Abstract
Inactivating mutations in the Staphylococcus aureus virulence regulator agr are associated with worse outcomes in bacteremic patients. However, whether agr dysfunction is primarily a cause or a consequence of early bacteremia is unknown. Analysis of 158 paired S. aureus clones from blood and nasal carriage sites in individual patients revealed that recovery of an agr-defective mutant from blood was usually predicted by the agr functionality of carriage isolates. Many agr-positive blood isolates produced low levels of hemolytic toxins, but levels were similar to those of colonizing strains within patients, suggesting that introduction into the blood did not select for mutations with minor functional effects. Evidently, the transition from commensalism to opportunism in S. aureus does not require full virulence in hospitalized patients. Furthermore, agr-defective mutants were found in uninfected nasal carriers in the same proportion as in carriers who develop bacteremia, suggesting low correlation between virulence and infectivity.
Primary control of the staphylococcal virulon is mediated by the accessory gene regulator (agr) quorum-sensing system [1]. agr-defective mutants are attenuated for virulence in animal models of acute infection, and agents that block agr and quorum-sensing exhibit antiinfective properties [2]. However, agr-defective mutants are frequently recovered from patients with bacteremia, where the mutants are associated with persistent infection and poor outcome [3, 4]. The frequent recovery of agr− and mixtures of agr+ and agr− phenotypes from patients supports the idea that agr variation is selected for in vivo. However, it is unknown whether and to what extent loss-of-function mutations arise during the initial invasive stage of blood infection in human hosts.
In the present work, we sought to determine the frequency of a within-host shift in agr functionality during infection by screening for mutational changes in the agr locus among 158 Staphylococcus aureus clone pairs obtained from blood and nasal sites of individual patients [5]. Additionally, when a clonal S. aureus isolate was recovered from the clinically presumed focus of infection, strains were characterized to determine their role as a reservoir for agr-defective mutants. To address the directionality of colonization and infection, we included in the analysis carriage strains isolated both before and after the detection of bacteremia. Finally, to determine whether fully virulent, agr+ strains were more likely to cause invasive disease than agr− strains, we compared the frequency of agr dysfunction in colonizing strains from bacteremic patients and uninfected controls.
We report that agr− strains were frequently recovered from patients with S. aureus bacteremia and that clones having the same inactivating agr mutation were recovered from colonizing sites within the same individual. Furthermore, agr− strain frequencies were similar among carriers who developed bacteremia and controls who did not, providing evidence that variation in agr was not correlated with progression to disease. Taken together, these results support the importance of variables other than agr functionality, such as host risk factors, the mode of bloodstream invasion, and possibly inoculum size, in determining the invasiveness of S. aureus in hospitalized patients.
MATERIALS AND METHODS
Staphylococcus aureus Isolates
Staphylococcus aureus isolates were obtained from 2 patient populations as part of previous study [5]. The first population consisted of 158 pairs of genotypically isogenic blood and nasal isolates from single patients, collected in a German multicenter study that included general and intensive care units at 32 university and community hospitals. Nasal specimens were obtained for culture immediately after the isolation of S. aureus from the blood. Twenty-two pairs of isolates from an original set of 180 were lost. In 48 of the 158 patients, an additional clonal S. aureus isolate was recovered from the clinically presumed focus of infection. Sites and types of infection included intravenous catheter–related infections, osteomyelitis, skin and soft tissue infections, and lower-respiratory-tract infections (Supplementary Table 1). The second population consisted of 221 single-patient nasal isolates prospectively collected over a 6-year period at a single tertiary-care hospital in Germany (Supplementary Table 2). In addition to the nasal isolates, 12 clonal S. aureus blood isolates were collected from colonized patients who subsequently developed S. aureus bacteremia (Supplementary Table 1). None of the other 209 nasally colonized patients developed S. aureus bacteremia during the study. Staphylococcus aureus from 14 patients (8.2%) with bacteremia from the 2 parts of the study and from 6 uninfected nasal carriers (2.7%) harbored methicillin-resistant S. aureus strains, previously confirmed by testing for the mecA gene by polymerase chain reaction (PCR) [5].
Screening for Hemolytic Activity as an Indicator of agr Function
Hemolysin production in S. aureus can be used to approximate agr activity because δ-hemolysin is a translation product of agr RNAIII and because α-hemolysin and the phenol-soluble modulins (PSMs; a family of peptides that include δ-hemolysin) are upregulated by RNAIII and agrA, respectively [6, 7]. Production of δ-hemolysin and other PSMs can be semiquantitatively assayed on sheep blood agar (SBA) by virtue of their synergism with β-hemolysin [8, 9]. Individual colonies can be analyzed for these toxins by cross-streaking against RN4220, which produces only β-hemolysin. However, because cross-streaking of unfractionated cultures obscures heterogeneity, we first plated diluted cultures onto SBA and scored for α-hemolysin production, which causes direct hemolysis. In the case of α-hemolysin–negative populations, SBA plates were coated with a β-hemolysin–containing culture supernatant, as described elsewhere [10]. A total of 50–100 colonies per sample were surveyed from each culture to ensure the phenotypes reported were those that dominated in the original sample. Cultures were provisionally scored as hemolytic, nonhemolytic, or, when >30% of colonies were hemolytically distinct, mixed.
Exoprotein Profiles
Cultures were grown in trypticase soy broth (TSB), and 1.5-mL aliquots were centrifuged to remove bacteria. Culture supernatants were precipitated with a 10% volume of 50% trichloroacetic acid, and the pellet was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis according to the method of Laemmli [11].
RNA Extraction and Quantitative Nucleic Acid Sequence–Based Amplification (NASBA) Assay
Cultures were grown in TSB for 4 hours (optical density, approximately 0.9) at 37°C with shaking, and cell pellets were treated with lysostaphin (AMBI) for 30 minutes at 37°C. RNA was purified using the Qiagen RNeasy kit, and its integrity was checked by agarose gel electrophoresis [12]. RNAIII expression was quantified using a real-time NASBA assay, as previously described [13]. NASBA reactions were performed on a NucliSens EasyQ analyzer (BioMérieux), using the NucliSENS EasyQ Basic Kit Version 2 (BioMérieux). Standard curves were made using 10-fold serial dilutions of in vitro transcribed RNAIII and gyrB targets (Stratagene, Agilent Technologies). Time to positivity (TTP) was defined as the time at which the fluorescence signal rose 1.25-fold above the background emitted over the course of the first 5 measurements [13]. The RNAIII expression levels for RNA preparations were calculated from the standard curves and expressed as TTP ratios (TTPgyrB/TTPRNAIII). A TTPgyrB/TTPRNAIII ratio of 1.10 was used as a cutoff to define the lower limit of agr functionality, as previously described [13].
Genotyping
The relatedness of strains from the blood, nares, and presumed foci of the same patients, as well as variants from mixed agr+ and agr− cultures, was confirmed by DNA sequence analysis of the protein A gene variable repeat region (spa typing) [14]. spa types were used to identify multilocus sequence typing (MLST) clonal complexes, using the Ridom SpaServer database (available at: http://spa.ridom.de/mlst.shtml) [15].
Staphylococcus aureus isolates can be divided into 4 predominant agr groups on the basis of the specificity of the autoinducing peptide, encoded within agrD, for its membrane sensor (AgrC). All isolates were previously assigned to one of the 4 major agr groups by PCR-based assays [16].
agr Sequencing
Nucleotide sequences were determined for the agrA and agrC genes and for the entire locus when mutations were found to be absent in these genes by PCR and automated DNA sequencing, as described elsewhere [10, 17]. Sequences were compared with the agr sequences of strains from the appropriate agr specificity group—NCTC 8325, D302, Not266, and RN9107 (agr group I); D61, JH1, and C126 (agr group II); MRSA252 and MW2 (agr group III); and RN4850 (agr group IV)—by means of a sequence analysis suite (DNAStar). When genotypically isogenic mixtures of agr+ and agr− organisms were recovered, we sequenced the agr+ component. When an agr− mutant was characterized by a single amino acid substitution resulting in a missense mutation and the strain was not from a mixed culture, agr sequences were compared to a genotypically isogenic agr+ clone from a different site in the same patient or from a different patient in the same collection. Missense substitutions that were predicted to affect protein function were identified by the SIFT (sorting intolerant from tolerant) algorithm [18].
Statistical Analysis
Analyses were performed in the R statistical environment (http://www.R-project.org). Categorical variables were compared using a 2-tailed Fisher exact test. spa type diversity was measured by the Simpson index of diversity (1-D), using the “vegan” package for R (http://CRAN.R-project.org/package=vegan) [19]. A P value of < .05 was considered statistically significant, and no adjustments were made for multiple comparisons.
RESULTS
When considering the within-host dynamics of agr function, we hypothesize that agr+ strains are more likely to initiate the change from benign colonization to active infection and that the subsequent state of infection, in this case bacteremia, provides the necessary milieu for the emergence of mutations in the agr locus. To test this hypothesis, we screened for hemolysin activity, which is widely used to distinguish agr+ from agr− staphylococci [3, 4, 10, 20, 21], among clones from multiple sites in individual patients with bacteremia. Two strain populations from a previous study were analyzed [5]. The first consisted of 364 S. aureus strains, isolated from 158 patients, from whom 158 strains were isolated from the blood, 158 from the anterior nares, and 48 from the clinically presumed focus of infection. The clonality of isolates from individual patients was confirmed in the original analysis by pulsed-field gel electrophoresis (PFGE) and here by spa typing (Supplementary Table 1). Strains were considered clonal if they had identical PFGE banding patterns and spa types. Fifteen of 158 blood isolates (9.5%) were found to be purely agr defective (ie, negative for synergistic hemolysis) (Table 1 and Supplementary Table 1). agr Dysfunction was detected with a similar prevalence among nares and foci isolates: 14 nares isolates (8.8%) and 4 foci isolates (8.3%) were either agr defective or, in one case, contained a mixture of agr+ and agr− organisms (patient 158, foci strain 652; Supplementary Table 1). In the case of the mixture, spa typing confirmed that subpopulations were naturally occurring variants from the same progenitor strain, indicating that the allelic split occurred very recently, probably within the patient.
Table 1.
Characteristics of agr-Defective Staphylococcus aureus Strains Recovered From Patients
| Genotype |
Blood |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Patient No. | spa Type | spa Motif | CCa | Reference Sequence | agr Group | Strain No. | δ-Hemolysin | agr Mutationb | Predicted Result |
| 1 | 16 | 4 | YHFGFMBQBLO | 8 | NCTC8325 | 1 | 4 | − | 126-IS256 | Frameshift-truncated AgrC |
| 1 | 24 | 21 | UJGBBGGJAGJ | 15 | D61 | 2 | 13 | − | g517a | Ala>Thr at aa 173 in AgrAc |
| 1 | 30 | 532 | WGKAKAOM | 30 | MRSA252 | 3 | 19 | − | No mutation found | None |
| 1 | 34 | 576 | ZDGMDMGMMM | 101 | Not266 | 1 | 26 | − | a409 (−1 bp), g52a | Frameshift-truncated AgrC, Ala>Thr at aa 18 in AgrAd |
| 1 | 46 | 2 | TJMBMDMGMK | 5 | JH1 | 2 | 46 | Weak | No mutation found | None |
| 1 | 53 | 363 | YGFMBQBLO | 8 | NCTC8325 | 1 | 54 | − | g1133a | Gly>Asp at aa 378 in AgrCe |
| 1 | 60 | 385 | TO2MBMDMGMK | 5 | JH1 | 2 | 61 | − | c136t | Gln>Stop at aa 46 in AgrA |
| 1 | 67 | 852 | UJGJAGJ | 15 | D61 | 2 | 78 | − | c771t, c583t | Pro>Pro at aa 316 in AgrC, Arg>Cys at aa 195 in AgrAc |
| 1 | 73 | 33 | WGKAKAOMQQ | 30 | MRSA252 | 3 | 85 | − | t224a | Ile>Asn at aa 75 in AgrAe |
| 1 | 77 | 755 | UJGPLM | 12 | C126 | 2 | 91 | − | t313 (+1 bp), a662 g | Frameshift-truncated AgrC, Tyr>Cys at aa 221 in AgrAc |
| 1 | 84 | 1 | YHGFMBQBLO | 8 | NCTC8325 | 1 | 99 | + | … | … |
| 1 | 110 | 33 | WGKAKAOMQQ | 30 | MRSA252 | 3 | 130 | − | t485c | Phe>Ser at aa 162 in AgrCe |
| 1 | 117 | 44 | WGMQ | 30 | MRSA252 | 3 | 143 | − | 955–985 (−30 bp) | Frameshift-truncated AgrC |
| 1 | 119 | 16 | WGKAKAOMQQQ | 30 | MRSA252 | 3 | 146 | + | … | … |
| 1 | 120 | 664 | YC2BQBLO | 8 | NCTC8325 | 1 | 147 | − | a86c | Asp>Ala at aa 29 in AgrAe |
| 1 | 123 | 1365 | ZFFGU2DMGGM | 25 | D302 | 1 | 150 | + | … | … |
| 1 | 135 | 53 | YGFMBQBLQBLPO | 8 | NCTC8325 | 1 | 165 | − | 121-IS256 | Frameshift- truncated AgrC |
| 1 | 136 | 3 | WGKAOMQ | 8 | NCTC8325 | 1 | 166 | + | … | … |
| 1 | 140 | 184 | ZFGU2DMGGM | 25 | D302 | 1 | 171 | − | t349 (+1 bp), g543t | Frameshift-truncated AgrC, Gln>His at aa 181 in AgrCc |
| 1 | 146 | 385 | TO2MBMDMGMK | 5 | JH1 | 2 | 180 | − | g739a | Glu>Lys at aa 247 in AgrCc |
| 2 | 5 | 1405 | UJJKBPE | 30 | MW2 | 3 | 14 | − | c254a, a514g | Thr>Asn at aa 85 AgrCc, Ile>Val at aa 172 AgrAc |
| 2 | 7 | 43 | WGKAKAOMQ | 30 | MRSA252 | 3 | 18 | − | 667–677 (−11 bp) | Frameshift-truncated AgrC |
| Nasal |
Foci |
|||||||||
| Group | Patient No. | Strain No. | δ-Hemolysin | agr Mutationb | Predicted Result | Strain No. | Specimen Source | δ-Hemolysin | agr Mutationb | Predicted Result |
| 1 | 16 | 13 | − | 126-IS256 | Frameshift-truncated AgrC | … | … | … | … | … |
| 1 | 24 | 39 | − | t826 (−1 bp) | Frameshift-truncated AgrC | … | … | … | … | … |
| 1 | 30 | 54 | − | No mutation found | None | … | … | … | … | … |
| 1 | 34 | 75 | − | a409 (−1 bp), g52a | Frameshift-truncated AgrC, Ala>Thr at aa 18 in AgrAd | … | … | … | … | … |
| 1 | 46 | 138 | − | g1177t | Gly>Cys at 393 aa in AgrCe | … | … | … | … | … |
| 1 | 53 | 164 | + | … | … | … | … | … | … | … |
| 1 | 60 | 191 | + | … | … | … | … | … | … | … |
| 1 | 67 | 253 | − | c771t, c583t | Pro>Pro at aa 316 in AgrC, Arg>Cys at aa 195 in AgrAc | … | … | … | … | … |
| 1 | 73 | 277 | + | … | … | … | … | … | … | … |
| 1 | 77 | 295 | − | t313 (+1 bp), a662 g | Frameshift-truncated AgrC, Tyr>Cys at aa 221 in AgrAc | 296 | Catheter | − | t313 (+1 bp), a662 g | Frameshift-truncated AgrC, Tyr>Cys at aa 221 in AgrAc |
| 1 | 84 | 326 | − | t1107 (+1 bp) | Frameshift-truncated AgrC | … | … | … | … | … |
| 1 | 110 | 430 | + | … | … | … | … | … | … | … |
| 1 | 117 | 466 | + | … | … | 468 | CSF | + | … | … |
| 1 | 119 | 479 | − | t47a | Lys>Met at aa 16 in AgrA | … | … | … | … | … |
| 1 | 120 | 484 | − | a86c | Asp>Ala at aa 29 in AgrAe | … | … | … | … | … |
| 1 | 123 | 494 | − | t2c | Val>Ala at aa 1 in AgrCe | … | … | … | … | … |
| 1 | 135 | 545 | + | … | … | 546 | Wound | − | 121-IS256 | Frameshift-truncated AgrC |
| 1 | 136 | 549 | − | 78–84 (−7 bp) | Frameshift-truncated AgrA | 550 | Catheter | + | … | … |
| 1 | 140 | 564 | − | t349 (+1 bp), g543t | Frameshift-truncated AgrC, Gln>His at aa 181 in AgrCc | 566 | Catheter | − | t349 (+1 bp), g543t | Frameshift-truncated AgrC, Gln>His at aa 181 in AgrCc |
| 1 | 146 | 595 | − | g739a | Glu>Lys at aa 247 in AgrCc | … | … | … | … | … |
| 2 | 5 | 13 | − | c254a, a514 g | Thr>Asn at aa 85 AgrCc, Ile>Val at aa 172 AgrAc | … | … | … | … | … |
| 2 | 7 | 17 | − | 667–677 (−11 bp)g | Frameshift-truncated AgrC | … | … | … | … | … |
Group 1 indicates retrospectively collected strain pairs from patients with bacteremia, and group 2 (in bold) indicates prospectively collected strain pairs from patients who subsequently developed bacteremia (see text).
Abbreviations: −, negative, +, positive; aa, amino acid; bp, base pair; CSF, cerebral spinal fluid.
a spa-type deduced clonal complex (CC).
b Designation corresponds to the region in agrA or agrC of the genome sequenced isolate of the appropriate agr group (Materials and Methods).
c Tolerated by the sorting intolerant from tolerant (SIFT) algorithm.
d Insignificant polymorphism found in closely related agr-positive clone.
e Not tolerated by the SIFT algorithm.
f Not tolerated by the SIFT algorithm (low confidence prediction).
g The indicated mutation was identified in a subpopulation of the culture, which contained additional agr-defective species.
Among the 158 patients with S. aureus bacteremia, only 11 were nasally colonized with a strain of S. aureus that differed in agr functionality, indicating that a shift in agr function was uncommon in hosts with S. aureus bacteremia. However, among the 15 patients with agr-defective bacteremia, 5 (33%) did not have an agr-defective isolate in their nares or in their foci (see patient 135), whereas of the 143 patients with agr-positive bacteremia, only 5 (3.5%) were nasally colonized with an agr-defective strain (P < .001 by the Fisher exact test). Thus, a shift to agr positivity in bloodstream isolates occurs less frequently than does the reverse.
Identity of mutations among agr-defective isolates from different sites implies a clonal origin. We therefore determined the basis of agr dysfunction in all hemolysin-negative strains by nucleotide sequencing of the agrA and agrC region of the locus. Previous work comparing functional and nonfunctional staphylococcal strains indicates that inactivating mutations are localized to these regions of the locus in most cases [10, 20, 21]. Indeed, putative inactivating mutations were identified in the coding region of these genes for all but 1 of the 15 agr− blood strains. The variants were characterized by frameshift indels (insertions/deletions; n = 6), nonsense mutations (n = 1), and nonsynonymous changes (n = 12). Thus, a variety of types of mutation, including a high frequency of insertion and deletion events, contributed to the production of the agr variants, consistent with the results of previous studies [20–22].
Examination of agr-defective clones from blood and nares indicated that in all but one case the putative inactivating agr mutation of isolates from the blood was the same as that from the nares of the same patients (7 of 8 agr− blood and nasal mutant pairs). Additionally, agr mutants were isolated from 3 foci; all 3 patients had the same mutations in the blood, and 2 patients had the same agr mutations at all 3 sites. Thus, although agr diversification occurs at a sufficient frequency in vivo to result in mixed infections, most patients possess a predominant population of 1 mutant (derived from single organisms).
It is possible that agr mutants originating during wound infection and subsequent bacteremia could result in subsequent self-inoculation of the nares. To address the issue, a second population was studied in which nasal swabs were prospectively collected from 221 patients (Supplementary Table 2), 12 of whom subsequently developed bacteremia (Table 1 and Supplementary Table 1). In the 2 cases of agr-defective bacteremia, nasal colonization with an agr-defective S. aureus preceded the recovery of a clonally identical mutant in the blood (Table 1). Thus, the results of the multicenter study were confirmed by the prospective study. Together the 2 showed that 70.6% of the patients with agr-defective S. aureus bacteremia were colonized in the anterior nares or infected at foci by the same mutant. Apparently, agr dysfunction primarily precedes new-onset bloodstream infection and therefore is not required for subsequent development of bacteremia.
The apparent lack of a within-host shift in agr function could have been due to inadequate screening. For example, closer comparison of colonizing and invasive organisms may reveal more subtle adaptive changes rather than discrete agr+ or agr− phenotypes, perhaps owing to partial loss-of-function mutations in the locus that reduce but do not eliminate agr activation. To address this possibility, hemolytic toxin levels were assayed among agr+ strain pairs from blood and nasal sites in the 2 study populations. Strikingly, in each case, toxin levels among strains were indistinguishable. Thus, selection for mutants with attenuated (or enhanced) virulence in the colonizing population did not correlate with the transition from commensal to pathologic habitats.
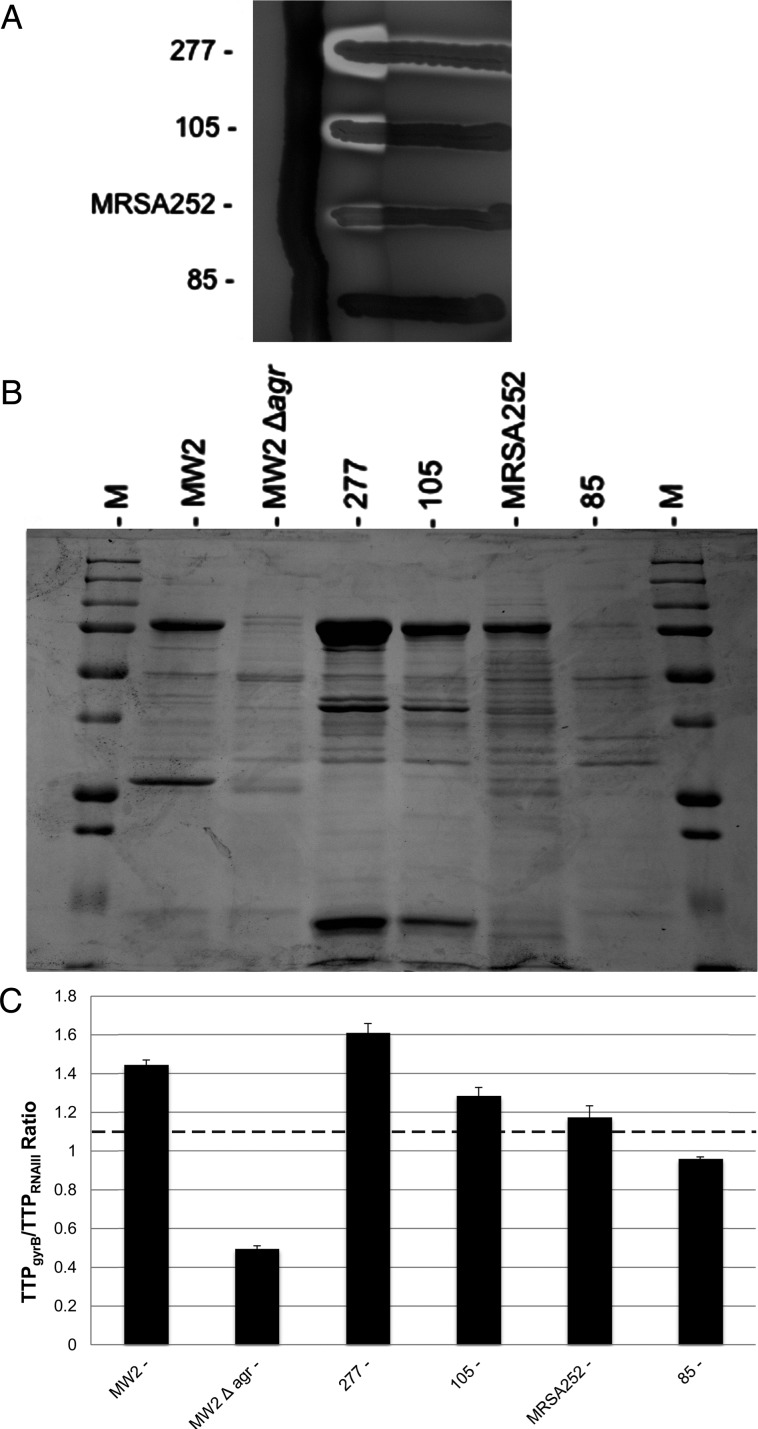
Analysis of hemolytic toxin levels revealed that production was weak but not absent in many strain pairs, the majority of which belonged to the clonal complex 30 (CC30) genotype (Supplementary Table 3). DeLeo and colleagues recently reported that contemporary CC30 strains have a mutation in agrC, resulting in an amino acid change at Gly55 (AgrC G55R) that attenuates agr function [23]. While this substitution was present in all CC30 strains with reduced hemolysin production, several CC30 strains in which it was present showed robust hemolysis and RNAIII expression (Table 2 and Figure 1). This distribution of phenotypes to strong and weak agr activity may be explained by plasticity of mutational effects in different genetic backgrounds. In support of this hypothesis, phenotypically agr-defective CC30 strains did not consistently harbor secondary mutations that would be predicted to fully inactivate the locus (Table 2).
Table 2.
Characteristics of Clonal Complex (CC) 30 Blood Isolates
| Patient No. | Strain No. | spa Type | spa Motif | CCa | δ-Hemolysin | agr Mutationb | Predicted Result |
|---|---|---|---|---|---|---|---|
| 167 | 211 | 43 | WGKAKAOMQ | 30 | + | No mutation found | None |
| 70 | 81 | 33 | WGKAKAOMQQ | 30 | + | No mutation found | None |
| 119 | 146 | 16 | WGKAKAOMQQQ | 30 | + | No mutation found | None |
| 56 | 57 | 19 | XKAKAOMQ | 30 | + | No mutation found | None |
| 122 | 149 | 1384 | WGKAKAOLOMQQ | 30 | Weak | g272a | Ser>Asn at aa 91 AgrCc |
| 163 | 204 | 43 | WGKAKAOMQ | 30 | Weak | t487g | Ser>Ala at aa 163 AgrCd |
| 33 | 22 | 43 | WGKAKAOMQ | 30 | Weak | No mutation found | None |
| 80 | 95 | 43 | WGKAKAOMQ | 30 | Weak | No mutation found | None |
| 89 | 109 | 43 | WGKAKAOMQ | 30 | Weak | No mutation found | None |
| 92 | 105 | 43 | WGKAKAOMQ | 30 | Weak | No mutation found | None |
| 15 | 3 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 72 | 84 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 75 | 88 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 87 | 102 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 94 | 112 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 103 | 122 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 109 | 129 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 118 | 144 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 131 | 159 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 154 | 190 | 33 | WGKAKAOMQQ | 30 | Weak | No mutation found | None |
| 40 | 40 | 16 | WGKAKAOMQQQ | 30 | Weak | No mutation found | None |
| 96 | 114 | 16 | WGKAKAOMQQQ | 30 | Weak | No mutation found | None |
| 157 | 194 | 19 | XKAKAOMQ | 30 | Weak | No mutation found | None |
| 30 | 19 | 532 | WGKAKAOM | 30 | − | No mutation found | None |
| 73 | 85 | 33 | WGKAKAOMQQ | 30 | − | t224a | Ile>Asn at aa 75 in AgrAc |
| 110 | 130 | 33 | WGKAKAOMQQ | 30 | − | t485c | Phe>Ser at aa 162 in AgrCc |
Abbreviations: −, negative; +, positive; aa, amino acid.
a spa-type-deduced CC. Primary grouping of strains is by agr functionality, followed by polymorphism of spa.
b Designation corresponds to the region in agrC or agrA of genome-sequenced CC30 prototype strain MRSA252, which has an AgrC G55R amino acid change that attenuates agr function (see text).
c Not tolerated by the sorting intolerant from tolerant (SIFT) algorithm.
d Tolerated by the SIFT algorithm.
Figure 1.
A representative subset of clonal complex 30 strains showing various levels of agr activity. A, Cross-streaking alongside the β-hemolysin producing strain RN4220; differentiation of the various hemolytic activities in Staphylococcus aureus can be scored on sheep blood agar by virtue of their synergism with β-hemolysin. Strain 277, strain 105, and weakly agr-positive reference strain MRSA252 have identical agr sequences, including an AgrC G55R amino acid change that attenuates agr function (Table 2) [23]. Strain 85 has the same G55R amino acid change but is agr defective, likely owing to an additional mutation in agrA (Table 1 and Table 2). B, Exoprotein profiles prepared from culture supernatants analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. M, protein ladder; MW2 and MW2 Δagr, control strains. C, Relative quantitation of RNAIII by quantitative nucleic acid sequence–based amplification analysis and the time-to-positivity (TTP) ratio (TTPgyrB/TTPRNAIII). Data are representative of 3 separate experiments. Dotted line, TTP ratio of 1.10, indicating the cutoff for agr functionality; MW2 and MW2 Δagr, control strains.
Although S. aureus strains with low virulence are apparently capable of invading host tissues, reduced virulence may slow the rate of invasion. To address this issue, we compared the frequency of agr-defective mutants among nasal isolates from bacteremic patients with nasal isolates from patients in the second study who never developed infection. agr dysfunction was present in 10.0% of nasal isolates (17 of 170) from bacteremic patients and in 9.6% (20 of 209) from uninfected carriers. Thus, agr-defective mutants were not more likely to be found in the nares of asymptomatic carriers than those of bacteremic patients, suggesting that agr-defective mutants possess the same infectivity as fully virulent strains. Comparison of spa type diversity among nasal isolates from bacteremic patients with those of uninfected patients demonstrated that both groups were nonclonal and equally diverse (Table 3). Thus, there was no evidence of clustering of genotypes by invasiveness, and infections did not reflect dissemination of an outbreak strain, which could skew results.
Table 3.
Diversity of Staphylococcus aureus Genotypes Among 170 Bacteremic Patients and 209 Colonized Uninfected Controls
| Major spa Typea | spa Motif | CCb | Bacteremic | Colonized Uninfected | P Valuec |
|---|---|---|---|---|---|
| 2 | TJMBMDMGMK | 5 | 4.7 (8) | 1 (2) | .05 |
| 21 | UJGBBGGJAGJ | 15 | 9.4 (16) | 9.1 (19) | 1 |
| 43 | WGKAKAOMQ | 30 | 4.1 (7) | 3.4 (7) | .79 |
| 33 | WGKAKAOMQQ | 30 | 7.6 (13) | 7.2 (15) | 1 |
| 73 | XKAKBEMBKB | 45 | 3.5 (6) | 4.8 (10) | .62 |
| 1 | YHGFMBQBLO | 8 | 3.5 (6) | 1.9 (4) | .35 |
| 201 | ZDGMDMGMM | 101 | 3.5 (6) | 1.9 (4) | .35 |
| 184 | ZFGU2DMGGM | 25 | 3.5 (6) | 4.3 (9) | .79 |
| 113 | TJEJNCMOMOKR | 22 | 0.6 (1) | 2.4 (5) | .23 |
| Otherd | … | … | 75.3 (128) | 70.8 (148) | .35 |
| SID | … | … | 0.98 | 0.98 |
Data are percentage (occurrence), unless otherwise indicated.
Abbreviations: CC, clonal complex; SID, Simpson index of diversity.
a Defined as ≥5 strains with the same spa type.
b Isolates broadly grouped according to their evolutionary relatedness.
c By the Fisher exact test.
d Minor spa types. Two colonizing isolates were non–spa typeable.
Because molecular typing can be used to reconstruct transmission networks, we sought evidence for transmission of agr mutations among clones from the 2 study populations. We found that identical agr variants were present in each of 2 groups of genotypically related strains selected from among all 329 patients. The first group of 2 strains, isolates W9 and 103, came from the colonized but uninfected study population (Supplementary Table 2). They were obtained from the nares of 2 patients from the same hospital. The second group of 4 strains, isolates 12, 78, 91, and 135, came from 4 patients, 3 of whom were uninfected S. aureus carriers from the same hospital (patients 12, 85, and 125; Supplementary Table 2), and 1 of whom was from a patient with bacteremia from a different hospital (patient 67; Table 1 and Supplementary Table 1). The recovery of identical agr-defective alleles in different hosts suggests nosocomial cross-infection, with secondary transmission indicated in the case of the agr mutant that was found in multiple cultures. Consistent with this hypothesis, agr alleles in both strains of the first group and in one of the strains from the second group (isolate 78) were able to circulate long enough to accumulate subsequent mutations. Such changes would be expected if agr genes are not transcribed and their sequences are no longer subjected to selection. Furthermore, strains in the second group were distinct but closely related to each other by spa repeat region organization, indicating genotypic diversification of the clones subsequent to acquisition of their respective agr mutations. Collectively, these results suggest that while continuous, indefinite circulation of specific agr-defective alleles in natural populations may be uncommon [20], they can circulate long enough to be detected in epidemiologically unlinked hosts.
DISCUSSION
agr is a global regulator of staphylococcal virulence that, in vitro, coordinates a switch from an establishment mode, in which genes for adhesins and protective surface proteins are expressed, to an invasive mode, in which genes for factors that promote cell and tissue destruction are activated [1]. However, agr-defective mutants are frequently isolated from clinical material and laboratory cultures [3, 10, 20, 21, 24, 25]. Recent reports indicate that recovery of agr-defective strains from bacteremic patients is associated with persistent infection and worse outcomes [3, 4], perhaps owing to increased production of cell surface proteins that facilitate biofilm formation and immune evasion. However, lack of knowledge regarding the timing of changes in agr function during or prior to bacteremic infections complicates the interpretation of such data.
We report that among patients with S. aureus bacteremia, the agr function of paired strains cultured from blood and nares was most frequently identical, regardless of whether the nasal culture was obtained before or after the detection of bacteremia. This result suggests that attenuation in agr during the transition to bloodstream infection occurs in a minority of patients. The results suggest that factors such as hospitalization and intravenous catheter use permit S. aureus lacking full virulence to cause infection. Bacteria such as these express fewer and different virulence factors than fully virulent organisms, such as community-acquired MRSA strains, which are almost always agr+ [26, 27] and able to infect structurally and functionally immune competent hosts. The data also show that agr+ colonization frequencies were not greater in the noses of infected patients than in those of uninfected controls. Thus, lack of secreted toxins did not adversely affect invasiveness.
In a previous report, we showed that genotypic diversification subsequent to agr inactivation is uncommon among S. aureus isolated from clinical infections, indicating that agr-defective mutants are often short-lived [20]. At the same time, the success with which the CC30 clone has established itself in natural populations of S. aureus indicates that mutations with more moderate or weak functional effects, such as the amino acid change at Gly55 [23], can demonstrate distinctive population dynamics. Indeed, the differential populational stability of agr mutations appears to be inversely proportional to the degree of functional effect induced. This pattern may result from functional trade-offs, wherein completely inactivating mutations may promote survival in certain host niches in the short term but represent a liability to clones over the longer term.
The main strategy adopted for silencing the agr locus usually relies on mutations in agrCA genes. However, the ability of the G55R amino acid change to inactivate the locus appears to depend on an appropriate genetic background, either through direct effects on agr genes themselves or the "fine-tuning" of the interaction between genetic background and agr response. This interaction may provide the CC30 lineage a unique mechanism to resolve adaptive conflicts that arise from having alleles under negative selection in one environment but under positive selection in others.
Our study has a number of limitations. First, study data may not be generalizable to nonstudy areas. Second, correlations between agr functionality and bacteremia may reflect factors we did not account for, such as duration of hospitalization. Third, our data are limited to bacteremia, so agr functionality patterns between colonizing and infecting sites in different syndromes might be different. Finally, the data do not capture cases of undiagnosed bacteremia. Antibiotic treatment prior to sample collection would both decrease the rate of overall disease detected and potentially increase the proportion of agr-positive strains that may be linked to more severe disease.
In summary, the results shed light on the role of agr dysfunction in the causal pathway leading to complicated disease and poor outcomes with which these strains are associated [3, 4]. A better understanding of the biological detail of such pathways is needed before assuming the general applicability of attempts to block infection by interfering with agr-mediated virulence. A critical question is to determine which steps during infection involve agr evolution. In this connection, future work will determine whether complicated S. aureus bacteremia is more likely to have developed as a consequence of being infected with an agr− isolate or whether an agr− isolate is more likely to develop in a patient who presents with bacteremia complicated by deep-seated infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgment. We thank Karl Drlica for critical comments on the manuscript.
Financial support. This work was supported by an American Heart Association Fellow-to-Faculty Transition Award (to B. S.), a grant from the Cystic Fibrosis Foundation (to B. S.), the National Institutes of Health (grant R01-AI30138 to R. P. N.), and the Bundesministerium für Bildung und Forschung (grant 01KI1009A to K. B. and G. P. and grant 0315832A to K. B.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–49. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 2.Wright JS, 3rd, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci U S A. 2005;102:1691–6. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler VG, Jr, Sakoulas G, McIntyre LM, et al. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis. 2004;190:1140–9. doi: 10.1086/423145. [DOI] [PubMed] [Google Scholar]
- 4.Schweizer ML, Furuno JP, Sakoulas G, et al. Increased mortality with accessory gene regulator (agr) dysfunction in Staphylococcus aureus among bacteremic patients. Antimicrob Agents Chemother. 2011;55:1082–7. doi: 10.1128/AAC.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001;344:11–6. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 6.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 7.Queck SY, Jameson-Lee M, Villaruz AE, et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–8. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung GY, Duong AC, Otto M. Direct and synergistic hemolysis caused by Staphylococcus phenol-soluble modulins: implications for diagnosis and pathogenesis. Microbes Infect. 2012;14:380–6. doi: 10.1016/j.micinf.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elek SD, Levy E. Distribution of haemolysins in pathogenic and non-pathogenic staphylococci. J Pathol Bacteriol. 1950;62:541–54. doi: 10.1002/path.1700620405. [DOI] [PubMed] [Google Scholar]
- 10.Traber KE, Lee E, Benson S, et al. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265–74. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Shopsin B, Zhao Y, et al. Real-time nucleic acid sequence-based amplification (NASBA) assay for rapid detection and quantification of agr functionality in clinical Staphylococcus aureus isolates. J Clin Microbiol. 2012;50:657–61. doi: 10.1128/JCM.06253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shopsin B, Gomez M, Montgomery SO, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–63. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmsen D, Claus H, Witte W, Rothganger J, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–8. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Eiff C, Friedrich AW, Peters G, Becker K. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis. 2004;49:157–62. doi: 10.1016/j.diagmicrobio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Traber K, Novick R. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol Microbiol. 2006;59:1519–30. doi: 10.1111/j.1365-2958.2006.04986.x. [DOI] [PubMed] [Google Scholar]
- 18.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–6. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shopsin B, Eaton C, Wasserman GA, et al. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2010;202:1593–9. doi: 10.1086/656915. [DOI] [PubMed] [Google Scholar]
- 21.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004;190:1498–505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- 22.Shopsin B, Drlica-Wagner A, Mathema B, Adhikari RP, Kreiswirth BN, Novick RP. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J Infect Dis. 2008;198:1171–4. doi: 10.1086/592051. [DOI] [PubMed] [Google Scholar]
- 23.Deleo FR, Kennedy AD, Chen L, et al. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc Natl Acad Sci U S A. 2011;108:18091–6. doi: 10.1073/pnas.1111084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boles BR, Horswill AR. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Somerville GA, Beres SB, Fitzgerald JR, et al. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J Bacteriol. 2002;184:1430–7. doi: 10.1128/JB.184.5.1430-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakoulas G. The accessory gene regulator (agr) in methicillin-resistant Staphylococcus aureus: role in virulence and reduced susceptibility to glycopeptide antibiotics. Drug Discovery Today. 2006;3:287–94. [Google Scholar]
- 27.Tsuji BT, MacLean RD, Dresser LD, McGavin MJ, Simor AE. Impact of accessory gene regulator (agr) dysfunction on vancomycin pharmacodynamics among Canadian community and health-care associated methicillin-resistant Staphylococcus aureus. Ann Clin Microbiol Antimicrob. 2011;10:20. doi: 10.1186/1476-0711-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.