borovjagin AV. Greagg MA. Jeffrey IW. Russell P. Laing KG. Lee M. Clemens MJ. The mRNA of the translationally controlled tumor protein P23/TCTP is a highly structured RNA, which activates the dsRNA-dependent protein kinase PKR. RNA. 2002;8:478–496. doi: 10.1017/s1355838202022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonilla E. Estevez J. Suarez H. Morales LM. Chacin de Bonilla L. Villalobos R. Davila JO. Serum ferritin deficiency in Huntington's disease patients. Neurosci Lett. 1991;129:22–24. doi: 10.1016/0304-3940(91)90711-2. [DOI] [PubMed] [Google Scholar]
- 46.Bonini NM. Giasson BI. Snaring the function of alpha-synuclein. Cell. 2005;123:359–361. doi: 10.1016/j.cell.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 47.Bosco DA. Morfini G. Karabacak NM. Song Y. Gros-Louis F. Pasinelli P. Goolsby H. Fontaine BA. Lemay N. McKenna-Yasek D. Frosch MP. Agar JN. Julien JP. Brady ST. Brown RH., Jr. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosetti F. Brizzi F. Barogi S. Mancuso M. Siciliano G. Tendi EA. Murri L. Rapoport SI. Solaini G. Cytochrome c oxidase and mitochondrial F1F0-ATPase (ATP synthase) activities in platelets and brain from patients with Alzheimer's disease. Neurobiol Aging. 2002;23:371–376. doi: 10.1016/s0197-4580(01)00314-1. [DOI] [PubMed] [Google Scholar]
- 49.Bowling AC. Schulz JB. Brown RH., Jr. Beal MF. Superoxide dismutase activity, oxidative damage, and mitochondrial energy metabolism in familial and sporadic amyotrophic lateral sclerosis. J Neurochem. 1993;61:2322–2325. doi: 10.1111/j.1471-4159.1993.tb07478.x. [DOI] [PubMed] [Google Scholar]
- 50.Boyd-Kimball D. Castegna A. Sultana R. Poon HF. Petroze R. Lynn BC. Klein JB. Butterfield DA. Proteomic identification of proteins oxidized by Abeta(1–42) in synaptosomes: implications for Alzheimer's disease. Brain Res. 2005;1044:206–215. doi: 10.1016/j.brainres.2005.02.086. [DOI] [PubMed] [Google Scholar]
- 51.Bradley MA. Markesbery WR. Lovell MA. Increased levels of 4-hydroxynonenal and acrolein in the brain in preclinical Alzheimer disease. Free Radic Biol Med. 2010;48:1570–1576. doi: 10.1016/j.freeradbiomed.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Browne SE. Mitochondria and Huntington's disease pathogenesis: insight from genetic and chemical models. Ann N Y Acad Sci. 2008;1147:358–382. doi: 10.1196/annals.1427.018. [DOI] [PubMed] [Google Scholar]
- 53.Browne SE. Beal MF. Oxidative damage in Huntington's disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 54.Browne SE. Bowling AC. Baik MJ. Gurney M. Brown RH., Jr. Beal MF. Metabolic dysfunction in familial, but not sporadic, amyotrophic lateral sclerosis. J Neurochem. 1998;71:281–287. doi: 10.1046/j.1471-4159.1998.71010281.x. [DOI] [PubMed] [Google Scholar]
- 55.Browne SE. Bowling AC. MacGarvey U. Baik MJ. Berger SC. Muqit MM. Bird ED. Beal MF. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 56.Browne SE. Ferrante RJ. Beal MF. Oxidative stress in Huntington's disease. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bruening W. Roy J. Giasson B. Figlewicz DA. Mushynski WE. Durham HD. Up-regulation of protein chaperones preserves viability of cells expressing toxic Cu/Zn-superoxide dismutase mutants associated with amyotrophic lateral sclerosis. J Neurochem. 1999;72:693–699. doi: 10.1046/j.1471-4159.1999.0720693.x. [DOI] [PubMed] [Google Scholar]
- 58.Bubber P. Haroutunian V. Fisch G. Blass JP. Gibson GE. Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann Neurol. 2005;57:695–703. doi: 10.1002/ana.20474. [DOI] [PubMed] [Google Scholar]
- 59.Buee L. Bussiere T. Buee-Scherrer V. Delacourte A. Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 60.Burkhard PR. Sanchez JC. Landis T. Hochstrasser DF. CSF detection of the 14-3-3 protein in unselected patients with dementia. Neurology. 2001;56:1528–1533. doi: 10.1212/wnl.56.11.1528. [DOI] [PubMed] [Google Scholar]
- 61.Busciglio J. Pelsman A. Wong C. Pigino G. Yuan M. Mori H. Yankner BA. Altered metabolism of the amyloid beta precursor protein is associated with mitochondrial dysfunction in Down's syndrome. Neuron. 2002;33:677–688. doi: 10.1016/s0896-6273(02)00604-9. [DOI] [PubMed] [Google Scholar]
- 62.Butterfield D. Castegna A. Pocernich C. Drake J. Scapagnini G. Calabrese V. Nutritional approaches to combat oxidative stress in Alzheimer's disease. J Nutr Biochem. 2002;13:444. doi: 10.1016/s0955-2863(02)00205-x. [DOI] [PubMed] [Google Scholar]
- 63.Butterfield DA. beta-Amyloid-associated free radical oxidative stress and neurotoxicity: implications for Alzheimer's disease. Chem Res Toxicol. 1997;10:495–506. doi: 10.1021/tx960130e. [DOI] [PubMed] [Google Scholar]
- 64.Butterfield DA. Gnjec A. Poon HF. Castegna A. Pierce WM. Klein JB. Martins RN. Redox proteomics identification of oxidatively modified brain proteins in inherited Alzheimer's disease: An initial assessment. J Alzheimers Dis. 2006;10:391–397. doi: 10.3233/jad-2006-10407. [DOI] [PubMed] [Google Scholar]
- 65.Butterfield DA. Abdul HM. Opii W. Newman SF. Joshi G. Ansari MA. Sultana R. Pin1 in Alzheimer's disease. J Neurochem. 2006;98:1697–1706. doi: 10.1111/j.1471-4159.2006.03995.x. [DOI] [PubMed] [Google Scholar]
- 66.Butterfield DA. Bader Lange ML. Sultana R. Involvements of the lipid peroxidation product, HNE, in the pathogenesis and progression of Alzheimer's disease. Biochim Biophys Acta. 2010;1801:924–929. doi: 10.1016/j.bbalip.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butterfield DA. Boyd-Kimball D. The critical role of methionine 35 in Alzheimer's amyloid beta-peptide (1–42)-induced oxidative stress and neurotoxicity. Biochim Biophys Acta. 2005;1703:149–156. doi: 10.1016/j.bbapap.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 68.Butterfield DA. Castegna A. Proteomics for the identification of specifically oxidized proteins in brain: technology and application to the study of neurodegenerative disorders. Amino Acids. 2003;25:419–425. doi: 10.1007/s00726-003-0027-7. [DOI] [PubMed] [Google Scholar]
- 69.Butterfield DA. Drake J. Pocernich C. Castegna A. Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid beta-peptide. Trends Mol Med. 2001;7:548–554. doi: 10.1016/s1471-4914(01)02173-6. [DOI] [PubMed] [Google Scholar]
- 70.Butterfield DA. Galvan V. Lange MB. Tang H. Sowell RA. Spilman P. Fombonne J. Gorostiza O. Zhang J. Sultana R. Bredesen DE. In vivo oxidative stress in brain of Alzheimer disease transgenic mice: requirement for methionine 35 in amyloid beta-peptide of APP. Free Radic Biol Med. 2010;48:136–144. doi: 10.1016/j.freeradbiomed.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Butterfield DA. Hardas SS. Lange ML. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer's disease: many pathways to neurodegeneration. J Alzheimers Dis. 2010;20:369–393. doi: 10.3233/JAD-2010-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Butterfield DA. Hensley K. Cole P. Subramaniam R. Aksenov M. Aksenova M. Bummer PM. Haley BE. Carney JM. Oxidatively induced structural alteration of glutamine synthetase assessed by analysis of spin label incorporation kinetics: relevance to Alzheimer's disease. J Neurochem. 1997;68:2451–2457. doi: 10.1046/j.1471-4159.1997.68062451.x. [DOI] [PubMed] [Google Scholar]
- 73.Butterfield DA. Lange ML. Multifunctional roles of enolase in Alzheimer's disease brain: beyond altered glucose metabolism. J Neurochem. 2009;111:915–933. doi: 10.1111/j.1471-4159.2009.06397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Butterfield DA. Lauderback CM. Lipid peroxidation and protein oxidation in Alzheimer's disease brain: potential causes and consequences involving amyloid beta-peptide-associated free radical oxidative stress. Free Radic Biol Med. 2002;32:1050–1060. doi: 10.1016/s0891-5849(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 75.Butterfield DA. Perluigi M. Sultana R. Oxidative stress in Alzheimer's disease brain: new insights from redox proteomics. Eur J Pharmacol. 2006;545:39–50. doi: 10.1016/j.ejphar.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 76.Butterfield DA. Poon HF. St. Clair D. Keller JN. Pierce WM. Klein JB. Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer's disease. Neurobiol Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 77.Butterfield DA. Reed T. Newman SF. Sultana R. Roles of amyloid beta-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer's disease and mild cognitive impairment. Free Radic Biol Med. 2007;43:658–677. doi: 10.1016/j.freeradbiomed.2007.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butterfield DA. Reed T. Perluigi M. De Marco C. Coccia R. Cini C. Sultana R. Elevated protein-bound levels of the lipid peroxidation product, 4-hydroxy-2-nonenal, in brain from persons with mild cognitive impairment. Neurosci Lett. 2006;397:170–173. doi: 10.1016/j.neulet.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 79.Butterfield DA. Stadtman ER. Protein oxidation processes in aging brain. Adv Cell Aging Gerontol. 1997;2:161–191. [Google Scholar]
- 80.Butterfield DA. Sultana R. Redox proteomics identification of oxidatively modified brain proteins in Alzheimer's disease and mild cognitive impairment: insights into the progression of this dementing disorder. J Alzheimers Dis. 2007;12:61–72. doi: 10.3233/jad-2007-12107. [DOI] [PubMed] [Google Scholar]
- 81.Calabrese V. Highlight Commentary on Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice—a model of familial amyotrophic lateral sclerosis. Free Radic Biol Med. 2007;43:160–162. doi: 10.1016/j.freeradbiomed.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 82.Calabrese V. Mancuso C. Calvani M. Rizzarelli E. Butterfield DA. Stella AM. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 83.Calabrese V. Scapagnini G. Colombrita C. Ravagna A. Pennisi G. Giuffrida Stella AM. Galli F. Butterfield DA. Redox regulation of heat shock protein expression in aging and neurodegenerative disorders associated with oxidative stress: a nutritional approach. Amino Acids. 2003;25:437–444. doi: 10.1007/s00726-003-0048-2. [DOI] [PubMed] [Google Scholar]
- 84.Calabrese V. Scapagnini G. Ravagna A. Colombrita C. Spadaro F. Butterfield DA. Giuffrida Stella AM. Increased expression of heat shock proteins in rat brain during aging: relationship with mitochondrial function and glutathione redox state. Mech Ageing Dev. 2004;125:325–335. doi: 10.1016/j.mad.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 85.Canet-Aviles RM. Wilson MA. Miller DW. Ahmad R. McLendon C. Bandyopadhyay S. Baptista MJ. Ringe D. Petsko GA. Cookson MR. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci U S A. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Caperna TJ. Shannon AE. Blomberg le A. Garrett WM. Ramsay TG. Identification of protein carbonyls in serum of the fetal and neonatal pig. Comp Biochem Physiol B Biochem Mol Biol. 2010;156:189–196. doi: 10.1016/j.cbpb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 87.Casado A. Encarnacion Lopez-Fernandez M. Concepcion Casado M. de La Torre R. Lipid peroxidation and antioxidant enzyme activities in vascular and Alzheimer dementias. Neurochem Res. 2008;33:450–458. doi: 10.1007/s11064-007-9453-3. [DOI] [PubMed] [Google Scholar]
- 88.Casagrande S. Bonetto V. Fratelli M. Gianazza E. Eberini I. Massignan T. Salmona M. Chang G. Holmgren A. Ghezzi P. Glutathionylation of human thioredoxin: a possible crosstalk between the glutathione and thioredoxin systems. Proc Natl Acad Sci U S A. 2002;99:9745–9749. doi: 10.1073/pnas.152168599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Casoni F. Basso M. Massignan T. Gianazza E. Cheroni C. Salmona M. Bendotti C. Bonetto V. Protein nitration in a mouse model of familial amyotrophic lateral sclerosis: possible multifunctional role in the pathogenesis. J Biol Chem. 2005;280:16295–16304. doi: 10.1074/jbc.M413111200. [DOI] [PubMed] [Google Scholar]
- 90.Cassina AM. Hodara R. Souza JM. Thomson L. Castro L. Ischiropoulos H. Freeman BA. Radi R. Cytochrome c nitration by peroxynitrite. J Biol Chem. 2000;275:21409–21415. doi: 10.1074/jbc.M909978199. [DOI] [PubMed] [Google Scholar]
- 91.Castegna A. Aksenov M. Aksenova M. Thongboonkerd V. Klein JB. Pierce WM. Booze R. Markesbery WR. Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 92.Castegna A. Aksenov M. Thongboonkerd V. Klein JB. Pierce WM. Booze R. Markesbery WR. Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 93.Castegna A. Thongboonkerd V. Klein JB. Lynn B. Markesbery WR. Butterfield DA. Proteomic identification of nitrated proteins in Alzheimer's disease brain. J Neurochem. 2003;85:1394–1401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 94.Caudle WM. Pan S. Shi M. Quinn T. Hoekstra J. Beyer RP. Montine TJ. Zhang J. Proteomic identification of proteins in the human brain: towards a more comprehensive understanding of neurodegenerative disease. Proteomics Clin Appl. 2008;2:1484–1497. doi: 10.1002/prca.200800043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cenini G. Dowling ALS. Beckett T. Barone E. Mancuso C. Murphy MP. Levine H., III Schmitt FA. Butterfield DA. Head E. Association between frontal cortex oxidative damage and beta-amyloid neuropathology as a function of age in Down syndrome. Biochem Biophys Acta. 2011;1822:130–138. doi: 10.1016/j.bbadis.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Charkoudian LK. Franz KJ. Fe(III)-coordination properties of neuromelanin components: 5,6-dihydroxyindole and 5,6-dihydroxyindole-2-carboxylic acid. Inorg Chem. 2006;45:3657–3664. doi: 10.1021/ic060014r. [DOI] [PubMed] [Google Scholar]
- 97.Chavez J. Chung WG. Miranda CL. Singhal M. Stevens JF. Maier CS. Site-specific protein adducts of 4-hydroxy-2(E)-nonenal in human THP-1 monocytic cells: protein carbonylation is diminished by ascorbic acid. Chem Res Toxicol. 2010;23:37–47. doi: 10.1021/tx9002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chavez J. Wu J. Han B. Chung WG. Maier CS. New role for an old probe: affinity labeling of oxylipid protein conjugates by N'-aminooxymethylcarbonylhydrazino d-biotin. Anal Chem. 2006;78:6847–6854. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- 99.Chen Q. Surmeier DJ. Reiner A. NMDA and non-NMDA receptor-mediated excitotoxicity are potentiated in cultured striatal neurons by prior chronic depolarization. Exp Neurol. 1999;159:283–296. doi: 10.1006/exnr.1999.7135. [DOI] [PubMed] [Google Scholar]
- 100.Chen ZH. Saito Y. Yoshida Y. Sekine A. Noguchi N. Niki E. 4-Hydroxynonenal induces adaptive response and enhances PC12 cell tolerance primarily through induction of thioredoxin reductase 1 via activation of Nrf2. J Biol Chem. 2005;280:41921–41927. doi: 10.1074/jbc.M508556200. [DOI] [PubMed] [Google Scholar]
- 101.Chiappetta G. Corbo C. Palmese A. Galli F. Piroddi M. Marino G. Amoresano A. Quantitative identification of protein nitration sites. Proteomics. 2009;9:1524–1537. doi: 10.1002/pmic.200800493. [DOI] [PubMed] [Google Scholar]
- 102.Choi DE. Jeong JY. Lim BJ. Chung S. Chang YK. Lee SJ. Na KR. Kim SY. Shin YT. Lee KW. Pretreatment of sildenafil attenuates ischemia-reperfusion renal injury in rats. Am J Physiol Renal Physiol. 2009;297:F362–F370. doi: 10.1152/ajprenal.90609.2008. [DOI] [PubMed] [Google Scholar]
- 103.Choi J. Levey AI. Weintraub ST. Rees HD. Gearing M. Chin LS. Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 104.Cimini A. Moreno S. D'Amelio M. Cristiano L. D'Angelo B. Falone S. Benedetti E. Carrara P. Fanelli F. Cecconi F. Amicarelli F. Ceru MP. Early biochemical and morphological modifications in the brain of a transgenic mouse model of Alzheimer's disease: a role for peroxisomes. J Alzheimers Dis. 2009;18:935–952. doi: 10.3233/JAD-2009-1199. [DOI] [PubMed] [Google Scholar]
- 105.Clementi ME. Pezzotti M. Orsini F. Sampaolese B. Mezzogori D. Grassi C. Giardina B. Misiti F. Alzheimer's amyloid beta-peptide (1–42) induces cell death in human neuroblastoma via bax/bcl-2 ratio increase: an intriguing role for methionine 35. Biochem Biophys Res Commun. 2006;342:206–213. doi: 10.1016/j.bbrc.2006.01.137. [DOI] [PubMed] [Google Scholar]
- 106.Cleveland DW. Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 107.Coleman PD. Flood DG. Neuron numbers and dendritic extent in normal aging and Alzheimer's disease. Neurobiol Aging. 1987;8:521–545. doi: 10.1016/0197-4580(87)90127-8. [DOI] [PubMed] [Google Scholar]
- 108.Connor JR. Snyder BS. Beard JL. Fine RE. Mufson EJ. Regional distribution of iron and iron-regulatory proteins in the brain in aging and Alzheimer's disease. J Neurosci Res. 1992;31:327–335. doi: 10.1002/jnr.490310214. [DOI] [PubMed] [Google Scholar]
- 109.Crichton RR. Pierre JL. Old iron, young copper: from Mars to Venus. Biometals. 2001;14:99–112. doi: 10.1023/a:1016710810701. [DOI] [PubMed] [Google Scholar]
- 110.Cruthirds DL. Novak L. Akhi KM. Sanders PW. Thompson JA. MacMillan-Crow LA. Mitochondrial targets of oxidative stress during renal ischemia/reperfusion. Arch Biochem Biophys. 2003;412:27–33. doi: 10.1016/s0003-9861(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 111.Curti D. Malaspina A. Facchetti G. Camana C. Mazzini L. Tosca P. Zerbi F. Ceroni M. Amyotrophic lateral sclerosis: oxidative energy metabolism and calcium homeostasis in peripheral blood lymphocytes. Neurology. 1996;47:1060–1064. doi: 10.1212/wnl.47.4.1060. [DOI] [PubMed] [Google Scholar]
- 112.D'Alessandro A. Rinalducci S. Zolla L. Redox proteomics and drug development. J Proteomics. 2011;74:2575–2595. doi: 10.1016/j.jprot.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 113.Dalle-Donne I. Familial amyotrophic lateral sclerosis (FALS): Emerging hints from redox proteomics. Highlight commentary on: “Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice—a model of familial amyotrophic lateral sclerosis”. Free Radic Biol Med. 2007;43:157–159. doi: 10.1016/j.freeradbiomed.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 114.Dalle-Donne I. Giustarini D. Colombo R. Rossi R. Milzani A. Protein carbonylation in human diseases. Trends Mol Med. 2003;9:169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 115.Dalle-Donne I. Scaloni A. Butterfield DA. Redox Proteomics: From Protein Modifications to Cellular Dysfunction and Diseases. Hoboken, NJ: John Wiley and Sons; 2006. [DOI] [PubMed] [Google Scholar]
- 116.Daly NL. Hoffmann R. Otvos L., Jr. Craik DJ. Role of phosphorylation in the conformation of tau peptides implicated in Alzheimer's disease. Biochemistry. 2000;39:9039–9046. doi: 10.1021/bi0004807. [DOI] [PubMed] [Google Scholar]
- 117.David S. Shoemaker M. Haley BE. Abnormal properties of creatine kinase in Alzheimer's disease brain: correlation of reduced enzyme activity and active site photolabeling with aberrant cytosol-membrane partitioning. Brain Res Mol Brain Res. 1998;54:276–287. doi: 10.1016/s0169-328x(97)00343-4. [DOI] [PubMed] [Google Scholar]
- 118.Davies KJ. Shringarpure R. Preferential degradation of oxidized proteins by the 20S proteasome may be inhibited in aging and in inflammatory neuromuscular diseases. Neurology. 2006;66:S93–S96. doi: 10.1212/01.wnl.0000192308.43151.63. [DOI] [PubMed] [Google Scholar]
- 119.Davies P. Moualla D. Brown DR. Alpha-synuclein is a cellular ferrireductase. PLoS One. 2011;6:e15814. doi: 10.1371/journal.pone.0015814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.De Iuliis A. Grigoletto J. Recchia A. Giusti P. Arslan P. A proteomic approach in the study of an animal model of Parkinson's disease. Clin Chim Acta. 2005;357:202–209. doi: 10.1016/j.cccn.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 121.Demicheli V. Quijano C. Alvarez B. Radi R. Inactivation and nitration of human superoxide dismutase (SOD) by fluxes of nitric oxide and superoxide. Free Radic Biol Med. 2007;42:1359–1368. doi: 10.1016/j.freeradbiomed.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 122.Dexter DT. Jenner P. Schapira AH. Marsden CD. Alterations in levels of iron, ferritin, and other trace metals in neurodegenerative diseases affecting the basal ganglia. The Royal Kings and Queens Parkinson's Disease Research Group. Ann Neurol. 1992;32(Suppl):S94–S100. doi: 10.1002/ana.410320716. [DOI] [PubMed] [Google Scholar]
- 123.Di Domenico F. Sultana R. Tiu GF. Scheff NN. Perluigi M. Cini C. Butterfield DA. Protein levels of heat shock proteins 27, 32, 60, 70, 90 and thioredoxin-1 in amnestic mild cognitive impairment: an investigation on the role of cellular stress response in the progression of Alzheimer disease. Brain Res. 2010;1333:72–81. doi: 10.1016/j.brainres.2010.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Di Stasi AM. Mallozzi C. Macchia G. Petrucci TC. Minetti M. Peroxynitrite induces tryosine nitration and modulates tyrosine phosphorylation of synaptic proteins. J Neurochem. 1999;73:727–735. doi: 10.1046/j.1471-4159.1999.0730727.x. [DOI] [PubMed] [Google Scholar]
- 125.Diamandis EP. Christopoulos TK. The biotin-(strept)avidin system: principles and applications in biotechnology. Clin Chem. 1991;37:625–636. [PubMed] [Google Scholar]
- 126.DiFiglia M. Sapp E. Chase KO. Davies SW. Bates GP. Vonsattel JP. Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 127.Ding B. Chen KM. Ling HW. Sun F. Li X. Wan T. Chai WM. Zhang H. Zhan Y. Guan YJ. Correlation of iron in the hippocampus with MMSE in patients with Alzheimer's disease. J Magn Reson Imaging. 2009;29:793–798. doi: 10.1002/jmri.21730. [DOI] [PubMed] [Google Scholar]
- 128.Ding Q. Markesbery WR. Cecarini V. Keller JN. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer's disease. Neurochem Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- 129.Doorn JA. Maser E. Blum A. Claffey DJ. Petersen DR. Human carbonyl reductase catalyzes reduction of 4-oxonon-2-enal. Biochemistry. 2004;43:13106–13114. doi: 10.1021/bi049136q. [DOI] [PubMed] [Google Scholar]
- 130.Doorn JA. Petersen DR. Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chem Biol Interact. 2003;143–144:93–100. doi: 10.1016/s0009-2797(02)00178-3. [DOI] [PubMed] [Google Scholar]
- 131.Drake J. Link CD. Butterfield DA. Oxidative stress precedes fibrillar deposition of Alzheimer's disease amyloid beta-peptide (1–42) in a transgenic Caenorhabditis elegans model. Neurobiol Aging. 2003;24:415–420. doi: 10.1016/s0197-4580(02)00225-7. [DOI] [PubMed] [Google Scholar]
- 132.Duan Z. Lamendola DE. Yusuf RZ. Penson RT. Preffer FI. Seiden MV. Overexpression of human phosphoglycerate kinase 1 (PGK1) induces a multidrug resistance phenotype. Anticancer Res. 2002;22:1933–1941. [PubMed] [Google Scholar]
- 133.Duce JA. Tsatsanis A. Cater MA. James SA. Robb E. Wikhe K. Leong SL. Perez K. Johanssen T. Greenough MA. Cho HH. Galatis D. Moir RD. Masters CL. McLean C. Tanzi RE. Cappai R. Barnham KJ. Ciccotosto GD. Rogers JT. Bush AI. Iron-export ferroxidase activity of beta-amyloid precursor protein is inhibited by zinc in Alzheimer's disease. Cell. 2010;142:857–867. doi: 10.1016/j.cell.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dumont M. Wille E. Stack C. Calingasan NY. Beal MF. Lin MT. Reduction of oxidative stress, amyloid deposition, and memory deficit by manganese superoxide dismutase overexpression in a transgenic mouse model of Alzheimer's disease. FASEB J. 2009;23:2459–2466. doi: 10.1096/fj.09-132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Duyckaerts C. Delatour B. Potier MC. Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 2009;118:5–36. doi: 10.1007/s00401-009-0532-1. [DOI] [PubMed] [Google Scholar]
- 136.Eismann T. Huber N. Shin T. Kuboki S. Galloway E. Wyder M. Edwards MJ. Greis KD. Shertzer HG. Fisher AB. Lentsch AB. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266–G274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.El Tannir El Tayara N. Delatour B. Le Cudennec C. Guegan M. Volk A. Dhenain M. Age-related evolution of amyloid burden, iron load, and MR relaxation times in a transgenic mouse model of Alzheimer's disease. Neurobiol Dis. 2006;22:199–208. doi: 10.1016/j.nbd.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 138.Eliuk SM. Renfrow MB. Shonsey EM. Barnes S. Kim H. Active site modifications of the brain isoform of creatine kinase by 4-hydroxy-2-nonenal correlate with reduced enzyme activity: mapping of modified sites by Fourier transform-ion cyclotron resonance mass spectrometry. Chem Res Toxicol. 2007;20:1260–1268. doi: 10.1021/tx7000948. [DOI] [PubMed] [Google Scholar]
- 139.Esterbauer H. Schaur RJ. Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 140.Estevez AG. Good science shows the way. Highlight Commentary on “Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice—a model of familial amyotrophic lateral sclerosis”. Free Radic Biol Med. 2007;43:163–164. doi: 10.1016/j.freeradbiomed.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fagni L. Bockaert J. Effects of nitric oxide on glutamate-gated channels and other ionic channels. J Chem Neuroanat. 1996;10:231–240. doi: 10.1016/0891-0618(95)00140-9. [DOI] [PubMed] [Google Scholar]
- 142.Fan Q. Yang XC. Cao XB. Wang SY. Yang SL. Liu XL. Gao F. Glutathione reverses peroxynitrite-mediated deleterious effects of nitroglycerin on ischemic rat hearts. J Cardiovasc Pharmacol. 2006;47:405–412. doi: 10.1097/01.fjc.0000210073.48991.bf. [DOI] [PubMed] [Google Scholar]
- 143.Faucheux BA. Martin ME. Beaumont C. Hunot S. Hauw JJ. Agid Y. Hirsch EC. Lack of up-regulation of ferritin is associated with sustained iron regulatory protein-1 binding activity in the substantia nigra of patients with Parkinson's disease. J Neurochem. 2002;83:320–330. doi: 10.1046/j.1471-4159.2002.01118.x. [DOI] [PubMed] [Google Scholar]
- 144.Ferrante RJ. Browne SE. Shinobu LA. Bowling AC. Baik MJ. MacGarvey U. Kowall NW. Brown RH., Jr. Beal MF. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 145.Ferrante RJ. Shinobu LA. Schulz JB. Matthews RT. Thomas CE. Kowall NW. Gurney ME. Beal MF. Increased 3-nitrotyrosine and oxidative damage in mice with a human copper/zinc superoxide dismutase mutation. Ann Neurol. 1997;42:326–334. doi: 10.1002/ana.410420309. [DOI] [PubMed] [Google Scholar]
- 146.Ferrer I. Differential expression of phosphorylated translation initiation factor 2 alpha in Alzheimer's disease and Creutzfeldt-Jakob's disease. Neuropathol Appl Neurobiol. 2002;28:441–451. doi: 10.1046/j.1365-2990.2002.t01-1-00410.x. [DOI] [PubMed] [Google Scholar]
- 147.Ferrer I. Gomez-Isla T. Puig B. Freixes M. Ribe E. Dalfo E. Avila J. Current advances on different kinases involved in tau phosphorylation, and implications in Alzheimer's disease and tauopathies. Curr Alzheimer Res. 2005;2:3–18. doi: 10.2174/1567205052772713. [DOI] [PubMed] [Google Scholar]
- 148.Ferrer I. Martinez A. Blanco R. Dalfo E. Carmona M. Neuropathology of sporadic Parkinson disease before the appearance of parkinsonism: preclinical Parkinson disease. J Neural Transm. 2011;118:821–839. doi: 10.1007/s00702-010-0482-8. [DOI] [PubMed] [Google Scholar]
- 149.Filipovic MR. Stanic D. Raicevic S. Spasic M. Niketic V. Consequences of MnSOD interactions with nitric oxide: nitric oxide dismutation and the generation of peroxynitrite and hydrogen peroxide. Free Radic Res. 2007;41:62–72. doi: 10.1080/10715760600944296. [DOI] [PubMed] [Google Scholar]
- 150.Fishman-Jacob T. Reznichenko L. Youdim MB. Mandel SA. A sporadic Parkinson disease model via silencing of the ubiquitin-proteasome/E3 ligase component SKP1A. J Biol Chem. 2009;284:32835–32845. doi: 10.1074/jbc.M109.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fitzmaurice PS. Shaw IC. Kleiner HE. Miller RT. Monks TJ. Lau SS. Mitchell JD. Lynch PG. Evidence for DNA damage in amyotrophic lateral sclerosis. Muscle Nerve. 1996;19:797–798. [PubMed] [Google Scholar]
- 152.Flint DH. Tuminello JF. Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 153.Floor E. Wetzel MG. Increased protein oxidation in human substantia nigra pars compacta in comparison with basal ganglia and prefrontal cortex measured with an improved dinitrophenylhydrazine assay. J Neurochem. 1998;70:268–275. doi: 10.1046/j.1471-4159.1998.70010268.x. [DOI] [PubMed] [Google Scholar]
- 154.Forrest GL. Gonzalez B. Carbonyl reductase. Chem Biol Interact. 2000;129:21–40. doi: 10.1016/s0009-2797(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 155.Foy CJ. Passmore AP. Vahidassr MD. Young IS. Lawson JT. Plasma chain-breaking antioxidants in Alzheimer's disease, vascular dementia and Parkinson's disease. QJM. 1999;92:39–45. doi: 10.1093/qjmed/92.1.39. [DOI] [PubMed] [Google Scholar]
- 156.Francis PT. Palmer AM. Snape M. Wilcock GK. The cholinergic hypothesis of Alzheimer's disease: a review of progress. J Neurol Neurosurg Psychiatry. 1999;66:137–147. doi: 10.1136/jnnp.66.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fratelli M. Demol H. Puype M. Casagrande S. Eberini I. Salmona M. Bonetto V. Mengozzi M. Duffieux F. Miclet E. Bachi A. Vandekerckhove J. Gianazza E. Ghezzi P. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc Natl Acad Sci U S A. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Frautschy SA. Baird A. Cole GM. Effects of injected Alzheimer beta-amyloid cores in rat brain. Proc Natl Acad Sci U S A. 1991;88:8362–8366. doi: 10.1073/pnas.88.19.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fucci L. Oliver CN. Coon MJ. Stadtman ER. Inactivation of key metabolic enzymes by mixed-function oxidation reactions: possible implication in protein turnover and ageing. Proc Natl Acad Sci U S A. 1983;80:1521–1525. doi: 10.1073/pnas.80.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fukuyama H. Ogawa M. Yamauchi H. Yamaguchi S. Kimura J. Yonekura Y. Konishi J. Altered cerebral energy metabolism in Alzheimer's disease: a PET study. J Nucl Med. 1994;35:1–6. [PubMed] [Google Scholar]
- 161.Gabbita SP. Aksenov MY. Lovell MA. Markesbery WR. Decrease in peptide methionine sulfoxide reductase in Alzheimer's disease brain. J Neurochem. 1999;73:1660–1666. doi: 10.1046/j.1471-4159.1999.0731660.x. [DOI] [PubMed] [Google Scholar]
- 162.Gadoni E. Olivero A. Miglietta A. Bocca C. Gabriel L. Cytoskeletal modifications induced by 4-hydroxynonenal. Cytotechnology. 1993;11(Suppl 1):S62–S64. [PubMed] [Google Scholar]
- 163.Ganea E. Chaperone-like activity of alpha-crystallin and other small heat shock proteins. Curr Protein Pept Sci. 2001;2:205–225. doi: 10.2174/1389203013381107. [DOI] [PubMed] [Google Scholar]
- 164.Gardner PR. Nguyen DD. White CW. Aconitase is a sensitive and critical target of oxygen poisoning in cultured mammalian cells and in rat lungs. Proc Natl Acad Sci U S A. 1994;91:12248–12252. doi: 10.1073/pnas.91.25.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Geddes JW. Pang Z. Wiley DH. Hippocampal damage and cytoskeletal disruption resulting from impaired energy metabolism. Implications for Alzheimer disease. Mol Chem Neuropathol. 1996;28:65–74. doi: 10.1007/BF02815206. [DOI] [PubMed] [Google Scholar]
- 166.George AJ. Holsinger RM. McLean CA. Tan SS. Scott HS. Cardamone T. Cappai R. Masters CL. Li QX. Decreased phosphatidylethanolamine binding protein expression correlates with Abeta accumulation in the Tg2576 mouse model of Alzheimer's disease. Neurobiol Aging. 2006;27:614–623. doi: 10.1016/j.neurobiolaging.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 167.Giambattistelli F. Bucossi S. Salustri C. Panetta V. Mariani S. Siotto M. Ventriglia M. Vernieri F. Dell'acqua ML. Cassetta E. Rossini PM. Squitti R. Effects of hemochromatosis and transferrin gene mutations on iron dyshomeostasis, liver dysfunction and on the risk of Alzheimer's disease. Neurobiol Aging in press. 2011 doi: 10.1016/j.neurobiolaging.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 168.Giasson BI. Ischiropoulos H. Lee VM. Trojanowski JQ. The relationship between oxidative/nitrative stress and pathological inclusions in Alzheimer's and Parkinson's diseases. Free Radic Biol Med. 2002;32:1264–1275. doi: 10.1016/s0891-5849(02)00804-3. [DOI] [PubMed] [Google Scholar]
- 169.Giovannini MG. Cerbai F. Bellucci A. Melani C. Grossi C. Bartolozzi C. Nosi D. Casamenti F. Differential activation of mitogen-activated protein kinase signalling pathways in the hippocampus of CRND8 transgenic mouse, a model of Alzheimer's disease. Neuroscience. 2008;153:618–633. doi: 10.1016/j.neuroscience.2008.02.061. [DOI] [PubMed] [Google Scholar]
- 170.Glabe CG. Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol Aging. 2006;27:570–575. doi: 10.1016/j.neurobiolaging.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 171.Gokulrangan G. Zaidi A. Michaelis ML. Schoneich C. Proteomic analysis of protein nitration in rat cerebellum: effect of biological aging. J Neurochem. 2007;100:1494–1504. doi: 10.1111/j.1471-4159.2006.04334.x. [DOI] [PubMed] [Google Scholar]
- 172.Golej J. Hoeger H. Radner W. Unfried G. Lubec G. Oral administration of methylglyoxal leads to kidney collagen accumulation in the mouse. Life Sci. 1998;63:801–807. doi: 10.1016/s0024-3205(98)00336-1. [DOI] [PubMed] [Google Scholar]
- 173.Gomez A. Ferrer I. Increased oxidation of certain glycolysis and energy metabolism enzymes in the frontal cortex in Lewy body diseases. J Neurosci Res. 2009;87:1002–1013. doi: 10.1002/jnr.21904. [DOI] [PubMed] [Google Scholar]
- 174.Goshima Y. Nakamura F. Strittmatter P. Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to Unc-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- 175.Gotz ME. Kunig G. Riederer P. Youdim MB. Oxidative stress: free radical production in neural degeneration. Pharmacol Ther. 1994;63:37–122. doi: 10.1016/0163-7258(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 176.Grundke-Iqbal I. Iqbal K. Quinlan M. Tung YC. Zaidi MS. Wisniewski HM. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- 177.Gulesserian T. Seidl R. Hardmeier R. Cairns N. Lubec G. Superoxide dismutase SOD1, encoded on chromosome 21, but not SOD2 is overexpressed in brains of patients with Down syndrome. J Investig Med. 2001;49:41–46. doi: 10.2310/6650.2001.34089. [DOI] [PubMed] [Google Scholar]
- 178.Gurney ME. The use of transgenic mouse models of amyotrophic lateral sclerosis in preclinical drug studies. J Neurol Sci. 1997;152(Suppl 1):S67–S73. doi: 10.1016/s0022-510x(97)00247-5. [DOI] [PubMed] [Google Scholar]
- 179.Hague SM. Klaffke S. Bandmann O. Neurodegenerative disorders: Parkinson's disease and Huntington's disease. J Neurol Neurosurg Psychiatry. 2005;76:1058–1063. doi: 10.1136/jnnp.2004.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Halliwell B. Oxidative stress and neurodegeneration: where are we now? J Neurochem. 2006;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- 181.Halliwell B. Proteasomal dysfunction: a common feature of neurodegenerative diseases? Implications for the environmental origins of neurodegeneration. Antioxid Redox Signal. 2006;8:2007–2019. doi: 10.1089/ars.2006.8.2007. [DOI] [PubMed] [Google Scholar]
- 182.Hand CK. Rouleau GA. Familial amyotrophic lateral sclerosis. Muscle Nerve. 2002;25:135–159. doi: 10.1002/mus.10001. [DOI] [PubMed] [Google Scholar]
- 183.Hara MR. Snyder SH. Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell Mol Neurobiol. 2006;26:527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hauptmann S. Keil U. Scherping I. Bonert A. Eckert A. Muller WE. Mitochondrial dysfunction in sporadic and genetic Alzheimer's disease. Exp Gerontol. 2006;41:668–673. doi: 10.1016/j.exger.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 185.Hayes JP. Tipton KF. Interactions of the neurotoxin 6-hydroxydopamine with glyceraldehyde-3-phosphate dehydrogenase. Toxicol Lett. 2002;128:197–206. doi: 10.1016/s0378-4274(02)00013-9. [DOI] [PubMed] [Google Scholar]
- 186.Healy DG. Abou-Sleiman PM. Wood NW. Genetic causes of Parkinson's disease: UCHL-1. Cell Tissue Res. 2004;318:189–194. doi: 10.1007/s00441-004-0917-3. [DOI] [PubMed] [Google Scholar]
- 187.Hellberg K. Grimsrud PA. Kruse AC. Banaszak LJ. Ohlendorf DH. Bernlohr DA. X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal. Prot Sci. 2010;19:1480–1489. doi: 10.1002/pro.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Helman M. Givol D. Isolation of nitrotyrosine-containing peptides by using an insoluble-antibody column. Biochem J. 1971;125:971–974. doi: 10.1042/bj1250971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hensley K. Fedynyshyn J. Ferrell S. Floyd RA. Gordon B. Grammas P. Hamdheydari L. Mhatre M. Mou S. Pye QN. Stewart C. West M. West S. Williamson KS. Message and protein-level elevation of tumor necrosis factor alpha (TNF alpha) and TNF alpha-modulating cytokines in spinal cords of the G93A-SOD1 mouse model for amyotrophic lateral sclerosis. Neurobiol Dis. 2003;14:74–80. doi: 10.1016/s0969-9961(03)00087-1. [DOI] [PubMed] [Google Scholar]
- 190.Hensley K. Hall N. Subramaniam R. Cole P. Harris M. Aksenov M. Aksenova M. Gabbita SP. Wu JF. Carney JM, et al. Brain regional correspondence between Alzheimer's disease histopathology and biomarkers of protein oxidation. J Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 191.Hilditch-Maguire P. Trettel F. Passani LA. Auerbach A. Persichetti F. MacDonald ME. Huntingtin: an iron-regulated protein essential for normal nuclear and perinuclear organelles. Hum Mol Genet. 2000;9:2789–2797. doi: 10.1093/hmg/9.19.2789. [DOI] [PubMed] [Google Scholar]
- 192.Hillmer AS. Putcha P. Levin J. Hogen T. Hyman BT. Kretzschmar H. McLean PJ. Giese A. Converse modulation of toxic alpha-synuclein oligomers in living cells by N'-benzylidene-benzohydrazide derivates and ferric iron. Biochem Biophys Res Commun. 2010;391:461–466. doi: 10.1016/j.bbrc.2009.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hink U. Oelze M. Kolb P. Bachschmid M. Zou MH. Daiber A. Mollnau H. August M. Baldus S. Tsilimingas N. Walter U. Ullrich V. Munzel T. Role for peroxynitrite in the inhibition of prostacyclin synthase in nitrate tolerance. J Am Coll Cardiol. 2003;42:1826–1834. doi: 10.1016/j.jacc.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 194.Hirai K. Aliev G. Nunomura A. Fujioka H. Russell RL. Atwood CS. Johnson AB. Kress Y. Vinters HV. Tabaton M. Shimohama S. Cash AD. Siedlak SL. Harris PL. Jones PK. Petersen RB. Perry G. Smith MA. Mitochondrial abnormalities in Alzheimer's disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hirano M. Quinzii CM. Mitsumoto H. Hays AP. Kirk Roberts J. Richard P. Rowland LP. Senataxin mutations and amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2010;12:223–227. doi: 10.3109/17482968.2010.545952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Holzer M. Gartner U. Stobe A. Hartig W. Gruschka H. Bruckner MK. Arendt T. Inverse association of Pin1 and tau accumulation in Alzheimer's disease hippocampus. Acta Neuropathol. 2002;104:471–481. doi: 10.1007/s00401-002-0581-1. [DOI] [PubMed] [Google Scholar]
- 197.Hoyer S. Brain glucose and energy metabolism abnormalities in sporadic Alzheimer disease. Causes and consequences: an update. Exp Gerontol. 2000;35:1363–1372. doi: 10.1016/s0531-5565(00)00156-x. [DOI] [PubMed] [Google Scholar]
- 198.Hoyer S. Glucose metabolism and insulin receptor signal transduction in Alzheimer disease. Eur J Pharmacol. 2004;490:115–125. doi: 10.1016/j.ejphar.2004.02.049. [DOI] [PubMed] [Google Scholar]
- 199.Hsu JL. Chen SH. Li DT. Shi FK. Enhanced a1 fragmentation for dimethylated proteins and its applications for N-terminal identification and comparative protein quantitation. J Proteome Res. 2007;6:2376–2383. doi: 10.1021/pr060639n. [DOI] [PubMed] [Google Scholar]
- 200.Huang LL. Shang F. Nowell TR., Jr. Taylor A. Degradation of differentially oxidized alpha-crystallins in bovine lens epithelial cells. Exp Eye Res. 1995;61:45–54. doi: 10.1016/s0014-4835(95)80057-3. [DOI] [PubMed] [Google Scholar]
- 201.Hyson HC. Kieburtz K. Shoulson I. McDermott M. Ravina B. de Blieck EA. Cudkowicz ME. Ferrante RJ. Como P. Frank S. Zimmerman C. Ferrante K. Newhall K. Jennings D. Kelsey T. Walker F. Hunt V. Daigneault S. Goldstein M. Weber J. Watts A. Beal MF. Browne SE. Metakis LJ. Safety and tolerability of high-dosage coenzyme Q10 in Huntington's disease and healthy subjects. Mov Disord. 2010;25:1924–1928. doi: 10.1002/mds.22408. [DOI] [PubMed] [Google Scholar]
- 202.Iacono D. O'Brien R. Resnick SM. Zonderman AB. Pletnikova O. Rudow G. An Y. West MJ. Crain B. Troncoso JC. Neuronal hypertrophy in asymptomatic Alzheimer disease. J Neuropathol Exp Neurol. 2008;67:578–589. doi: 10.1097/NEN.0b013e3181772794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Iannello RC. Crack PJ. de Haan JB. Kola I. Oxidative stress and neural dysfunction in Down syndrome. J Neural Transm Suppl. 1999;57:257–267. doi: 10.1007/978-3-7091-6380-1_17. [DOI] [PubMed] [Google Scholar]
- 204.Ignarro LJ. Nitric oxide. A novel signal transduction mechanism for transcellular communication. Hypertension. 1990;16:477–483. doi: 10.1161/01.hyp.16.5.477. [DOI] [PubMed] [Google Scholar]
- 205.Ihara Y. Nobukuni K. Takata H. Hayabara T. Oxidative stress and metal content in blood and cerebrospinal fluid of amyotrophic lateral sclerosis patients with and without a Cu, Zn-superoxide dismutase mutation. Neurol Res. 2005;27:105–108. doi: 10.1179/016164105X18430. [DOI] [PubMed] [Google Scholar]
- 206.Ince PG. Shaw PJ. Candy JM. Mantle D. Tandon L. Ehmann WD. Markesbery WR. Iron, selenium and glutathione peroxidase activity are elevated in sporadic motor neuron disease. Neurosci Lett. 1994;182:87–90. doi: 10.1016/0304-3940(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 207.Ishihara K. Amano K. Takaki E. Ebrahim AS. Shimohata A. Shibazaki N. Inoue I. Takaki M. Ueda Y. Sago H. Epstein CJ. Yamakawa K. Increased lipid peroxidation in Down's syndrome mouse models. J Neurochem. 2009;110:1965–1976. doi: 10.1111/j.1471-4159.2009.06294.x. [DOI] [PubMed] [Google Scholar]
- 208.Jang BG. Yun SM. Ahn K. Song JH. Jo SA. Kim YY. Kim DK. Park MH. Han C. Koh YH. Plasma carbonic anhydrase II protein is elevated in Alzheimer's disease. J Alzheimers Dis JAD. 2010;21:939–945. doi: 10.3233/JAD-2010-100384. [DOI] [PubMed] [Google Scholar]
- 209.Jenner P. Oxidative stress in Parkinson's disease. Ann Neurol. 2003;53(Suppl 3):S26–S36. doi: 10.1002/ana.10483. discussion S36–S38. [DOI] [PubMed] [Google Scholar]
- 210.Jeong SY. Rathore KI. Schulz K. Ponka P. Arosio P. David S. Dysregulation of iron homeostasis in the CNS contributes to disease progression in a mouse model of amyotrophic lateral sclerosis. J Neurosci. 2009;29:610–619. doi: 10.1523/JNEUROSCI.5443-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jiang H. Luan Z. Wang J. Xie J. Neuroprotective effects of iron chelator Desferal on dopaminergic neurons in the substantia nigra of rats with iron-overload. Neurochem Int. 2006;49:605–609. doi: 10.1016/j.neuint.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 212.Joguchi A. Otsuka I. Minagawa S. Suzuki T. Fujii M. Ayusawa D. Overexpression of VDUP1 mRNA sensitizes HeLa cells to paraquat. Biochem Biophys Res Commun. 2002;293:293–297. doi: 10.1016/S0006-291X(02)00208-5. [DOI] [PubMed] [Google Scholar]
- 213.Jouvenceau A. Dutar P. Billard JM. Alteration of NMDA receptor-mediated synaptic responses in CA1 area of the aged rat hippocampus: contribution of GABAergic and cholinergic deficits. Hippocampus. 1998;8:627–637. doi: 10.1002/(SICI)1098-1063(1998)8:6<627::AID-HIPO5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 214.Jovanovic SV. Clements D. MacLeod K. Biomarkers of oxidative stress are significantly elevated in Down syndrome. Free Radic Biol Med. 1998;25:1044–1048. doi: 10.1016/s0891-5849(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 215.Kahle PJ. Neumann M. Ozmen L. Muller V. Odoy S. Okamoto N. Jacobsen H. Iwatsubo T. Trojanowski JQ. Takahashi H. Wakabayashi K. Bogdanovic N. Riederer P. Kretzschmar HA. Haass C. Selective insolubility of alpha-synuclein in human Lewy body diseases is recapitulated in a transgenic mouse model. Am J Pathol. 2001;159:2215–2225. doi: 10.1016/s0002-9440(10)63072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kamat CD. Gadal S. Mhatre M. Williamson KS. Pye QN. Hensley K. Antioxidants in central nervous system diseases: preclinical promise and translational challenges. J Alzheimers Dis. 2008;15:473–493. doi: 10.3233/jad-2008-15314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kanski J. Alterman MA. Schoneich C. Proteomic identification of age-dependent protein nitration in rat skeletal muscle. Free Radic Biol Med. 2003;35:1229–1239. doi: 10.1016/s0891-5849(03)00500-8. [DOI] [PubMed] [Google Scholar]
- 218.Kanski J. Hong SJ. Schoneich C. Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J Biol Chem. 2005;280:24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 219.Kapogiannis D. Mattson MP. Disrupted energy metabolism and neuronal circuit dysfunction in cognitive impairment and Alzheimer's disease. Lancet Neurol. 2011;10:187–198. doi: 10.1016/S1474-4422(10)70277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kato S. Hayashi H. Nakashima K. Nanba E. Kato M. Hirano A. Nakano I. Asayama K. Ohama E. Pathological characterization of astrocytic hyaline inclusions in familial amyotrophic lateral sclerosis. Am J Pathol. 1997;151:611–620. [PMC free article] [PubMed] [Google Scholar]
- 221.Keller JN. Hanni KB. Markesbery WR. Impaired proteasome function in Alzheimer's disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 222.Keller JN. Schmitt FA. Scheff SW. Ding Q. Chen Q. Butterfield DA. Markesbery WR. Evidence of increased oxidative damage in subjects with mild cognitive impairment. Neurology. 2005;64:1152–1156. doi: 10.1212/01.WNL.0000156156.13641.BA. [DOI] [PubMed] [Google Scholar]
- 223.Kida K. Nishio T. Nagai K. Matsuda H. Nakagawa H. Gluconeogenesis in the kidney in vivo in fed rats. Circadian change and substrate specificity. J Biochem (Tokyo) 1982;91:755–760. doi: 10.1093/oxfordjournals.jbchem.a133762. [DOI] [PubMed] [Google Scholar]
- 224.Kienzl E. Jellinger K. Stachelberger H. Linert W. Iron as catalyst for oxidative stress in the pathogenesis of Parkinson's disease? Life Sci. 1999;65:1973–1976. doi: 10.1016/s0024-3205(99)00458-0. [DOI] [PubMed] [Google Scholar]
- 225.Kieran D. Kalmar B. Dick JR. Riddoch-Contreras J. Burnstock G. Greensmith L. Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat Med. 2004;10:402–405. doi: 10.1038/nm1021. [DOI] [PubMed] [Google Scholar]
- 226.Kim M. Jung Y. Lee K. Kim C. Identification of the calcium binding sites in translationally controlled tumor protein. Arch Pharm Res. 2000;23:633–636. doi: 10.1007/BF02975253. [DOI] [PubMed] [Google Scholar]
- 227.Kim SH. Fountoulakis M. Cairns N. Lubec G. Protein levels of human peroxiredoxin subtypes in brains of patients with Alzheimer's disease and Down syndrome. J Neural Transm Suppl. 2001;(61):223–235. doi: 10.1007/978-3-7091-6262-0_18. [DOI] [PubMed] [Google Scholar]
- 228.Klose J. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik. 1975;26:231–243. doi: 10.1007/BF00281458. [DOI] [PubMed] [Google Scholar]
- 229.Knyushko TV. Sharov VS. Williams TD. Schoneich C. Bigelow DJ. 3-Nitrotyrosine modification of SERCA2a in the aging heart: a distinct signature of the cellular redox environment. Biochemistry. 2005;44:13071–13081. doi: 10.1021/bi051226n. [DOI] [PubMed] [Google Scholar]
- 230.Koeppen AH. The history of iron in the brain. J Neurol Sci. 1995;134(Suppl 1) doi: 10.1016/0022-510x(95)00202-d. [DOI] [PubMed] [Google Scholar]
- 231.Kondrikov D. Elms S. Fulton D. Su Y. eNOS-beta-actin interaction contributes to increased peroxynitrite formation during hyperoxia in pulmonary artery endothelial cells and mouse lungs. J Biol Chem. 2010;285:35479–35487. doi: 10.1074/jbc.M110.140269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Korenberg JR. Bradley C. Disteche CM. Down syndrome: molecular mapping of the congenital heart disease and duodenal stenosis. Am J Hum Genet. 1992;50:294–302. [PMC free article] [PubMed] [Google Scholar]
- 233.Korolainen MA. Goldsteins G. Alafuzoff I. Koistinaho J. Pirttila T. Proteomic analysis of protein oxidation in Alzheimer's disease brain. Electrophoresis. 2002;23:3428–3433. doi: 10.1002/1522-2683(200210)23:19<3428::AID-ELPS3428>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 234.Korolainen MA. Goldsteins G. Nyman TA. Alafuzoff I. Koistinaho J. Pirttila T. Oxidative modification of proteins in the frontal cortex of Alzheimer's disease brain. Neurobiol Aging. 2006;27:42–53. doi: 10.1016/j.neurobiolaging.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 235.LaFontaine MA. Mattson MP. Butterfield DA. Oxidative stress in synaptosomal proteins from mutant presenilin-1 knock-in mice: implications for familial Alzheimer's disease. Neurochem Res. 2002;27:417–421. doi: 10.1023/a:1015560116208. [DOI] [PubMed] [Google Scholar]
- 236.Lamprecht R. LeDoux J. Structural plasticity and memory. Nat Rev Neurosci. 2004;5:45–54. doi: 10.1038/nrn1301. [DOI] [PubMed] [Google Scholar]
- 237.Landino LM. Skreslet TE. Alston JA. Cysteine oxidation of tau and microtubule-associated protein-2 by peroxynitrite: modulation of microtubule assembly kinetics by the thioredoxin reductase system. J Biol Chem. 2004;279:35101–35105. doi: 10.1074/jbc.M405471200. [DOI] [PubMed] [Google Scholar]
- 238.Lauderback CM. Hackett JM. Huang FF. Keller JN. Szweda LI. Markesbery WR. Butterfield DA. The glial glutamate transporter, GLT-1, is oxidatively modified by 4-hydroxy-2-nonenal in the Alzheimer's disease brain: the role of Abeta1-42. J Neurochem. 2001;78:413–416. doi: 10.1046/j.1471-4159.2001.00451.x. [DOI] [PubMed] [Google Scholar]
- 239.Layfield R. Fergusson J. Aitken A. Lowe J. Landon M. Mayer RJ. Neurofibrillary tangles of Alzheimer's disease brains contain 14-3-3 proteins. Neurosci Lett. 1996;209:57–60. doi: 10.1016/0304-3940(96)12598-2. [DOI] [PubMed] [Google Scholar]
- 240.Ledesma MD. Da Silva JS. Crassaerts K. Delacourte A. De Strooper B. Dotti CG. Brain plasmin enhances APP alpha-cleavage and Abeta degradation and is reduced in Alzheimer's disease brains. EMBO Rep. 2000;1:530–535. doi: 10.1093/embo-reports/kvd107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lee S. Young NL. Whetstone PA. Cheal SM. Benner WH. Lebrilla CB. Meares CF. Method to site-specifically identify and quantitate carbonyl end products of protein oxidation using oxidation-dependent element coded affinity tags (O-ECAT) and nanoliquid chromatography Fourier transform mass spectrometry. J Prot Res. 2006;5:539–547. doi: 10.1021/pr050299q. [DOI] [PubMed] [Google Scholar]
- 242.Lee SH. Takahashi R. Goto T. Oe T. Mass spectrometric characterization of modifications to angiotensin II by lipid peroxidation products, 4-oxo-2(E)-nonenal and 4-hydroxy-2(E)-nonenal. Chem Res Toxicol. 2010;23:1771–1785. doi: 10.1021/tx100228q. [DOI] [PubMed] [Google Scholar]
- 243.Leigh PN. Whitwell H. Garofalo O. Buller J. Swash M. Martin JE. Gallo JM. Weller RO. Anderton BH. Ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis. Morphology, distribution, and specificity. Brain. 1991;114(Pt 2):775–788. doi: 10.1093/brain/114.2.775. [DOI] [PubMed] [Google Scholar]
- 244.Lemieux N. Malfoy B. Forrest GL. Human carbonyl reductase (CBR) localized to band 21q22.1 by high-resolution fluorescence in situ hybridization displays gene dosage effects in trisomy 21 cells. Genomics. 1993;15:169–172. doi: 10.1006/geno.1993.1024. [DOI] [PubMed] [Google Scholar]
- 245.Lesage S. Janin S. Lohmann E. Leutenegger AL. Leclere L. Viallet F. Pollak P. Durif F. Thobois S. Layet V. Vidailhet M. Agid Y. Durr A. Brice A. Bonnet AM. Borg M. Broussolle E. Damier P. Destee A. Martinez M. Penet C. Rasco O. Tison F. Tranchan C. Verin M. LRRK2 exon 41 mutations in sporadic Parkinson disease in Europeans. Arch Neurol. 2007;64:425–430. doi: 10.1001/archneur.64.3.425. [DOI] [PubMed] [Google Scholar]
- 246.Leveugle B. Spik G. Perl DP. Bouras C. Fillit HM. Hof PR. The iron-binding protein lactotransferrin is present in pathologic lesions in a variety of neurodegenerative disorders: a comparative immunohistochemical analysis. Brain Res. 1994;650:20–31. doi: 10.1016/0006-8993(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 247.Levine RL. Wehr N. Williams JA. Stadtman ER. Shacter E. Determination of carbonyl groups in oxidized proteins. Methods Mol Biol. 2000;99:15–24. doi: 10.1385/1-59259-054-3:15. [DOI] [PubMed] [Google Scholar]
- 248.Li YF. Wang Y. Channon KM. Schultz HD. Zucker IH. Patel KP. Manipulation of neuronal nitric oxide synthase within the paraventricular nucleus using adenovirus and antisense technology. Methods Mol Med. 2005;112:59–79. doi: 10.1385/1-59259-879-x:059. [DOI] [PubMed] [Google Scholar]
- 249.Lillig CH. Holmgren A. Thioredoxin and related molecules—from biology to health and disease. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 250.Lin TK. Hughes G. Muratovska A. Blaikie FH. Brookes PS. Darley-Usmar V. Smith RA. Murphy MP. Specific modification of mitochondrial protein thiols in response to oxidative stress: a proteomics approach. J Biol Chem. 2002;277:17048–17056. doi: 10.1074/jbc.M110797200. [DOI] [PubMed] [Google Scholar]
- 251.Liochev SI. Fridovich I. Copper- and zinc-containing superoxide dismutase can act as a superoxide reductase and a superoxide oxidase. J Biol Chem. 2000;275:38482–38485. doi: 10.1074/jbc.M007891200. [DOI] [PubMed] [Google Scholar]
- 252.Liou YC. Ryo A. Huang HK. Lu PJ. Bronson R. Fujimori F. Uchida T. Hunter T. Lu KP. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci U S A. 2002;99:1335–1340. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Liu D. Wen J. Liu J. Li L. The roles of free radicals in amyotrophic lateral sclerosis: reactive oxygen species and elevated oxidation of protein, DNA, and membrane phospholipids. FASEB J. 1999;13:2318–2328. doi: 10.1096/fasebj.13.15.2318. [DOI] [PubMed] [Google Scholar]
- 254.Liu R. Althaus JS. Ellerbrock BR. Becker DA. Gurney ME. Enhanced oxygen radical production in a transgenic mouse model of familial amyotrophic lateral sclerosis. Ann Neurol. 1998;44:763–770. doi: 10.1002/ana.410440510. [DOI] [PubMed] [Google Scholar]
- 255.Lott IT. Head E. Doran E. Busciglio J. Beta-amyloid, oxidative stress and down syndrome. Curr Alzheimer Res. 2006;3:521–528. doi: 10.2174/156720506779025305. [DOI] [PubMed] [Google Scholar]
- 256.Lourette N. Smallwood H. Wu S. Robinson EW. Squier TC. Smith RD. Pasa-Tolic L. A top-down LC-FTICR MS-based strategy for characterizing oxidized calmodulin in activated macrophages. J Am Soc Mass Spectrom. 2010;21:930–939. doi: 10.1016/j.jasms.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 257.Lovell MA. Xie C. Gabbita SP. Markesbery WR. Decreased thioredoxin and increased thioredoxin reductase levels in Alzheimer's disease brain. Free Radic Biol Med. 2000;28:418–427. doi: 10.1016/s0891-5849(99)00258-0. [DOI] [PubMed] [Google Scholar]
- 258.Lovell MA. Xiong S. Markesbery WR. Lynn BC. Quantitative proteomic analysis of mitochondria from primary neuron cultures treated with amyloid beta peptide. Neurochem Res. 2005;30:113–122. doi: 10.1007/s11064-004-9692-5. [DOI] [PubMed] [Google Scholar]
- 259.Lu B. Motoyama A. Ruse C. Venable J. Yates JR., 3rd Improving protein identification sensitivity by combining MS and MS/MS information for shotgun proteomics using LTQ-Orbitrap high mass accuracy data. Anal Chem. 2008;80:2018–2025. doi: 10.1021/ac701697w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lubec G. Nonaka M. Krapfenbauer K. Gratzer M. Cairns N. Fountoulakis M. Expression of the dihydropyrimidinase related protein 2 (DRP-2) in Down syndrome and Alzheimer's disease brain is downregulated at the mRNA and dysregulated at the protein level. J Neural Transm Suppl. 1999;57:161–177. doi: 10.1007/978-3-7091-6380-1_10. [DOI] [PubMed] [Google Scholar]
- 261.MacMillan-Crow LA. Crow JP. Kerby JD. Beckman JS. Thompson JA. Nitration and inactivation of manganese superoxide dismutase in chronic rejection of human renal allografts. Proc Natl Acad Sci U S A. 1996;93:11853–11858. doi: 10.1073/pnas.93.21.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Macmillan-Crow LA. Cruthirds DL. Invited review: manganese superoxide dismutase in disease. Free Radic Res. 2001;34:325–336. doi: 10.1080/10715760100300281. [DOI] [PubMed] [Google Scholar]
- 263.Madian AG. Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Magrane J. Smith RC. Walsh K. Querfurth HW. Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed beta-amyloid in neurons. J Neurosci. 2004;24:1700–1706. doi: 10.1523/JNEUROSCI.4330-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mandel SA. Fishman-Jacob T. Youdim MB. Modeling sporadic Parkinson's disease by silencing the ubiquitin E3 ligase component, SKP1A. Parkinsonism Relat Disord. 2009;15(Suppl 3):S148–S151. doi: 10.1016/S1353-8020(09)70803-X. [DOI] [PubMed] [Google Scholar]
- 266.Masliah E. Alford M. DeTeresa R. Mallory M. Hansen L. Deficient glutamate transport is associated with neurodegeneration in Alzheimer's disease. Ann Neurol. 1996;40:759–766. doi: 10.1002/ana.410400512. [DOI] [PubMed] [Google Scholar]
- 267.Masliah E. Mallory M. Hansen L. DeTeresa R. Alford M. Terry R. Synaptic and neuritic alterations during the progression of Alzheimer's disease. Neurosci Lett. 1994;174:67–72. doi: 10.1016/0304-3940(94)90121-x. [DOI] [PubMed] [Google Scholar]
- 268.Matsuura I. Chiang KN. Lai CY. He D. Wang G. Ramkumar R. Uchida T. Ryo A. Lu K. Liu F. Pin1 promotes transforming growth factor-beta-induced migration and invasion. J Biol Chem. 2010;285:1754–1764. doi: 10.1074/jbc.M109.063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mattson MP. Pedersen WA. Duan W. Culmsee C. Camandola S. Cellular and molecular mechanisms underlying perturbed energy metabolism and neuronal degeneration in Alzheimer's and Parkinson's diseases. Ann N Y Acad Sci. 1999;893:154–175. doi: 10.1111/j.1749-6632.1999.tb07824.x. [DOI] [PubMed] [Google Scholar]
- 270.Meier-Ruge W. Bertoni-Freddari C. Iwangoff P. Changes in brain glucose metabolism as a key to the pathogenesis of Alzheimer's disease. Gerontology. 1994;40:246–252. doi: 10.1159/000213592. [DOI] [PubMed] [Google Scholar]
- 271.Meier-Ruge W. Iwangoff P. Reichlmeier K. Neurochemical enzyme changes in Alzheimer's and Pick's disease. Arch Gerontol Geriatr. 1984;3:161–165. doi: 10.1016/0167-4943(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 272.Messier C. Gagnon M. Glucose regulation and cognitive functions: relation to Alzheimer's disease and diabetes. Behav Brain Res. 1996;75:1–11. doi: 10.1016/0166-4328(95)00153-0. [DOI] [PubMed] [Google Scholar]
- 273.Mihm MJ. Schanbacher BL. Wallace BL. Wallace LJ. Uretsky NJ. Bauer JA. Free 3-nitrotyrosine causes striatal neurodegeneration in vivo. J Neurosci. 2001;21:RC149. doi: 10.1523/JNEUROSCI.21-11-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mirzaei H. Baena B. Barbas C. Regnier F. Identification of oxidized proteins in rat plasma using avidin chromatography and tandem mass spectrometry. Proteomics. 2008;8:1516–1527. doi: 10.1002/pmic.200700363. [DOI] [PubMed] [Google Scholar]
- 275.Mirzaei H. Regnier F. Affinity chromatographic selection of carbonylated proteins followed by identification of oxidation sites using tandem mass spectrometry. Anal Chem. 2005;77:2386–2392. doi: 10.1021/ac0484373. [DOI] [PubMed] [Google Scholar]
- 276.Mirzaei H. Regnier F. Identification and quantification of protein carbonylation using light and heavy isotope labeled Girard's P reagent. J Chromatogr A. 2006;1134:122–133. doi: 10.1016/j.chroma.2006.08.096. [DOI] [PubMed] [Google Scholar]
- 277.Molina JA. de Bustos F. Jimenez-Jimenez FJ. Benito-Leon J. Gasalla T. Orti-Pareja M. Vela L. Bermejo F. Martin MA. Campos Y. Arenas J. Respiratory chain enzyme activities in isolated mitochondria of lymphocytes from patients with Alzheimer's disease. Neurology. 1997;48:636–638. doi: 10.1212/wnl.48.3.636. [DOI] [PubMed] [Google Scholar]
- 278.Montine TJ. Quinn JF. Montine KS. Kaye JA. Breitner JC. Quantitative in vivo biomarkers of oxidative damage and their application to the diagnosis and management of Alzheimer's disease. J Alzheimers Dis. 2005;8:359–367. doi: 10.3233/jad-2005-8405. [DOI] [PubMed] [Google Scholar]
- 279.Morel P. Tallineau C. Pontcharraud R. Piriou A. Huguet F. Effects of 4-hydroxynonenal, a lipid peroxidation product, on dopamine transport and Na+/K+ ATPase in rat striatal synaptosomes. Neurochem Int. 1998;33:531–540. doi: 10.1016/s0197-0186(98)00062-x. [DOI] [PubMed] [Google Scholar]
- 280.Mouser PE. Head E. Ha KH. Rohn TT. Caspase-mediated cleavage of glial fibrillary acidic protein within degenerating astrocytes of the Alzheimer's disease brain. Am J Pathol. 2006;168:936–946. doi: 10.2353/ajpath.2006.050798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Muller WE. Eckert A. Kurz C. Eckert GP. Leuner K. Mitochondrial dysfunction: common final pathway in brain aging and Alzheimer's disease—therapeutic aspects. Mol Neurobiol. 2010;41:159–171. doi: 10.1007/s12035-010-8141-5. [DOI] [PubMed] [Google Scholar]
- 282.Nagy Z. Esiri MM. Cato AM. Smith AD. Cell cycle markers in the hippocampus in Alzheimer's disease. Acta Neuropathol. 1997;94:6–15. doi: 10.1007/s004010050665. [DOI] [PubMed] [Google Scholar]
- 283.Nakamura A. Goto S. Analysis of protein carbonyls with 2,4-dinitrophenyl hydrazine and its antibodies by immunoblot in two-dimensional gel electrophoresis. J Biochem. 1996;119:768–774. doi: 10.1093/oxfordjournals.jbchem.a021306. [DOI] [PubMed] [Google Scholar]
- 284.Neely MD. Boutte A. Milatovic D. Montine TJ. Mechanisms of 4-hydroxynonenal-induced neuronal microtubule dysfunction. Brain Res. 2005;1037:90–98. doi: 10.1016/j.brainres.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 285.Niatsetskaya Z. Basso M. Speer RE. McConoughey SJ. Coppola G. Ma TC. Ratan RR. HIF prolyl hydroxylase inhibitors prevent neuronal death induced by mitochondrial toxins: therapeutic implications for Huntington's disease and Alzheimer's disease. Antioxid Redox Signal. 2010;12:435–443. doi: 10.1089/ars.2009.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Nikov G. Bhat V. Wishnok JS. Tannenbaum SR. Analysis of nitrated proteins by nitrotyrosine-specific affinity probes and mass spectrometry. Anal Biochem. 2003;320:214–222. doi: 10.1016/s0003-2697(03)00359-2. [DOI] [PubMed] [Google Scholar]
- 287.Nishiyama A. Matsui M. Iwata S. Hirota K. Masutani H. Nakamura H. Takagi Y. Sono H. Gon Y. Yodoi J. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 288.Nunomura A. Perry G. Pappolla MA. Friedland RP. Hirai K. Chiba S. Smith MA. Neuronal oxidative stress precedes amyloid-beta deposition in Down syndrome. J Neuropathol Exp Neurol. 2000;59:1011–1017. doi: 10.1093/jnen/59.11.1011. [DOI] [PubMed] [Google Scholar]
- 289.O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 290.Odetti P. Angelini G. Dapino D. Zaccheo D. Garibaldi S. Dagna-Bricarelli F. Piombo G. Perry G. Smith M. Traverso N. Tabaton M. Early glycoxidation damage in brains from Down's syndrome. Biochem Biophys Res Commun. 1998;243:849–851. doi: 10.1006/bbrc.1998.8186. [DOI] [PubMed] [Google Scholar]
- 291.Ojika K. Tsugu Y. Mitake S. Otsuka Y. Katada E. NMDA receptor activation enhances the release of a cholinergic differentiation peptide (HCNP) from hippocampal neurons in vitro. Brain Res Dev Brain Res. 1998;106:173–180. doi: 10.1016/s0165-3806(98)00014-5. [DOI] [PubMed] [Google Scholar]
- 292.Oka S. Masutani H. Liu W. Horita H. Wang D. Kizaka-Kondoh S. Yodoi J. Thioredoxin-binding protein-2-like inducible membrane protein is a novel vitamin D3 and peroxisome proliferator-activated receptor (PPAR)gamma ligand target protein that regulates PPARgamma signaling. Endocrinology. 2006;147:733–743. doi: 10.1210/en.2005-0679. [DOI] [PubMed] [Google Scholar]
- 293.Op den Velde W. Stam FC. Some cerebral proteins and enzyme systems in Alzheimer's presenile and senile dementia. J Am Geriatr Soc. 1976;24:12–16. doi: 10.1111/j.1532-5415.1976.tb03247.x. [DOI] [PubMed] [Google Scholar]
- 294.Orrenius S. Burgess DH. Hampton MB. Zhivotovsky B. Mitochondria as the focus of apoptosis research. Cell Death Differ. 1997;4:427–428. doi: 10.1038/sj.cdd.4400272. [DOI] [PubMed] [Google Scholar]
- 295.Ostrerova-Golts N. Petrucelli L. Hardy J. Lee JM. Farer M. Wolozin B. The A53T alpha-synuclein mutation increases iron-dependent aggregation and toxicity. J Neurosci. 2000;20:6048–6054. doi: 10.1523/JNEUROSCI.20-16-06048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ou J. Fontana JT. Ou Z. Jones DW. Ackerman AW. Oldham KT. Yu J. Sessa WC. Pritchard KA., Jr. Heat shock protein 90 and tyrosine kinase regulate eNOS NO* generation but not NO* bioactivity. Am J Physiol Heart Circ Physiol. 2004;286:H561–H569. doi: 10.1152/ajpheart.00736.2003. [DOI] [PubMed] [Google Scholar]
- 297.Owen JB. Sultana R. Aluise CD. Erickson MA. Price TO. Bu G. Banks WA. Butterfield DA. Oxidative modification to LDL receptor-related protein 1 in hippocampus from subjects with Alzheimer disease: implications for Abeta accumulation in AD brain. Free Radic Biol Med. 2010;49:1798–1803. doi: 10.1016/j.freeradbiomed.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Palacino JJ. Sagi D. Goldberg MS. Krauss S. Motz C. Wacker M. Klose J. Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 299.Panov AV. Gutekunst CA. Leavitt BR. Hayden MR. Burke JR. Strittmatter WJ. Greenamyre JT. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 300.Pei JJ. Braak H. An WL. Winblad B. Cowburn RF. Iqbal K. Grundke-Iqbal I. Up-regulation of mitogen-activated protein kinases ERK1/2 and MEK1/2 is associated with the progression of neurofibrillary degeneration in Alzheimer's disease. Brain Res Mol Brain Res. 2002;109:45–55. doi: 10.1016/s0169-328x(02)00488-6. [DOI] [PubMed] [Google Scholar]
- 301.Perluigi M. di Domenico F. Fiorini A. Cocciolo A. Giorgi A. Foppoli C. Butterfield DA. Giorlandino M. Giorlandino C. Schinina ME. Coccia R. Oxidative stress occurs early in Down syndrome pregnancy: a redox proteomics analysis of amniotic fluid. Prot Clin Appl. 2011;5:167–178. doi: 10.1002/prca.201000121. [DOI] [PubMed] [Google Scholar]
- 302.Perluigi M. Fai Poon H. Hensley K. Pierce WM. Klein JB. Calabrese V. De Marco C. Butterfield DA. Proteomic analysis of 4-hydroxy-2-nonenal-modified proteins in G93A-SOD1 transgenic mice—a model of familial amyotrophic lateral sclerosis. Free Radic Biol Med. 2005;38:960–968. doi: 10.1016/j.freeradbiomed.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 303.Perluigi M. Poon HF. Maragos W. Pierce WM. Klein JB. Calabrese V. Cini C. De Marco C. Butterfield DA. Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: a model of Huntington disease. Mol Cell Proteomics. 2005;4:1849–1861. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 304.Perluigi M. Sultana R. Cenini G. Di Domenico F. Memo M. Pierce WM. Coccia R. Butterfield DA. Redox proteomics identification of HNE-modified brain proteins in Alzheimer's disease: role of lipid peroxidation in Alzheimer's disease pathogenesis. Proteomics Clin Appl. 2009;3:682–693. doi: 10.1002/prca.200800161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Perumal AS. Tordzro WK. Katz M. Jackson-Lewis V. Cooper TB. Fahn S. Cadet JL. Regional effects of 6-hydroxydopamine (6-OHDA) on free radical scavengers in rat brain. Brain Res. 1989;504:139–141. doi: 10.1016/0006-8993(89)91611-9. [DOI] [PubMed] [Google Scholar]
- 306.Pestova TV. Hellen CU. The structure and function of initiation factors in eukaryotic protein synthesis. Cell Mol Life Sci. 2000;57:651–674. doi: 10.1007/PL00000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Petersen A. Hansson O. Puschban Z. Sapp E. Romero N. Castilho RF. Sulzer D. Rice M. DiFiglia M. Przedborski S. Brundin P. Mice transgenic for exon 1 of the Huntington's disease gene display reduced striatal sensitivity to neurotoxicity induced by dopamine and 6-hydroxydopamine. Eur J Neurosci. 2001;14:1425–1435. doi: 10.1046/j.0953-816x.2001.01765.x. [DOI] [PubMed] [Google Scholar]
- 308.Petronis A. Alzheimer's disease and down syndrome: from meiosis to dementia. Exp Neurol. 1999;158:403–413. doi: 10.1006/exnr.1999.7128. [DOI] [PubMed] [Google Scholar]
- 309.Pettegrew JW. Panchalingam K. Klunk WE. McClure RJ. Muenz LR. Alterations of cerebral metabolism in probable Alzheimer's disease: a preliminary study. Neurobiol Aging. 1994;15:117–132. doi: 10.1016/0197-4580(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 310.Picklo MJ. Montine TJ. Amarnath V. Neely MD. Carbonyl toxicology and Alzheimer's disease. Toxicol Appl Pharmacol. 2002;184:187–197. doi: 10.1006/taap.2002.9506. [DOI] [PubMed] [Google Scholar]
- 311.Planel E. Miyasaka T. Launey T. Chui DH. Tanemura K. Sato S. Murayama O. Ishiguro K. Tatebayashi Y. Takashima A. Alterations in glucose metabolism induce hypothermia leading to tau hyperphosphorylation through differential inhibition of kinase and phosphatase activities: implications for Alzheimer's disease. J Neurosci. 2004;24:2401–2411. doi: 10.1523/JNEUROSCI.5561-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Pocernich CB. Lange ML. Sultana R. Butterfield DA. Nutritional approaches to modulate oxidative stress in Alzheimer's disease. Curr Alzheimer Res. 2011;8:452–469. doi: 10.2174/156720511796391908. [DOI] [PubMed] [Google Scholar]
- 313.Poon HF. Frasier M. Shreve N. Calabrese V. Wolozin B. Butterfield DA. Mitochondrial associated metabolic proteins are selectively oxidized in A30P alpha-synuclein transgenic mice—a model of familial Parkinson's disease. Neurobiol Dis. 2005;18:492–498. doi: 10.1016/j.nbd.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 314.Poon HF. Hensley K. Thongboonkerd V. Merchant ML. Lynn BC. Pierce WM. Klein JB. Calabrese V. Butterfield DA. Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice—a model of familial amyotrophic lateral sclerosis. Free Radic Biol Med. 2005;39:453–462. doi: 10.1016/j.freeradbiomed.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 315.Rakhit R. Cunningham P. Furtos-Matei A. Dahan S. Qi XF. Crow JP. Cashman NR. Kondejewski LH. Chakrabartty A. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J Biol Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 316.Ramakrishnan P. Dickson DW. Davies P. Pin1 colocalization with phosphorylated tau in Alzheimer's disease and other tauopathies. Neurobiol Dis. 2003;14:251–264. doi: 10.1016/s0969-9961(03)00109-8. [DOI] [PubMed] [Google Scholar]
- 317.Rapoport SI. In vivo PET imaging and postmortem studies suggest potentially reversible and irreversible stages of brain metabolic failure in Alzheimer's disease. Eur Arch Psychiatry Clin Neurosci. 1999;249(Suppl 3):46–55. doi: 10.1007/pl00014174. [DOI] [PubMed] [Google Scholar]
- 318.Rauniyar N. Prokai L. Detection and identification of 4-hydroxy-2-nonenal Schiff-base adducts along with products of Michael addition using data-dependent neutral loss-driven MS3 acquisition: method evaluation through an in vitro study on cytochrome c oxidase modifications. Proteomics. 2009;9:5188–5193. doi: 10.1002/pmic.200900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Reaume AG. Elliott JL. Hoffman EK. Kowall NW. Ferrante RJ. Siwek DF. Wilcox HM. Flood DG. Beal MF. Brown RH., Jr. Scott RW. Snider WD. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 320.Reed T. Perluigi M. Sultana R. Pierce WM. Klein JB. Turner DM. Coccia R. Markesbery WR. Butterfield DA. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer's disease. Neurobiol Dis. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 321.Reed TT. Pierce WM., Jr. Turner DM. Markesbery WR. Butterfield DA. Proteomic identification of nitrated brain proteins in early Alzheimer's disease inferior parietal lobule. J Cell Mol Med. 2009;13:2019–2029. doi: 10.1111/j.1582-4934.2008.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Reed TT. Pierce WM. Markesbery WR. Butterfield DA. Proteomic identification of HNE-bound proteins in early Alzheimer disease: insights into the role of lipid peroxidation in the progression of AD. Brain Res. 2009;1274:66–76. doi: 10.1016/j.brainres.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 323.Reiner M. Bloch W. Addicks K. Functional interaction of caveolin-1 and eNOS in myocardial capillary endothelium revealed by immunoelectron microscopy. J Histochem Cytochem. 2001;49:1605–1610. doi: 10.1177/002215540104901214. [DOI] [PubMed] [Google Scholar]
- 324.Rhee SG. Chae HZ. Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 325.Riederer P. Sofic E. Rausch WD. Schmidt B. Reynolds GP. Jellinger K. Youdim MB. Transition metals, ferritin, glutathione, and ascorbic acid in parkinsonian brains. J Neurochem. 1989;52:515–520. doi: 10.1111/j.1471-4159.1989.tb09150.x. [DOI] [PubMed] [Google Scholar]
- 326.Riviere S. Birlouez-Aragon I. Nourhashemi F. Vellas B. Low plasma vitamin C in Alzheimer patients despite an adequate diet. Int J Geriatr Psychiatry. 1998;13:749–754. doi: 10.1002/(sici)1099-1166(1998110)13:11<749::aid-gps860>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 327.Roe MR. McGowan TF. Thompson LV. Griffin TJ. Targeted 18O-labeling for improved proteomic analysis of carbonylated peptides by mass spectrometry. J Am Soc Mass Spectrom. 2010;21:1190–1203. doi: 10.1016/j.jasms.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Rossor MN. Iversen LL. Johnson AJ. Mountjoy CQ. Roth M. Cholinergic deficit in frontal cerebral cortex in Alzheimer's disease is age dependent. Lancet. 1981;2:1422. doi: 10.1016/s0140-6736(81)92836-1. [DOI] [PubMed] [Google Scholar]
- 329.Saigoh K. Wang YL. Suh JG. Yamanishi T. Sakai Y. Kiyosawa H. Harada T. Ichihara N. Wakana S. Kikuchi T. Wada K. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 330.Sajdel-Sulkowska EM. Marotta CA. Alzheimer's disease brain: alterations in RNA levels and in a ribonuclease-inhibitor complex. Science. 1984;225:947–949. doi: 10.1126/science.6206567. [DOI] [PubMed] [Google Scholar]
- 331.Salmon M. Dedessus Le Moutier J. Wenders F. Chiarizia S. Eliaers F. Remacle J. Royer V. Pascal T. Toussaint O. Role of the PLA2-independent peroxiredoxin VI activity in the survival of immortalized fibroblasts exposed to cytotoxic oxidative stress. FEBS Lett. 2004;557:26–32. doi: 10.1016/s0014-5793(03)01437-6. [DOI] [PubMed] [Google Scholar]
- 332.Sayre LM. Zelasko DA. Harris PL. Perry G. Salomon RG. Smith MA. 4-Hydroxynonenal-derived advanced lipid peroxidation end products are increased in Alzheimer's disease. J Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- 333.Schapira AH. Mitochondrial involvement in Parkinson's disease, Huntington's disease, hereditary spastic paraplegia and Friedreich's ataxia. Biochim Biophys Acta. 1999;1410:159–170. doi: 10.1016/s0005-2728(98)00164-9. [DOI] [PubMed] [Google Scholar]
- 334.Scheff SW. Price DA. Synaptic pathology in Alzheimer's disease: a review of ultrastructural studies. Neurobiol Aging. 2003;24:1029–1046. doi: 10.1016/j.neurobiolaging.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 335.Scheff SW. Price DA. Alzheimer's disease-related alterations in synaptic density: neocortex and hippocampus. J Alzheimers Dis. 2006;9:101–115. doi: 10.3233/jad-2006-9s312. [DOI] [PubMed] [Google Scholar]
- 336.Schilling G. Becher MW. Sharp AH. Jinnah HA. Duan K. Kotzuk JA. Slunt HH. Ratovitski T. Cooper JK. Jenkins NA. Copeland NG. Price DL. Ross CA. Borchelt DR. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 337.Schneider C. Porter NA. Brash AR. Autoxidative transformation of chiral omega6 hydroxy linoleic and arachidonic acids to chiral 4-hydroxy-2E-nonenal. Chem Res Toxicol. 2004;17:937–941. doi: 10.1021/tx049913n. [DOI] [PubMed] [Google Scholar]
- 338.Schulze PC. Yoshioka J. Takahashi T. He Z. King GL. Lee RT. Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. J Biol Chem. 2004;279:30369–30374. doi: 10.1074/jbc.M400549200. [DOI] [PubMed] [Google Scholar]
- 339.Schutkowski M. Bernhardt A. Zhou XZ. Shen M. Reimer U. Rahfeld JU. Lu KP. Fischer G. Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry. 1998;37:5566–5575. doi: 10.1021/bi973060z. [DOI] [PubMed] [Google Scholar]
- 340.Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 341.Sergeant N. Wattez A. Galvan-valencia M. Ghestem A. David JP. Lemoine J. Sautiere PE. Dachary J. Mazat JP. Michalski JC. Velours J. Mena-Lopez R. Delacourte A. Association of ATP synthase alpha-chain with neurofibrillary degeneration in Alzheimer's disease. Neurosci. 2003;117:293–303. doi: 10.1016/s0306-4522(02)00747-9. [DOI] [PubMed] [Google Scholar]
- 342.Shaw PJ. Ince PG. Falkous G. Mantle D. Oxidative damage to protein in sporadic motor neuron disease spinal cord. Ann Neurol. 1995;38:691–695. doi: 10.1002/ana.410380424. [DOI] [PubMed] [Google Scholar]
- 343.Sherer TB. Betarbet R. Greenamyre JT. Environment, mitochondria, and Parkinson's disease. Neuroscientist. 2002;8:192–197. doi: 10.1177/1073858402008003004. [DOI] [PubMed] [Google Scholar]
- 344.Shibata N. Nagai R. Uchida K. Horiuchi S. Yamada S. Hirano A. Kawaguchi M. Yamamoto T. Sasaki S. Kobayashi M. Morphological evidence for lipid peroxidation and protein glycoxidation in spinal cords from sporadic amyotrophic lateral sclerosis patients. Brain Res. 2001;917:97–104. doi: 10.1016/s0006-8993(01)02926-2. [DOI] [PubMed] [Google Scholar]
- 345.Shinder GA. Lacourse MC. Minotti S. Durham HD. Mutant Cu/Zn-superoxide dismutase proteins have altered solubility and interact with heat shock/stress proteins in models of amyotrophic lateral sclerosis. J Biol Chem. 2001;276:12791–12796. doi: 10.1074/jbc.M010759200. [DOI] [PubMed] [Google Scholar]
- 346.Shults CW. Oakes D. Kieburtz K. Beal MF. Haas R. Plumb S. Juncos JL. Nutt J. Shoulson I. Carter J. Kompoliti K. Perlmutter JS. Reich S. Stern M. Watts RL. Kurlan R. Molho E. Harrison M. Lew M. Effects of coenzyme Q10 in early Parkinson disease: evidence of slowing of the functional decline. Arch Neurol. 2002;59:1541–1550. doi: 10.1001/archneur.59.10.1541. [DOI] [PubMed] [Google Scholar]
- 347.Siciliano G. Piazza S. Carlesi C. Del Corona A. Franzini M. Pompella A. Malvaldi G. Mancuso M. Paolicchi A. Murri L. Antioxidant capacity and protein oxidation in cerebrospinal fluid of amyotrophic lateral sclerosis. J Neurol. 2007;254:575–580. doi: 10.1007/s00415-006-0301-1. [DOI] [PubMed] [Google Scholar]
- 348.Siems WG. Hapner SJ. van Kuijk FJ. 4-hydroxynonenal inhibits Na(+)-K(+)-ATPase. Free Radic Biol Med. 1996;20:215–223. doi: 10.1016/0891-5849(95)02041-1. [DOI] [PubMed] [Google Scholar]
- 349.Simmons DA. Casale M. Alcon B. Pham N. Narayan N. Lynch G. Ferritin accumulation in dystrophic microglia is an early event in the development of Huntington's disease. Glia. 2007;55:1074–1084. doi: 10.1002/glia.20526. [DOI] [PubMed] [Google Scholar]
- 350.Singh AK. Gupta S. Jiang Y. Oxidative stress and protein oxidation in the brain of water drinking and alcohol drinking rats administered the HIV envelope protein, gp120. J Neurochem. 2008;104:1478–1493. doi: 10.1111/j.1471-4159.2007.05094.x. [DOI] [PubMed] [Google Scholar]
- 351.Sly WS. Hewett-Emmett D. Whyte MP. Yu YS. Tashian RE. Carbonic anhydrase II deficiency identified as the primary defect in the autosomal recessive syndrome of osteopetrosis with renal tubular acidosis and cerebral calcification. Proc Natl Acad Sci U S A. 1983;80:2752–2756. doi: 10.1073/pnas.80.9.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Smith CD. Carney JM. Starkereed PE. Oliver CN. Stadtman ER. Floyd RA. Markesbery WR. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer-disease. Proc Natl Acad Sci U S A. 1991;88:10540–10543. doi: 10.1073/pnas.88.23.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Smith MA. Sayre LM. Anderson VE. Harris PL. Beal MF. Kowall N. Perry G. Cytochemical demonstration of oxidative damage in Alzheimer disease by immunochemical enhancement of the carbonyl reaction with 2,4-dinitrophenylhydrazine. J Histochem Cytochem. 1998;46:731–735. doi: 10.1177/002215549804600605. [DOI] [PubMed] [Google Scholar]
- 354.Smith PJ. Tappel AL. Chow CK. Glutathione peroxidase activity as a function of dietary selenomethionine. Nature. 1974;247:392–393. doi: 10.1038/247392a0. [DOI] [PubMed] [Google Scholar]
- 355.Smith RG. Henry YK. Mattson MP. Appel SH. Presence of 4-hydroxynonenal in cerebrospinal fluid of patients with sporadic amyotrophic lateral sclerosis. Ann Neurol. 1998;44:696–699. doi: 10.1002/ana.410440419. [DOI] [PubMed] [Google Scholar]
- 356.Snell RG. MacMillan JC. Cheadle JP. Fenton I. Lazarou LP. Davies P. MacDonald ME. Gusella JF. Harper PS. Shaw DJ. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 357.Sorolla MA. Reverter-Branchat G. Tamarit J. Ferrer I. Ros J. Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45:667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 358.Sousa LP. Silva BM. Brasil BS. Nogueira SV. Ferreira PC. Kroon EG. Kato K. Bonjardim CA. Plasminogen/plasmin regulates alpha-enolase expression through the MEK/ERK pathway. Biochem Biophys Res Commun. 2005;337:1065–1071. doi: 10.1016/j.bbrc.2005.09.154. [DOI] [PubMed] [Google Scholar]
- 359.Sperandio S. Poksay KS. Schilling B. Crippen D. Gibson BW. Bredesen DE. Identification of new modulators and protein alterations in non-apoptotic programmed cell death. J Cell Biochem. 2010;111:1401–1412. doi: 10.1002/jcb.22870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Stadtman ER. Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 361.Subramaniam R. Roediger F. Jordan B. Mattson MP. Keller JN. Waeg G. Butterfield DA. The lipid peroxidation product, 4-hydroxy-2-trans-nonenal, alters the conformation of cortical synaptosomal membrane proteins. J Neurochem. 1997;69:1161–1169. doi: 10.1046/j.1471-4159.1997.69031161.x. [DOI] [PubMed] [Google Scholar]
- 362.Sultana R. Boyd-Kimball D. Cai J. Pierce WM. Klein JB. Merchant M. Butterfield DA. Proteomics analysis of the Alzheimer's disease hippocampal proteome. J Alzheimers Dis. 2007;11:153–164. doi: 10.3233/jad-2007-11203. [DOI] [PubMed] [Google Scholar]
- 363.Sultana R. Boyd-Kimball D. Poon HF. Cai J. Pierce WM. Klein JB. Merchant M. Markesbery WR. Butterfield DA. Redox proteomics identification of oxidized proteins in Alzheimer's disease hippocampus and cerebellum: an approach to understand pathological and biochemical alterations in AD. Neurobiol Aging. 2006;27:1564–1576. doi: 10.1016/j.neurobiolaging.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 364.Sultana R. Butterfield DA. Oxidatively modified GST and MRP1 in Alzheimer's disease brain: implications for accumulation of reactive lipid peroxidation products. Neurochem Res. 2004;29:2215–2220. doi: 10.1007/s11064-004-7028-0. [DOI] [PubMed] [Google Scholar]
- 365.Sultana R. Butterfield DA. Oxidatively modified, mitochondria-relevant brain proteins in subjects with Alzheimer disease and mild cognitive impairment. J Bioenerg Biomembr. 2009;41:441–446. doi: 10.1007/s10863-009-9241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Sultana R. Butterfield DA. Identification of the oxidative stress proteome in the brain. Free Radic Biol Med. 2011;50:487–494. doi: 10.1016/j.freeradbiomed.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Sultana R. Mecocci P. Mangialasche F. Cecchetti R. Baglioni M. Butterfield DA. Increased protein and lipid oxidative damage in mitochondria isolated from lymphocytes from patients with Alzheimer's disease: insights into the role of oxidative stress in Alzheimer's disease and initial investigations into a potential biomarker for this dementing disorder. J Alzheimers Dis. 2011;24:77–84. doi: 10.3233/JAD-2011-101425. [DOI] [PubMed] [Google Scholar]
- 368.Sultana R. Perluigi M. Butterfield DA. Protein oxidation and lipid peroxidation in brain of subjects with Alzheimer's disease: insights into mechanism of neurodegeneration from redox proteomics. Antioxid Redox Signal. 2006;8:2021–2037. doi: 10.1089/ars.2006.8.2021. [DOI] [PubMed] [Google Scholar]
- 369.Sultana R. Perluigi M. Butterfield DA. Oxidatively modified proteins in Alzheimer's disease (AD), mild cognitive impairment and animal models of AD: role of Abeta in pathogenesis. Acta Neuropathol. 2009;118:131–150. doi: 10.1007/s00401-009-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Sultana R. Perluigi M. Newman SF. Pierce WM. Cini C. Coccia R. Butterfield DA. Redox proteomic analysis of carbonylated brain proteins in mild cognitive impairment and early Alzheimer's disease. Antioxid Redox Signal. 2010;12:327–336. doi: 10.1089/ars.2009.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Sultana R. Poon HF. Cai J. Pierce WM. Merchant M. Klein JB. Markesbery WR. Butterfield DA. Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 372.Sultana R. Reed T. Perluigi M. Coccia R. Pierce WM. Butterfield DA. Proteomic identification of nitrated brain proteins in amnestic mild cognitive impairment: a regional study. J Cell Mol Med. 2007;11:839–851. doi: 10.1111/j.1582-4934.2007.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Surmeli NB. Litterman NK. Miller AF. Groves JT. Peroxynitrite mediates active site tyrosine nitration in manganese superoxide dismutase. Evidence of a role for the carbonate radical anion. J Am Chem Soc. 2010;132:17174–17185. doi: 10.1021/ja105684w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Suzuki YJ. Carini M. Butterfield DA. Protein carbonylation. Antioxid Redox Signal. 2010;12:323–325. doi: 10.1089/ars.2009.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Szabo C. Multiple pathways of peroxynitrite cytotoxicity. Toxicol Lett. 2003;140–141:105–112. doi: 10.1016/s0378-4274(02)00507-6. [DOI] [PubMed] [Google Scholar]
- 376.Tamarit J. Cabiscol E. Ros J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem. 1998;273:3027–3032. doi: 10.1074/jbc.273.5.3027. [DOI] [PubMed] [Google Scholar]
- 377.Temple A. Yen TY. Gronert S. Identification of specific protein carbonylation sites in model oxidations of human serum albumin. J Am Soc Mass Spectrom. 2006;17:1172–1180. doi: 10.1016/j.jasms.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 378.Thaw P. Baxter NJ. Hounslow AM. Price C. Waltho JP. Craven CJ. Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones. Nat Struct Biol. 2001;8:701–704. doi: 10.1038/90415. [DOI] [PubMed] [Google Scholar]
- 379.Thompson JE. Hopkins MT. Taylor C. Wang TW. Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci. 2004;9:174–179. doi: 10.1016/j.tplants.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 380.Thompson KJ. Shoham S. Connor JR. Iron and neurodegenerative disorders. Brain Res Bull. 2001;55:155–164. doi: 10.1016/s0361-9230(01)00510-x. [DOI] [PubMed] [Google Scholar]
- 381.Trettel F. Rigamonti D. Hilditch-Maguire P. Wheeler VC. Sharp AH. Persichetti F. Cattaneo E. MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 382.Trujillo M. Folkes L. Bartesaghi S. Kalyanaraman B. Wardman P. Radi R. Peroxynitrite-derived carbonate and nitrogen dioxide radicals readily react with lipoic and dihydrolipoic acid. Free Radic Biol Med. 2005;39:279–288. doi: 10.1016/j.freeradbiomed.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 383.Tsujimoto KH. Kawai T. Matsumoto H. Oxidized protein quantitation method using isotope-substituted labeling reagent and mass spectrometry. pp 2007-JP55617 2007111193, 2020070320.
- 384.Uchida K. Stadtman ER. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra- and intermolecular cross-linking reaction. J Biol Chem. 1993;268:6388–6393. [PubMed] [Google Scholar]
- 385.Valencia CA. Ju W. Liu R. Matrin 3 is a Ca2+/calmodulin-binding protein cleaved by caspases. Biochem Biophys Res Commun. 2007;361:281–286. doi: 10.1016/j.bbrc.2007.06.156. [DOI] [PubMed] [Google Scholar]
- 386.Valente EM. Salvi S. Ialongo T. Marongiu R. Elia AE. Caputo V. Romito L. Albanese A. Dallapiccola B. Bentivoglio AR. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol. 2004;56:336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- 387.Valentine JS. Do oxidatively modified proteins cause ALS? Free Radic Biol Med. 2002;33:1314–1320. doi: 10.1016/s0891-5849(02)01080-8. [DOI] [PubMed] [Google Scholar]
- 388.Van Laar VS. Dukes AA. Cascio M. Hastings TG. Proteomic analysis of rat brain mitochondria following exposure to dopamine quinone: implications for Parkinson disease. Neurobiol Dis. 2008;29:477–489. doi: 10.1016/j.nbd.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Vana L. Kanaan NM. Hakala K. Weintraub ST. Binder LI. Peroxynitrite-induced nitrative and oxidative modifications alter tau filament formation. Biochemistry. 2011;50:1203–1212. doi: 10.1021/bi101735m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Vayssiere JL. Cordeau-Lossouarn L. Larcher JC. Basseville M. Gros F. Croizat B. Participation of the mitochondrial genome in the differentiation of neuroblastoma cells. In Vitro Cell Dev Biol. 1992;28A:763–772. doi: 10.1007/BF02631065. [DOI] [PubMed] [Google Scholar]
- 391.Vexler ZS. Wong A. Francisco C. Manabat C. Christen S. Tauber M. Ferriero DM. Gregory G. Fructose-1,6-bisphosphate preserves intracellular glutathione and protects cortical neurons against oxidative stress. Brain Res. 2003;960:90–98. doi: 10.1016/s0006-8993(02)03777-0. [DOI] [PubMed] [Google Scholar]
- 392.Vila A. Tallman KA. Jacobs AT. Liebler DC. Porter NA. Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Viner RI. Ferrington DA. Williams TD. Bigelow DJ. Schoneich C. Protein modification during biological aging: selective tyrosine nitration of the SERCA2a isoform of the sarcoplasmic reticulum Ca2+-ATPase in skeletal muscle. Biochem J. 1999;340(Pt 3):657–669. [PMC free article] [PubMed] [Google Scholar]
- 394.Wallimann T. Hemmer W. Creatine kinase in non-muscle tissues and cells. Mol Cell Biochem. 1994;133–134:193–220. doi: 10.1007/BF01267955. [DOI] [PubMed] [Google Scholar]
- 395.Wang J. Xu G. Gonzales V. Coonfield M. Fromholt D. Copeland NG. Jenkins NA. Borchelt DR. Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol Dis. 2002;10:128–138. doi: 10.1006/nbdi.2002.0498. [DOI] [PubMed] [Google Scholar]
- 396.Wang X. Moualla D. Wright JA. Brown DR. Copper binding regulates intracellular alpha-synuclein localisation, aggregation and toxicity. J Neurochem. 2010;113:704–714. doi: 10.1111/j.1471-4159.2010.06638.x. [DOI] [PubMed] [Google Scholar]
- 397.Wang XS. Lee S. Simmons Z. Boyer P. Scott K. Liu W. Connor J. Increased incidence of the Hfe mutation in amyotrophic lateral sclerosis and related cellular consequences. J Neurol Sci. 2004;227:27–33. doi: 10.1016/j.jns.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 398.Watanabe M. Dykes-Hoberg M. Culotta VC. Price DL. Wong PC. Rothstein JD. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 399.Weitzdoerfer R. Fountoulakis M. Lubec G. Aberrant expression of dihydropyrimidinase related proteins-2,-3 and 4 in fetal Down syndrome brain. J Neural Transm Suppl. 2001;61:95–107. doi: 10.1007/978-3-7091-6262-0_8. [DOI] [PubMed] [Google Scholar]
- 400.Winklhofer KF. The parkin protein as a therapeutic target in Parkinson's disease. Expert Opin Ther Targets. 2007;11:1543–1552. doi: 10.1517/14728222.11.12.1543. [DOI] [PubMed] [Google Scholar]
- 401.Winterbourn CC. Buss IH. Protein carbonyl measurement by enzyme-linked immunosorbent assay. Methods Enzymol. 1999;300:106–111. doi: 10.1016/s0076-6879(99)00118-4. [DOI] [PubMed] [Google Scholar]
- 402.Yamakura F. Matsumoto T. Fujimura T. Taka H. Murayama K. Imai T. Uchida K. Modification of a single tryptophan residue in human Cu,Zn-superoxide dismutase by peroxynitrite in the presence of bicarbonate. Biochim Biophys Acta. 2001;1548:38–46. doi: 10.1016/s0167-4838(01)00212-6. [DOI] [PubMed] [Google Scholar]
- 403.Yamashiro S. Yamakita Y. Ono S. Matsumura F. Fascin, an actin-bundling protein, induces membrane protrusions and increases cell motility of epithelial cells. Mol Biol Cell. 1998;9:993–1006. doi: 10.1091/mbc.9.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Yang L. Calingasan NY. Wille EJ. Cormier K. Smith K. Ferrante RJ. Beal MF. Combination therapy with coenzyme Q10 and creatine produces additive neuroprotective effects in models of Parkinson's and Huntington's diseases. J Neurochem. 2009;109:1427–1439. doi: 10.1111/j.1471-4159.2009.06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Yarian CS. Rebrin I. Sohal RS. Aconitase and ATP synthase are targets of malondialdehyde modification and undergo an age-related decrease in activity in mouse heart mitochondria. Biochem Biophys Res Commun. 2005;330:151–156. doi: 10.1016/j.bbrc.2005.02.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Yoo BS. Regnier FE. Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin-fluorescein affinity staining. Electrophoresis. 2004;25:1334–1341. doi: 10.1002/elps.200405890. [DOI] [PubMed] [Google Scholar]
- 407.Yoritaka A. Hattori N. Uchida K. Tanaka M. Stadtman ER. Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc Natl Acad Sci U S A. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Yoshida H. Watanabe A. Ihara Y. Collapsin response mediator protein-2 is associated with neurofibrillary tangles in Alzheimer's disease. J Biol Chem. 1998;273:9761–9768. doi: 10.1074/jbc.273.16.9761. [DOI] [PubMed] [Google Scholar]
- 409.Zeitlin S. Liu JP. Chapman DL. Papaioannou VE. Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 410.Zhang SM. Hernan MA. Chen H. Spiegelman D. Willett WC. Ascherio A. Intakes of vitamins E and C, carotenoids, vitamin supplements, and PD risk. Neurology. 2002;59:1161–1169. doi: 10.1212/01.wnl.0000028688.75881.12. [DOI] [PubMed] [Google Scholar]
- 411.Zhou J. Yu Q. Chng WJ. TXNIP (VDUP-1, TBP-2): a major redox regulator commonly suppressed in cancer by epigenetic mechanisms. Int J Biochem Cell Biol. 2011;43:1668–1673. doi: 10.1016/j.biocel.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 412.Zhou XZ. Kops O. Werner A. Lu PJ. Shen M. Stoller G. Kullertz G. Stark M. Fischer G. Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]