Abstract
Bone morphogenetic proteins (BMPs) are secreted signaling proteins – they transduce their signals by by assembling complexes comprised of one of three known type II receptors and one of four known type I receptors. BMP-9 binds and signals through the type I receptor Alk1, but not other Alks, while BMP-2, -4, and -7 bind and signal through Alk3, and the close homolog Alk6, but not Alk1. The present results, which include the determination of the Alk1 structure using NMR and identification of residues important for binding using SPR, show that the β-strand framework of Alk1 is highly similar to Alk3, yet there are significant differences in loops shown previously to be important for binding. The most pronounced difference is in the N-terminal portion of the β4-β5 loop, which is structurally ordered and includes a similarly placed but shorter helix in Alk1 compared to Alk3. The altered conformation of the β4-β5 loop, and to lesser extent β1-β2 loop, cause clashes when Alk1 is positioned onto BMP-9 in the manner that Alk3 is positioned onto BMP-2. This necessitates an alternative manner of binding, which is supported by a model of the BMP-9:Alk1 complex constructed using the program RosettaDock. The model shows that Alk1 is positioned similar to Alk3, but is rotated by 40 degrees. The alternate positioning allows Alk1 to bind BMP-9 through a large hydrophobic interface, consistent with mutational analysis that identified several residues in the central portion of the β4-β5 loop that contribute significantly to binding and are non-conservatively substituted relative to the corresponding residues in Alk3.
Bone morphogenetic proteins (BMPs) are small secreted signaling proteins that regulate embryonic patterning and organ development and maintain and regenerate tissues (1–3). They are present in both invertebrates and vertebrates and are the ancestors of an extended family of signaling proteins, known as the TGF-β superfamily (4). The other members of the superfamily include the closely related growth and differentiation factors (GDFs), which regulate cartilage and skeletal development, the activins and inhibins which regulate cell growth and the release of pituitary hormones, and the TGF-βs, which regulate cellular growth and differentiation. The superfamily has expanded as eukaryotes have diversified, with three known ligands in C. elegans, seven in D. melanogaster, and more than thirty, including more than fifteen known BMPs and GDFs, in humans.
BMPs and other proteins of the TGF-β superfamily transduce their signals by binding and bringing together two serine/threonine kinase receptors, known as type I and type II receptors (5). This initiates a phosphorylation cascade in which the constitutively active type II receptor phosphorylates conserved serines within a negative regulatory domain of the type I receptor (6). Activated type I receptor in turn phosphorylates downstream signaling messengers called receptor-regulated Smads, or R-Smads, to relay the signal from cell membrane to the nucleus (7). There are seven type I and five type II receptors in humans and other higher vertebrates. The type I receptors, which include activin-like kinases 1–7 (or Alk1–7), can be further divided into subgroups based on the ligand they bind and the R-Smad they activate. The type I receptors Alk1, Alk2, Alk3, and Alk6 principally bind BMPs and GDFs and activate R-Smads 1, 5, and 8, whereas Alk4, Alk5, and Alk7 bind ligands such as activins, myostatin and TGF-βs and activate R-Smads 2 and 3. Activated R-Smads form a complex with a common partner, Smad4, and translocate to the nucleus where they regulate transcription of target genes (7). R-Smads 2 and 3 and 1, 5, and 8 assemble distinct transcriptional complexes and target distinct genes, segregating the activities of BMPs and GDFs from activins, myostatin, and TGF-βs (8).
Alk1 and endoglin, an unrelated cell surface proteoglycan with a large ectodomain and a short non-catalytic cytoplasmic domain, are expressed mainly by endothelial cells and have been linked to hereditary hemorrhagic telangiectasia (HHT), a disease caused by the fragility of blood vessels (9). Alk1 and endoglin null mice have been further shown to have major defects in vasculogenesis and are embryonic lethal (10–12). Alk1 is required for the inhibition of endothelial cell proliferation and migration and balances the activity of factors, such as BMP-2 and BMP-7, which stimulate endothelial cell proliferation and migration (13). Alk1 was considered an orphan receptor for many years, although recently BMP-9, as well as the closely related ligand BMP-10, have been identified as high affinity ligands for both Alk1 (14) and endoglin (15). Isolated preparations of the Alk1 and endoglin extracellular domains have been further shown to non-competitively bind BMP-9 (16, 17), suggesting a common mechanism (i.e. BMP-9 binding) by which mutations in Alk1 and endoglin lead to HHT.
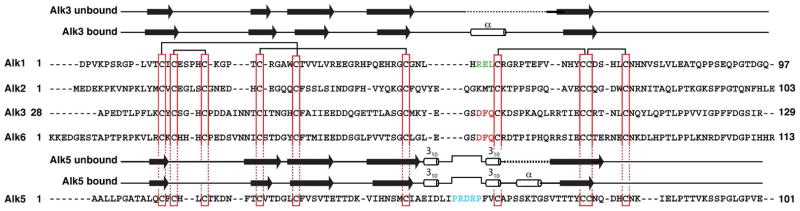
Alk1 binds BMP-9 with higher affinity than many other Alks, including Alk3 and its close homolog, Alk6 (14, 18). Alk3, in contrast, binds ligands such as BMP-2, -4, and -7 with greater affinity than Alk1 (14, 18). This suggests that these otherwise similar receptors have acquired local amino acid or structural differences that underlie their unique specificity. Structural studies have shown that BMP-2 binds Alk3 through a short solvent exposed α-helix that undergoes a disorder-to-order transition upon binding (19, 20). Sequence alignments show that two residues identified as critical for binding of Alk3, Phe85 and Gln86 (21), are replaced with a glutamate and a leucine, respectively, in Alk1 (Fig. 1). Structural analysis of Alk3 bound to BMP-2 has shown that the phenyl ring of Phe85 forms a knob that fits into a hole formed between two tryptophan residues found in most ligands of the superfamily (19), whereas the sidechain amide group of Gln86 forms a hydrogen bond to the backbone (21). The demonstrated importance of these residues for ligand binding, together with the demonstrated specificity differences between Alk1 and Alk3, suggests that the distinct specificity of Alk1 and Alk3 for different BMPs might arise from these amino acid differences.
Figure 1.
Sequence alignment of the Alk1 extracellular domain with the extracellular domains of the BMP type I receptors Alk2, Alk3 and Alk6 and the TGF-β type I receptor, Alk5. Conserved cysteines (vertical red boxes) and disulphide bonds (horizontal black bars) in the Alk3 (19, 20), Alk5 (39, 40), and Alk6 (41) structures that define the receptor three finger toxin fold are highlighted. Functionally important residues in the helical region of Alk3 and Alk6 are highlighted in red, while those in the pre-helical extension of Alk5 are highlighted in cyan. Proposed functionally important residues in Alk1 are highlighted in green. Unbound and bound secondary structural elements shown above the Alk1, Alk2, Alk3, and Alk6 sequences correspond to those from the solution structure of Alk3 (PDB 2K3G) and the crystal structure of Alk3 bound to BMP-2 (PDB 1REW); those above the Alk5 sequence correspond to those from the solution structure of Alk5 (2L5S) and the crystal structure of Alk5 bound to the TGF-β3:TβR-II complex (PDB 2PJY).
To investigate the underlying specificity differences between Alk1 and Alk3, the structure of the unbound form of the Alk1 extracellular domain was determined using nuclear magnetic resonance (NMR) spectroscopy. The secondary structure and overall fold of Alk1 were found to be highly similar to Alk3, however in contrast to Alk3, the helix in the binding region was shown to be both pre-formed and largely rigid on the ns-ps time scale. The pre-formed helix and rigidity in this region of Alk1 could account for its restricted binding compared to Alk3, with its Glu-Leu motif serving as a critical specificity determinant. This was evaluated by generating chimeras in which the Glu-Leu motif from Alk1 was swapped with the homologous Phe-Gln motif from Alk3.
Experimental Procedures
Chemicals and other reagents
Tris(hydroxymethyl)aminomethane (Tris), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), ethylenediaminetetraacetic acid (EDTA), sodium chloride, and oxidized and reduced glutathione were purchased from Sigma-Aldrich (St. Louis, MO). Tween 20, 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS) and CM-5 sensor chips were purchased from GE Healthcare (Piscataway, NJ). Human BMP-2 and human BMP-9 were purchased from R&D Systems (Minneapolis, MN). All other chemicals were reagent grade or better and were purchased from ThermoFischer Scientific (Waltham, MA).
Expression and purification of Alk1-ED
A DNA fragment corresponding to the full-length ectodomain of mature human Alk1 (22) was inserted between the XhoI and NdeI cleavage sites in pET15b (Novagen, Madison, WI). This construct, included in addition to residues 1–97 of the mature ectodomain, an N-terminal histidine tag followed by a thrombin cleavage site. The construct was overexpressed in E. coli BL21(DE3) cells cultured at 37 °C in LB medium containing 150 mg/L ampicillin. Protein expression was induced by adding 0.8 mM IPTG when the absorbance at 600 nm reached 0.6. Cells were harvested 6 – 8 hours after induction. Cell pellets from 6 L of culture were resuspended in 200 mL of lysis buffer (100 mM Tris, 10 mM EDTA, pH 8.3 with HCl) and sonicated. After sonication and three sequential washes with wash buffer (one time with 100 mM Tris-Cl, 10 mM EDTA, 1 M NaCl, pH 8.3 and two times with 100 mM Tris-Cl, 10 mM EDTA, 1% (v/v) TritonX-100, pH 8.3), the pellet was resuspended in 200 mL denaturing buffer (100 mM NaH2PO4, 10 mM Tris, 8 M Urea, pH 8.0) and stirred overnight at room temperature. The remaining insoluble material was removed by centrifugation and the supernatant was applied to 25 mL Ni-NTA resin (Qiagen, Valencia, CA) pre-equilibrated with 100 mL denaturing buffer followed by washes with 10 column volumes of denaturing buffer. The bound protein was eluted by applying 50 mL denaturing buffer containing 300 mM imidazole. The eluted protein was treated with reduced glutathione (final concentration 60 mM for 50 mL of protein sample) followed by stirring for 30 minutes at room temperature. The protein sample was diluted into 4 L pre-chilled refolding buffer (50 mM Tris, 5% glycerol, 0.5 mM oxidized glutathione, pH 9.0) and stirred for 24 – 36 hrs at 4 °C. The folding mixture was then applied to a bed of Ni-NTA resin pre-equilibrated with equilibration buffer (50 mM Tris, 5% glycerol, pH 8.3). Resin bound Alk1-ED was eluted with 50 mL equilibration buffer containing 300 mM imidazole. The eluted protein was incubated overnight with thrombin (4 units/mg of Alk1-ED) at 4° C to cleave the His-tag. After cleavage, the protein was applied to Source Q HPLC column and the bound protein was eluted with a 0 – 0.5 M linear NaCl gradient in 50 mM Tris-Cl, pH 8.5. Fractions were analyzed by non-reducing SDS-PAGE and those found to contain Alk1 monomers were pooled and applied to a 10 × 250 mm C18 reverse-phase HPLC column (Phenomenex, Torrance, CA) pre-equilibrated with 95% buffer “A” (99.9% v/v water, 0.1% v/v trifluoroacetic acid) and 5% buffer “B” (99.9% v/v acetonitrile, 0.1% v/v trifluoroacetic acid). Alk1-ED was eluted by stepping the buffer “B” up to 10% followed by a linear gradient from 10% B to 50% B at 2.5 mL/min over 15 column volumes. Fractions were analyzed by non-reducing SDS-PAGE and those found to contain Alk1 monomers were pooled and lyophilized.
Expression and purification of Alk3-ED
A DNA fragment corresponding to residues 28 – 129 of the mature of human Alk3 ectodomain (Alk3-ED) was inserted downstream of an expression cassette that included thioredoxin, a hexahistidine tag, and thrombin cleavage site in plasmid pET32a (Novagen, Madison, WI). The fusion protein was overexpressed in E. coli BL21(DE3) cells cultured at 37°C in LB medium containing 150 mg/L ampicillin; when the cells reached an absorbance at 600 nm of 0.5, the temperature was lowered to 20° C and grown for 30 minutes longer before inducing with 0.8 mM IPTG. The cells were incubated an additional 8 – 10 hours at 20 °C and then harvested by centrifugation. Cell pellets from 6 L of culture were resuspended in 200 mL of lysis buffer (25 mM sodium phosphate, 0.25 M NaCl and 7.5 mM imidazole, pH 8.0 and 1 mM PMSF) and sonicated. The lysate was centrifuged and the supernatant was loaded onto a column containing 25 mL of Ni-NTA resin equilibrated with lysis buffer. The column was washed with five column volumes of lysis buffer and the bound protein was eluted with a 0–300 mM linear gradient of imidazole in lysis buffer. The protein containing fractions were identified by SDS-PAGE and pooled. Solid urea was added to 6 M and after concentrating to 30 mL, glutathione was added to a final concentration of 30 mM. The reduced protein sample was slowly diluted into 900 mL of pre-chilled refolding buffer (100 mM Tris, 0.5 mM oxidized glutathione, pH 8.5) and stirred overnight at 4° C. The refolding mixture was concentrated to 75 mL, incubated with thrombin (3U/mg of protein) overnight at 4° C, dialyzed into 25 mM Tris-Cl, pH 8.0), and loaded onto a 25 mL Ni-NTA column that had been pre-equilibrated with Tris buffer. The column was washed with 100 mL of Tris buffer and the flow-through and wash, which contained Alk3-ED, were pooled and dialyzed three times against 4 L of 25 mM Hepes, pH 7.0. The dialyzed protein was loaded onto a Source Q anion-exchange HPLC column (GE Healthcare, Piscataway, NJ) and eluted with a 0.0 – 0.5 M linear NaCl gradient (buffer A - 25 mM Hepes, pH 7.0; buffer B - 25 mM Hepes, 0.5 M NaCl, pH 7.0). Fractions were analyzed by non-reducing SDS-PAGE and those found to contain Alk3-ED monomers were pooled and further fractionated on a C18 reverse phase column as for Alk1-ED.
Cell-based antagonist assays
Mouse NIH-3T3 fibroblasts were cultured to near confluency in DMEM and transiently transfected with a construct encoding full-length Alk1 or Alk3 under the control of a CMV promoter. Twelve hours post-transfection, the transfected cells transferred to serum free medium and cultured an additional six hours. The Alk1 transfected cells were then treated with 200 pM BMP-9 (Peprotech, Rocky Hill, NJ) with increasing concentrations of the purified Alk1 and Alk3 extracellular domains (40 nM, 400 nM, or 2000 nM for Alk1 and 40 nM, 400 nM, or 4000 nM for Alk3). The Alk3 transfected cells were treated with 800 pM BMP-2 (Peprotech, Rocky Hill, NJ) with increasing concentrations of the purified Alk1 and Alk3 extracellular domains (160 nM or 1600 nM for Alk1 and 160 nM, 1600 nM, or 16000 nM for Alk3). The cells were cultured an additional hour, isolated by centrifugation, and lysed in buffer containing protease and phosphatase inhibitors. The signaling activity was then assessed by subjecting 30 μg of protein from the soluble fraction to SDS-PAGE followed by Western blotting with a phospho-Smad1/5/8 antibody (Cell Signaling, Boston, MA). The total protein loaded and Smad expression levels were controlled by stripping the blot and probing with Smad5 (Abcam, Cambridge, MA) and GAPDH (Calbiochem, San Diego, CA) antibodies.
NMR samples and NMR data acquisition
Alk1-ED samples isotopically labeled with 15N or 15N and 13C for NMR were prepared by growing bacterial cells in M9 media containing 0.1 % (w/v) 15NH4Cl or 0.1 % (w/v) 15NH4Cl and 0.03% (w/v) 13C labeled glucose. All NMR samples were prepared in 25 mM sodium phosphate, 5% 2H2O, 0.02% w/v sodium azide at a protein concentration of 0.4 mM – 0.7 mM, pH 5.5 All NMR data was acquired at a sample temperature of 32 °C.
Backbone and side chain assignment
Backbone resonance assignments of Alk1-ED were obtained by collecting and analyzing sensitivity-enhanced triple-resonance data sets, including HNCACB (23), CBCA(CO)NH (24), C(CO)NH (25), HNCO (26), and HN(CA)CO (27). Aliphatic 1H and 13C assignments were obtained by collecting and analyzing HBHA(CO)NH (24), (H)CC(CO)NH (25), and H(CC)H-TOCSY (28) data sets.
Measurement of backbone 15N relaxation parameters
Backbone amide 15N T1, 15N T2, and {1H}-15N NOE relaxation parameters were measured in an interleaved manner at 300 °K at a 15N frequency of 60.8 MHz using 1H-detected pulse schemes previously described (29). The T1 and T2 data sets were each collected using 8 – 10 delay times, varying between 16 – 3200 ms and 16 – 192 ms, respectively. The T1 and T2 relaxation times were obtained by fitting relative peak intensities as a function of T1 or T2 delay time to a two parameter decaying exponential. {1H}-15N NOE values were obtained by taking the ratio of peak intensities from experiments performed with and without 1H presaturation and by applying a correction factor to account for the incomplete recovery of both 15N and 1H magnetization (30). All relaxation data were collected in duplicate and merged into single data set by averaging each value; error estimates were obtained from the corresponding standard deviations.
Analysis of backbone relaxation data
The overall rotational correlation time was determined by first using the criterion described by Barbato to identify any residues undergoing large amplitude motion on the ns-ps timescale or exchange (31). The trimmed data set (with these residues removed) was then analyzed using a conjugate gradient minimization procedure to identify the rotational correlation time by minimizing the difference between the calculated and experimental T1/T2 ratio (29). Internal dynamics were assessed by analyzing the experimental 15N relaxation parameters using the extended Model-Free formalism (32–34) with the overall correlation time derived from the analysis described above. Internal motional parameters were derived using the program ModelFree4, which employs F-statistics for model selection (35). Five different models for internal motion were considered, S2 (model 1), S2, τe (model 2), S2, Rex (model 3), S2, τe, Rex (model 4), and S2, S2f, τe (model 5).
Alk1 and Alk3 variants and characterization of their binding properties
Alk1-ED and Alk3-ED variants were generated by site directed mutagenesis of the corresponding expression plasmids using PCR (Quikchange, Stratagene, LaJolla, CA). All variants were expressed and purified as described for the corresponding wild type protein. Biacore 3000 biosensor instrument was used for quantitative measurement of the receptor-ligand interactions. BMP-9 and BMP-2 were diluted in 0.1 M acetic acid to a final concentration 2 – 6 μg/mL and coupled to the carboxymethylated dextran chip surface (CM5, GE Healthcare, Piscataway, NJ) using amine-coupling methods. The chip surface was first activated by injecting 35 μL of N-hydroxysuccinimide (NHS) and 1-ethyl-3′-(3-diethylaminopropyl) carbodiimide hydrochloride (EDC) followed by injection of 5 – 15 μL of the ligand to achieve the desired surface density (600–800 RU for BMP-9 and 2500 RU for BMP-2). Remaining activated groups were blocked by injecting 35 μL ethanolamine. One of the flow cells was activated and blocked without injecting any ligand as a reference surface. All injected samples were dialyzed against HBS-EP buffer (10 mM Hepes, 3 mM EDTA 500 mM NaCl, .002% surfactant P20, pH 7.4), centrifuged for 10 min at 50,000 x g, passed through a 0.2 mm filter prior to injection into the SPR instrument. Protein concentrations were determined based on the calculated extinction coefficient at 280 nm and the measured absorbance at this wavelength immediately prior to injection. Six or more concentrations of two-fold serial dilutions of the wild type and the variants of Alk1 and Alk3 were injected over the chip surface at a flow rate of 5 – 10 μL/min. All the injections were performed at room temperature were preceded by a brief injection of 2.5 M guanidine hydrochloride (20 μL/min for 12 seconds) to regenerate the surface. Instrument noise was removed by referencing the data against three or more buffer blank injections, while background signal was eliminated by referencing the data against a blank flow cell. Kds were determined by fitting the equilibrium binding response, Req, as a function of the injected receptor concentration, [R], to Req = (Rmax × [R])/(Kd + [R]) using the program Kaleidagraph (Synergy Software, Reading, PA).
Model of Alk1-BMP9 complex
A model of Alk1-BMP9 complex was generated using the program RosettaDock (36–38). Alk1 was initially positioned onto BMP-9 using the structure of the Alk3:BMP-2 complex as a template, followed by the introduction of a gap of 10 – 12 Å between Alk1 and BMP-9 to eliminate any bias in the initial steps of the docking. Alk1 was docked by performing an initial rigid body search, followed by optimization of side chain contacts. Alk3 was docked onto BMP-2 in a similar manner to evaluate the accuracy.
Data Deposition
Chemical shifts assignments for Alk1 ED have been deposited under BMRB 17628. Ten lowest energy structures satisfying all the experimental distance, dihedral angle, and RDC restraints have been deposited under PDB 2LCR.
Results
Isolation of recombinant Alk1-ED and Alk3-ED and initial characterization by NMR
The seven type I receptors of the TGF-β superfamily include a conserved pattern of ten cysteines (Fig. 1). These cysteines form an identical pattern of five disulfide bonds in the three type I receptor structures which are known, Alk3 (19, 20), Alk5 (39, 40), and Alk6 (41), suggesting that all type I receptors of superfamily share this common set of disulfides. The previously reported binding studies with the Alk1 extracellular domain were performed using protein produced in either CHO or HEK-293 cells (16–18, 42). Though these recombinant forms of Alk1 bound BMP-9 with high affinity, they could not be used for the proposed NMR studies due to the difficulty of producing fully 15N, 13C labeled samples for NMR. Thus, the bacterial expression and refolding procedure previously used to obtain the Alk5 extracellular domain (43) was used.
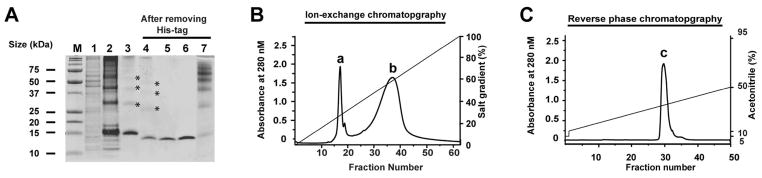
This entailed expression of the mature human Alk1 extracellular domain (residues 1 – 97) in E. coli, folding in a non-denaturing buffer in the presence of a glutathione redox couple, and purification by HPLC-based ion-exchange and reverse phase chromatography (Fig. 2A-C). The identity of the isolated Alk1 extracellular domain (Alk1-ED) was confirmed by electrospray ionization mass spectrometry, which yielded a mass of 11116.5 Da, just 0.2 Da greater than that calculated for the fully oxidized (i.e. five disulfide) form of the protein (Supplementary Material, Fig. 1A).
Figure 2.
Isolation and characterization of the Alk1 extracellular domain. (A) Non-reducing SDS gel from different stages of purification: Lane 1 – total lysate from non-induced cells, Lane 2 -solubilized inclusion bodies from induced cells, Lane 3 –refolded protein, Lane 4 - refolded protein after cleavage of the histidine tag, Lane 5 – monomer peak from ion exchange run (peak ‘a’ from panel B), Lane 6 – monomer peak from reverse phase chromatography run (peak ‘c’ from panel C), Lane 7 – multimer peak from ion exchange run (peak ‘b’ from panel B). Asterisks in lanes 3 and 4 identify disulfide-linked multimers that formed during the refolding reaction. (B) Fractionation of refolded thrombin-cleaved Alk1-ED on high resolution anion exchange column (Source Q, GE Healthcare) eluted with a linear 0 – 0.5 M NaCl gradient; peak ‘a’ corresponds to Alk1-ED monomers; peak ‘b’ corresponds to disulfide-linked multimers (C) Fractionation of peak ‘a’ from the ion-exchange run on a C18 reverse-phase HPLC column. The bound protein was eluted with a linear gradient (10–50%) of buffer B (acetonitrile+0.1% TFA) at 2.5 mL/min over 15 column volumes. Fractions eluted under peak ‘c’ were collected and lyophilized.
The Alk3 extracellular domain (Alk3-ED) was obtained using the previously reported E. coli expression and purification procedure (44), but modified to include a denaturation and renaturation step. This improved the recovery of native monomers and led to a highly homogenous sample suitable for NMR and binding studies (n.b. following the prior procedure, residues 1–27 of the Alk3 extracellular domain were excluded from the expression construct since this region is structurally disordered and not required for BMP binding (20)). The identity of the isolated Alk3 extracellular domain was confirmed by electrospray ionization mass spectrometry, which yielded a mass of 11560.9 Da, just 0.1 Da greater than that calculated for the fully oxidized (i.e. five disulfide) form of the protein (Supplementary Material, Fig. 1B).
The Alk1 extracellular domain was subsequently analyzed by NMR to assess folding and homogeneity. This was accomplished by recording a two-dimensional 1H-15N heteronuclear single quantum shift correlation (HSQC) spectrum of a 15N-labeled sample in phosphate buffer at pH 5.5 and 32 °C. The spectrum recorded under these conditions included 90 of the 91 expected backbone amide signals dispersed over a broad range of 1H chemical shifts (6.4 – 10.0 ppm) (Supplementary Material, Fig. 2). The Alk3 extracellular domain was analyzed in a similar manner, though in this case the spectrum was recorded at a somewhat higher pH (pH 6.3) and lower temperature (25 °C) to match the solution conditions under which it was previously studied (20). The spectrum recorded under these conditions had 94 of the 95 expected backbone amide signals (94/95) and closely matched that previously reported (Supplementary Material, Fig. 2). These data show that bacterial recombinant Alk1-ED and Alk3-ED are structurally ordered and homogenous.
Characterization of the BMP-2 and BMP-9 binding properties of Alk1-ED and Alk3-ED
The prior SPR binding studies showed that the monomeric Alk1 extracellular domain, as well as Fc-Alk1, a constitutive Alk1 dimer in which the Alk1 extracellular was fused to the Fc region of antibody, bound BMP-9 with Kds of 20 – 45 nM and 2 – 3 nM, respectively (16, 18, 42) (the higher affinity of the dimeric Alk1 is a result of multivalent binding, which has been previously observed with other receptors of the TGF-β superfamily (45)). This differs from other Alks, which appear to be incapable of binding BMP-9 either as Fc-Alk chimeras in SPR binding studies (42) or as receptors expressed on the cell surface of cultured cells (14). Moreover, this pattern is the opposite of BMP-2 and BMP-4, which bound Alk3 and Alk6 with high affinity, but not Alk1 (14).
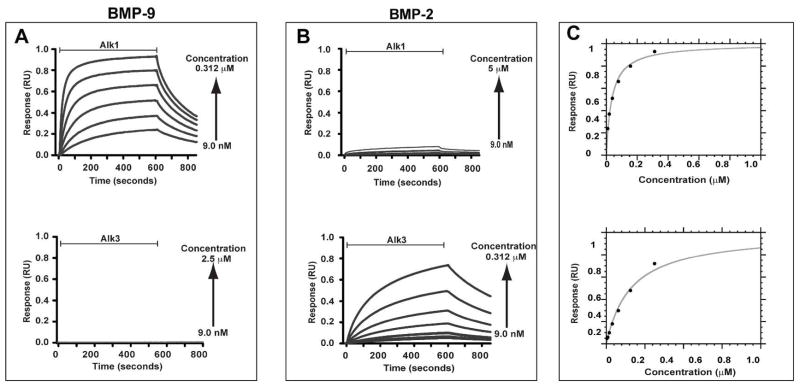
The functional activity of the bacterial recombinant Alk1-ED was evaluated by performing an equilibrium binding SPR experiment in which increasing concentrations of Alk1-ED were injected over either a control surface with no coupled ligand or over surfaces with BMP-2 and BMP-9 at a surface density of roughly 500 resonance units (RU) each. This yielded a robust concentration-dependent SPR response relative to the control when the bacterial recombinant Alk1-ED was injected over the BMP-9 surface, but not the BMP-2 (Fig. 3A, B). The bacterial recombinant Alk3-ED, in contrast, yielded the opposite result with a robust concentration-dependent response relative to the control when injected over the BMP-2 surface, but not the BMP-9 (Fig. 3A, B). The maximal response could be fitted as a function of concentration to derive the dissociation constant, Kd, and maximal response, Rmax, for Alk1-ED binding to BMP-9 and Alk3-ED binding to BMP-2, but not Alk1-ED binding to BMP-2 or Alk3-ED binding to BMP-9 due to the weak response (Fig. 3C, Table 1). The Kd values for Alk1-ED and Alk3-ED for their respective ligands fall in the low to mid nanomolar range (29 ± 5 nM and 330 ± 60 nM, respectively), indistinguishable from the SPR derived Kd for binding of monomeric HEK-293-derived Alk1-ED to immobilized BMP-9 (20 – 45 nM) (16) and comparable, though a bit higher than the SPR derived Kd for binding of monomeric bacterially derived Alk3-ED to immobilized BMP-2 (48 ± 20 nM) (46). This indicates that Alk1-ED and Alk3-ED are functionally active and native. To confirm this, the bacterially-derived Alk1-ED and Alk3-ED were evaluated in terms of their ability to compete against endogenously-expressed Alk1 and Alk3 in cultured NIH-3T3 fibroblasts. The results showed that Alk1-ED diminished the BMP-9 stimulated activation of phosphoSmad1/5/8 in a dose-dependent manner, whereas Alk3-ED did not, while Alk3-ED diminished the BMP-2 stimulated activation of phosphoSmad 1/5/8 in a dose-dependent manner, whereas Alk1-ED did not (Supplementary Material, Fig. 3). The Alk1-ED was further shown to fully attenuate the BMP-9 induced activation at a concentration of 2 μg/mL, which is roughly 5-fold lower than that reported by Alt, et al (16). This confirms that the bacterially-derived Alk1-ED and Alk3-ED are native and functionally active.
Figure 3.
SPR binding analysis of bacterial recombinant Alk1-ED and Alk3-ED. (A) SPR sensorgrams of serial two-fold dilutions of Alk1-ED and Alk3-ED over a BMP-9 surface. Sensorgrams shown have been corrected for background binding to a surface with no coupled ligand. (B) SPR sensorgrams as in panel A, except over a BMP-2 surface. (C) Saturation plots obtained by fitting the maximal binding response (circular data points) as a function of injected receptor concentration, [R], to Req = (Rmax[R]/(Kd + [R]). Data for Alk1 binding to BMP-9 and for Alk3 binding to BMP-2 are shown in the upper and lower panels, respectively.
Table 1.
Binding of Alk1 and Alk3 variants to BMP-2 and BMP-9
| Analyte | Injected concentration range (μM) | Kd (μM)1 |
|---|---|---|
| Binding to BMP-9 | ||
| Binding of Alk1 variants carrying alanine substitution | ||
| Alk1-WT | 0.009–0.312 | 0.029 ± 0.005 |
| Alk1-R26A | 0.009–0.312 | 0.031 ± 0.002 |
| Alk1-N50A | 0.009–5.000 | 0.26 ± 0.01 |
| Alk1-H52A | 0.009–5.000 | 0.25 ± 0.01 |
| Alk1-R53A | 0.009–2.500 | 0.087 ± 0.004 |
| Alk1-E54A | 0.039–5.000 | 0.49 ± 0.06 |
| Alk1-L55A | 0.009–5.000 | 0.21 ± 0.01 |
| Alk1-R59A | 0.009–5.000 | 0.30 ± 0.01 |
| Binding of Alk1 and Alk3 variants with residues swapped | ||
| Alk1-R53D | 0.039–5.000 | 0.43 ± 0.06 |
| Alk1-E54F | 0.039–0.156 | 0.020 ± 0.010 |
| Alk1-L55Q | 0.039–5.000 | 0.38 ± 0.020 |
| Alk1-DFQ | 0.009–5.000 | 5.7 ± 3.4 |
| Alk3 WT | 0.009–2.50 | n.d.2 |
| Alk3-REL | 0.019–10.00 | n.d.2 |
| Binding to BMP-2 | ||
| Alk1-WT | 0.009–5.0 | n.d.2 |
| Alk1-DFQ | 0.009–5.0 | n.d.2 |
| Alk3-WT | 0.009–0.312 | 0.33 ± .062 |
| Alk3-REL | 0.019–10.0 | n.d.2 |
The SPR-derived Kds and associated errors are the mean and standard deviations of two to three independent measurements for all the variants, except N50A, R53D, and L55Q, which were only measured once.
Not determined due to weak binding
NMR assignments and analysis of secondary shifts
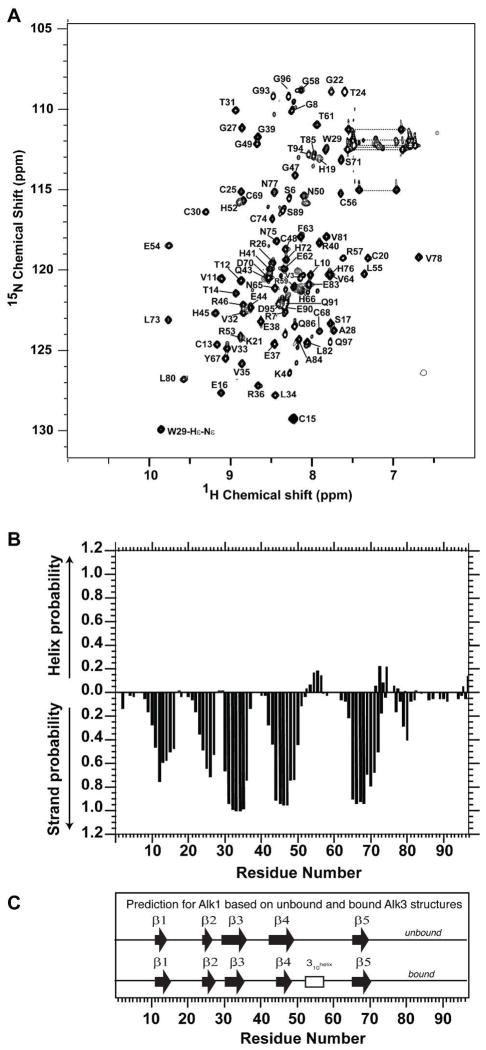
The backbone resonances of Alk1-ED were assigned by uniformly labeling it with 13C and 15N and by acquiring sensitivity-enhanced triple-resonance data sets with 0.4 – 0.7 mM samples in 25 mM sodium phosphate at pH 5.5 (Material and Methods). This enabled the sequence specific assignment of all the backbone amide signals of Alk1-ED, except for the first five residues on the N-terminus (four of which correspond to a GSHM tetrapeptide derived from the vector) and Leu51 (Fig. 4A). The sidechain 1H and 13C sidechain assignments were obtained by extending from the backbone using established methods (Experimental Procedures).
Figure 4.
Assigned 1H-15N HSQC spectrum and secondary structure probabilities of Alk1-ED. (A) 1H-15N HSQC spectrum of 0.4 mM 15N Alk1-ED in 25 mM sodium phosphate, 0.02% sodium azide, 5% 2H2O (pH 5.5) recorded at 32 °C at a magnetic field strength of 16.4 T (700 MHz 1H). Peaks are labeled according to their resonance assignments (residues are number as in Fig. 1). Horizontal dashed bars designate the side-chain amide groups of asparagine and glutamine. (B) Secondary structure probabilities based on an analysis of the NMR secondary shifts using the program PECAN (47). (C) Secondary structure predictions for Alk1 based on the structures of the unbound and bound forms of Alk3 (PDB 2K3G and 1REW, respectively) and an alignment of the Alk1 and Alk3 amino acid sequences.
The secondary shifts of Alk1-ED were analyzed using the program PECAN, which provides secondary structure probabilities on a residue-by-residue basis (47) (Fig. 4B). This showed that the secondary structure of Alk1-ED is comprised of five β-strands. The PECAN analysis also identified a short region with helical propensity (residue 54 – 56), although this was with reduced probability compared to the regions of β-strand. The positioning and length of the predicted β-strands coincide closely with those based on unbound and bound Alk3-ED structures (Fig 4C). The positioning and length of the helical region also closely coincides with that observed in Alk3, though as already noted, this α-helix was only observed in the BMP-2 bound form of Alk3-ED determined by crystallography (19), not the unbound form determined by NMR (20). The close correspondence of Alk1’s predicted secondary structure with that of Alk3 suggests that Alk1 shares the same disulfide-bonded three-finger toxin fold. The fact that the helical region can be detected based on the secondary shifts, though with reduced probability, suggests that this helix is at least partially formed in the unbound form of Alk1-ED.
Solution structure of Alk1-ED
The solution structure of Alk1-ED was determined using simulated annealing with torsion angle dynamics, as implemented in the program Aria 2.3 (48). The input data for the calculation consisted of 1816 experimental restraints, including 1612 NOE distance restraints, 88 TALOS-derived φ and ψ restraints (49), 24 3JHN-Hα restraints, and 67 1H-15N residual dipolar coupling restraints (RDCs) (Table 2). The ten lowest energy structures consistent with the experimental restraints are shown in Fig. 5A. The regions of secondary structure – β1 (residues 12–14), β2 (residues 24–26), β3 (residues 30–36), β4 (residues 42–48), 310 (residues 54–56), and β5 (residues 66–69) – correspond closely to those predicted based on the secondary shifts and were well-defined, with an overall backbone root mean square deviation (RMSD) of 0.33 Å. The structurally ordered core, which extends from residue 10–82 and includes several loop regions, had an overall backbone RMSD of 0.73 Å (Table 2). The terminal regions, residues 1–9 and 83–97 had no medium and long range NOEs and were disordered in the ensemble of calculated structures. The stereochemical quality, as assessed by the program PROCHECK (50), was typical of a well-refined structure, with 94% of the residues in the most favored or additionally allowed regions of the Ramachandran plot (Table 2). The residues in the generously allowed and disallowed regions of the Ramachandran plot were all in the terminal regions or loops.
Table 2.
Structural statistics for Alk1-EDa
| Restraints | ||
| NOE distance restraints | ||
| Intraresidue ((|i−j|=0) | 533 | |
| Sequential (|i−j|=1) | 440 | |
| Short Range (2≤|i−j|≤5) | 177 | |
| Long Range (|i−j|>5) | 462 | |
| Dihedral restraints | ||
| φ | 44 | |
| ψ | 44 | |
| RDC restraints | ||
| 1DNH | 67 | |
| Coupling restraints | ||
| 3JHNHa | 47 | |
| Total Restraints | 1814 | |
| Deviation Among Ensemble | ||
| Bonds (Å) | 0.0054 ± 0.0003 | |
| Angles (degrees) | 0.72 ± 0.04 | |
| Impropers (degrees) | 2.1±0.2 | |
| Dihedral restraints (degrees) | 0.71±0.22 | |
| RDC 1DNH (Hz) | 0.58 ± 0.12 | |
| JHNα restraints (Hz) | 0.71 ± 0.10 | |
| Ramachandran Plotb | ||
| Most favored (%) | 60.1 | |
| Additionally allowed (%) | 33.9 | |
| Generously allowed (%) | 4.1 | |
| Disallowed (%) | 1.9 | |
| Precision | ||
| Secondary | Backboned | 0.33 |
| Structurec | Heavyd | 0.73 |
| Ordered | Backboned | 0.75 |
| Residuesc | Heavyd | 1.25 |
Structural statistics are calculated for the ensemble of ten lowest energy structures; backbone atoms include NH, Cα, and CO; heavy includes all non-hydrogen atoms
Calculated using the program PROCHECK (50)
Ordered corresponds to residues 10–82 and secondary structure corresponds to 12–14, 24–26, 30–36, 42–48, 54–56, and 66–69
Backbone atoms include NH, Cα, and CO; heavy includes all non-hydrogen atoms
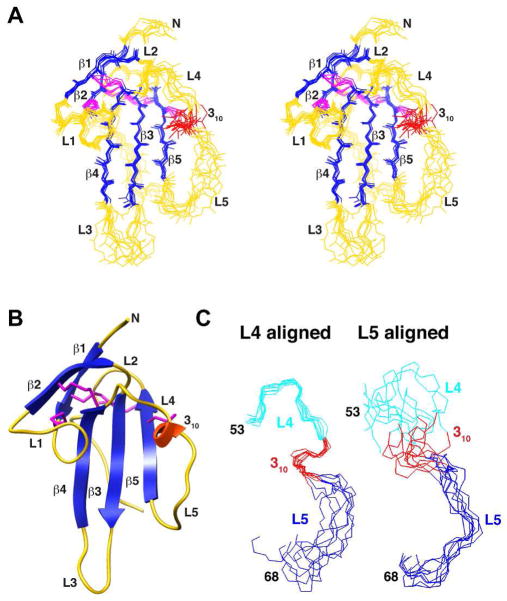
Figure 5.
Solution structure of Alk1. (A) Stereo view of the superimposition of the backbone atoms of the eight lowest energy structures consistent with the experimental restraints. β-strands are depicted in dark blue, the single 310 helix in red, and the loops in yellow. Disulfide bonds are depicted in magenta. Structures were aligned based on the regions of regular secondary structure. (B) Ribbon diagram of Alk1-ED highlighting the secondary structural elements and the five disulfide bonds that stabilize the receptor three-finger toxin fold. Secondary structural elements and disulfide bonds are colored as in panel A. (C) Segment from residue 49–63 bridging β-strands 4 and 5 aligned based on the backbone atoms of the N-terminal (residues 49–56, left panel) or C-terminal (residues 57–63) half. Residues in the N-terminal half are shaded cyan and red (310 helix); residues in the C-terminal half are shaded blue.
The helical region of Alk3 bearing the Phe85-Gln86 motif important for binding to BMPs 2, 4, and 7 resides in the extended segment connecting β-strands 4 and 5. The Alk1 ensemble has increased disorder in this segment, yet analysis of the calculated structure using the program DSSP (51) detects a 310 helix from residue 54–56 over all members of the ensemble (Fig. 5A, B). Though the extended segment bearing this 310 helix is clearly disordered over its length, the N-terminal portion is evidently less so as backbone overlays of this segment up to Cys56 have a considerably lower RMSD (0.47 Å) than overlays of the C-terminal portion (1.14 Å, respectively) (Fig. 5C). The structural basis for the apparent order in the N-terminal portion (hereafter designated L4) is probably a result of restrictions imposed by disulfide-bonded cysteines on the N- (Cys48) and C-terminal (Cys56) ends and the intrinsic propensity of residues 54–56 to form a 310 helix. The disorder in the C-terminal portion (hereafter designated L5) is probably result of the absence of an anchoring cysteine on the C-terminal end coupled with the lack of any internal structure or interactions with the underlying structured core.
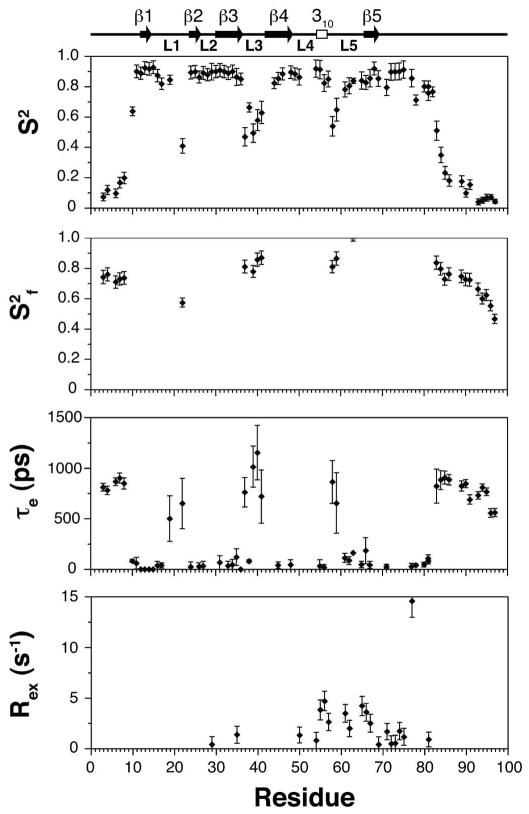
Internal dynamics of Alk1
The internal flexibility of Alk1-ED was modeled based on the measured 15N T1, 15N T2 and {1H}-15N-NOE parameters using the extended model free formalism as described in Experimental Procedures. The derived parameters show that the regions of regular secondary structure, including the 310 helix in the extended segment between β-strand 4 and 5, are rigid with a mean S2 of 0.89 ± 0.06 (Fig. 6). The N- and C-terminal regions from residues 1–10 and 82–97 are in contrast highly flexible, with S2 of 0.18 ± 0.16, consistent with the lack of medium and long range NOEs and high degree of disorder in these regions. The loops connecting β-strands 2 and 3 and β-strand 4 and the 310 helix are highly rigid, with S2 values of 0.89 ± 0.03. The loop connecting β-strands 1 and 2 is relatively rigid over most its length, except at the tip, which has an S2 of 0.42. This is probably because this loop has an internal disulfide bond (Cys15-Cys20), which restricts mobility within the loop. The loops connecting β-strands 3 and 4 and the 310 helix and β-strand 5 are flexible, with minimal S2 values of 0.4 – 0.5. The significant differences in flexibility between the N- and C-terminal halves of the segment connecting β-strands 4 and 5 are consistent with the differences in the degree of order among these regions in the calculated structures (Fig. 5C). Thus, the N-terminal half of the segment connecting β-strands 4 and 5, including the 310 helix from residue 54–56 is structurally ordered and rigid on the ns-ps timescale, but the C-terminal half is not.
Figure 6.
Model free parameters for Alk1-ED backbone amides derived by fitting 15N T1, 15N T2 and 15N-{1H} NOE data recorded at 15N frequency of 60.8 MHz. Lipari-Szabo S2, τe and S2f values are shown from top to bottom. Missing data points in the upper (S2) panel indicate that these residues could not be analyzed due to resonance overlap. Missing data points in the middle (τe) and lower (S2f) panels indicate that this parameter was not included in the model for those residues. Rex values were not required to fit the relaxation data for the missing residues. Secondary structure elements derived from DSSP analysis of the lowest energy Alk1 structures are shown along the top of the figure.
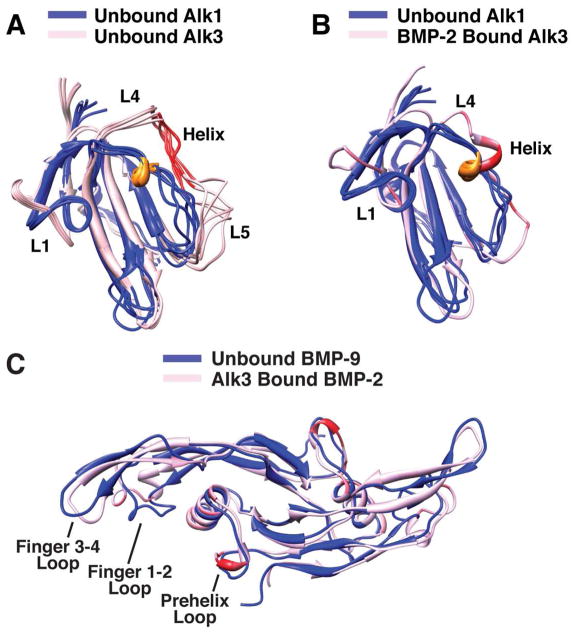
Structural comparison of Alk1 and Alk3
The superposition of the structure of the unbound form of Alk1 with the unbound and bound forms of Alk3 shows that the β-strand core superimposes closely, with backbone RMSDs of 0.89 and 0.83 Å, respectively (Fig. 7A, B). The largest difference is in the extended segment connecting β-strands 4 and 5 and is most pronounced for the unbound form of Alk3 where this region was shown to be flexible and structurally disordered (Fig. 7A). The difference is less pronounced for the bound form of Alk3 where this region is ordered and includes a six residue helix, the third, forth, and fifth residues of which, Phe85, Gln86 and Cys87, are positioned similarly to the three residues of Alk1’s 310 helix, Glu54, Leu55 and Cys56 (Fig. 7B). The similar spatial positioning of the helices in Alk1 and Alk3, coupled with the established importance of Alk3 Phe85 and Gln86 for binding of BMP-2 and other related ligands, suggests that residue differences in this region might be responsible for the restricted binding of these two type I receptors to their respective ligands. The other major structural difference between Alk1 and Alk3 is the conformation of loop 1. This loop includes an internal disulfide bond in both Alk1 and Alk3, though the two cysteines that form this disulfide are separated by four residues Alk1 and three in Alk3 (Fig. 1). This is probably responsible for the different conformation of the two loops as a disulfide cannot be accommodated if either of the cysteines are shifted toward one another in the Alk1 loop, or away from one another in the Alk3 loop.
Figure 7.
Structural comparison of Alk1 and Alk3 and BMP-2 and BMP-9. (A) An overlay of ensemble of the unbound forms of Alk1 (blue) and Alk3 (light pink) are shown. The structured 310 -helix in Alk1 is colored brown and the corresponding disordered α-helix region in Alk3 is colored red. (B) Cartoon representation of ligand bound Alk3 (pink/red) superimposed on Alk1 ensemble (blue). The extent of red coloring for Alk3 corresponds to fraction of total surface area buried in the Alk3:BMP-2 crystal structure. (C) Cartoon representation of Alk3-bound BMP-2 (pink/red) superimposed on unbound BMP-9 (blue). The extent of red coloring for Alk3-bound BMP-2 corresponds to fraction of total surface area buried in the Alk3:BMP-2 crystal structure.
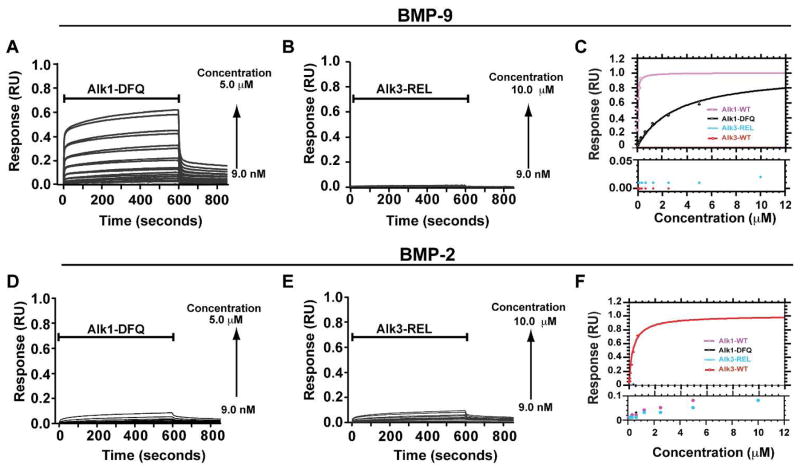
Role of Alk1 REL and Alk3 DFQ tripeptide motifs on BMP binding
To evaluate whether these residue differences might underlie the differences in specificity, Alk1 and Alk3 variants were generated in which Asp84, Phe85, and Gln86 in Alk3 and Arg53, Glu54, and Leu55 in Alk1 were swapped. These variants, designated Alk1-DFQ and Alk3-REL, were isolated as before and characterized in terms their binding using SPR. The SPR data showed that the affinity of Alk1-DFQ for its cognate ligand, BMP-9, was diminished approximately 200-fold compared to wild type Alk1 (Fig 8A,C, Table 1). Alk3-REL was similarly diminished in its affinity for its cognate ligand, BMP-2, compared to wild type Alk3, though the precise amount could not be determined due to weak binding (Fig. 8E, Table 1). SPR further showed that the Alk1 and Alk3 variants with swapped tripeptides did not lead to a detectable increase in the binding of the receptors to their non-cognate ligands, BMP-2 for Alk1 and BMP-9 for Alk3 (Fig. 8B, D). Although the precise kinetic parameters were not determined, visual inspection of the sensorgrams indicates that the effects of the substitutions are to increase to disassociation rate, consistent with the disruption of interactions that normally stabilize the complex.
Figure 8.
SPR binding analysis of Alk1 and Alk3 variants to BMP-9 and BMP-2. (A-C) SPR sensorgrams of serial two-fold dilutions of Alk1-DFQ (A) and Alk3-REL (B) over a BMP-9 surface and corresponding saturation plots (C). (D-F) SPR sensorgrams of serial two-fold dilutions of Alk1-DFQ (D) and Alk3-REL (E) over a BMP-2 surface and corresponding saturation plots (F). Sensorgrams shown have been corrected for background binding to a surface with no coupled ligand. Saturation plots were obtained by fitting the maximal binding response (circular data points) as a function of injected receptor concentration. Binding of Alk3 and Alk3-REL over BMP-9 and Alk1, Alk1-DFQ, and Alk3-REL over BMP-2 were not fitted due to weak responses.
The contribution of individual residues within Alk1’s tripeptide motif were assessed by substituting them with the corresponding residues from Alk3, yielding the Alk1 variants R53D, E54F, and L55Q. The R53D and L55Q variants were diminished 15- and 13-fold compared to wild type, while the E54F variant was unaffected (Table 1). On the other hand, when these residues were substituted with alanine, E54A was diminished to the greatest extent (17-fold), L55A was diminished to an intermediate extent (7-fold), and R53A the least (3-fold). Four residues outside the tripeptide motif were also substituted with alanine; the first three, N50A, H52A, and R59A, are within extended segment connecting β-strands 4 and 5 and lie within the predicted binding interface (presuming that Alk1 binds BMP-9 in the same manner that Alk3 binds BMP-2), while the fourth, R26A, is in a loop on the opposite side of the protein and lies outside of the predicted binding interface. The N50A, H52A, and R59A substitutions diminished BMP-9 binding between 9- and 10-fold, while the R26A substitution had no detectable effect (Table 1). The precise kinetic parameters were again not determined, but visual inspection of the sensorgrams indicates that the substitutions that most strongly disrupted binding are characterized by increased disassociation rates, consistent with the disruption of interactions that normally stabilize the complex. To determine whether any of the substitutions might have altered the folding or overall structure, the one-dimensional 1H NMR spectra of the variants were recorded using a WATERGATE solvent suppression scheme (Supplementary Material, Figs. S2 and S3). This showed that all of the variants had similar spectral patterns compared to their wild type counterparts in both the methyl and amide regions, indicating that they were unperturbed in terms of their folding. Taken together, these results show that the tripeptide motifs of these receptors are necessary for high affinity binding to their cognate ligands, but that other residues and structural features are required for high affinity binding to their non-cognate ligands.
Model of Alk1:BMP-9 complex
The results of the mutagenesis studies were interpreted by constructing a model of the Alk1:BMP-9 complex using the program RosettaDock (36–38). The criteria used to assess the docking were whether a consistent pattern of binding was observed among the lowest energy docked structures (designated as a ‘docking funnel’) and whether these were consistent with the mutagenesis data. The accuracy of the method was assessed by performing 1000 trial dockings to build the structure of the Alk3:BMP-2 complex (19) from its two component structures; this yielded a pronounced docking funnel with the 10 lowest energy structures closely clustered to one another (RMSD 0.15 Å) and the Alk3:BMP-2 crystal structure (RMSD 0.29 Å). The subsequent docking runs were performed with the solution structure of Alk1 and the crystal structure of BMP-9; this yielded a defined, though less pronounced docking funnel, with 6 of the 10 lowest energy structures clustered around a common position (RMSD 0.76 Å) with Alk1 bound to BMP-9 through the extended segment bridging β-strands 4 and 5.
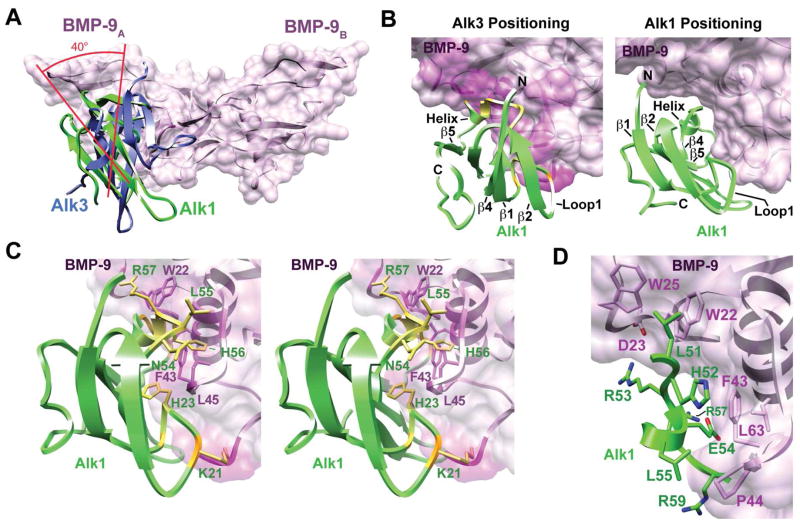
The overall positioning of Alk1 at the wrist epitope in the modeled complex is similar to that of Alk3 in the Alk3:BMP-2 complex, but differs in that the receptor is rotated by roughly 40 degrees relative to the ligand (Fig. 9A). This rotation is driven by steric clashes that prevent Alk1 from binding BMP-9 in an Alk3-like manner – this is illustrated in Figure 9B where the positioning of Alk1 in the modeled complex is shown to have few if any steric clashes (right panel), while Alk1 positioned onto BMP-9 in an Alk3-like manner leads to significant clashes (left panel). The first of these occur between residues in the putative ligand binding loop of Alk1, Glu54, Leu55, His56, Arg57, Glu58, and Arg59, and bulky residues in BMP-9, including Trp22, Phe43, Leu45, and Leu60 (Fig. 9C). The most severe of these is between the sidechain guanidinium group of Arg57 and the indole ring of Trp22 and occurs because of differences in the finger 1 – 2 loop of the ligands, which is shifted toward the wrist region by 2 – 3 Å in BMP-9 compared to BMP-2 (Fig. 7C). The clashes are also partly caused by differences in the N-terminal portion of the Alk1 β4-β5 loop, which is shifted toward β-strand 5 by about 3 Å in Alk1 compared to Alk3 (Fig. 7B) and causes Leu55 and His56 to clash with Phe43 and Leu45 on BMP-9 (Fig. 9C). The second of these occur between Lys21 and His23 in loop 1 of Alk1 and between Gly5 and Leu45 in BMP-9. This clash occurs because of differences in the conformation of loop 1, which is less extended in Alk1 compared to Alk3 (Fig. 7A). Similar, steric clashes are also observed when Alk1-ED is positioned onto BMP-2, or other ligands, such as BMP-7, GDF-5, and TGF-β1, in an Alk3-like manner (not shown), suggesting that the distinct conformation of the β1-β2 and β4-β5 loops that prevent Alk1 from binding BMP-9 in an Alk3-like manner BMP-7, also serves to prevent interactions with other ligands of the superfamily.
Figure 9.
Model of Alk1 bound to BMP-9 (A) Alk1 (green) bound to BMP-9 (pink ribbon with surface). Alk3 (purple) is overlaid according to the manner by which it binds to BMP-2 in the Alk3:BMP-2 crystal structure (19). (B) Model of Alk1 (green) bound to BMP-9 (pink) as determined using RosettaDock (right) or by positioning Alk1 as Alk3 is positioned onto BMP-2 (left). Extent of pink shading on the ligand surface and yellow color on the receptor ribbon corresponds to the severity of the clashes. (C) Stereoview of Alk1:BMP-9 complex in which Alk1 is bound in an Alk3-like manner (same as panel B, left). Extent of pink shading on BMP-9 and yellow color on Alk1 corresponds to the severity of the clashes. Sidechains of Alk1 (green labels) and BMP-9 (magenta labels) which clash are shown. (D) Interface of the RosettaDock derived Alk1:BMP-9 complex. Alk1 residues are shaded green; BMP-9 residues are shaded pink. Clashes depicted in panel B were calculated using the clash function within the program Chimera (58).
The interactions that stabilize Alk1 at the alternative interface are shown in Figure 9C. These are hydrophobic in nature and include interactions between Leu51, His52, Leu55, and the aliphatic portion of the sidechains of Glu54 and Arg59 in Alk3 with Trp22 and Trp25 (from the A-monomer) and Phe43, Pro44, and Leu63 (from the B-monomer) in BMP-9. The interactions are consistent with the mutational data – for example, Leu55 is surrounded by hydrophobic residues from the wrist region of BMP-9 (P44, L45 and A46) and is consistent with the greater loss in binding due to substitution with glutamine (16-fold) compared to alanine (8-fold). The model shows that the sidechain carboxylate of Glu54 forms an internal hydrogen-bonded ion-pair with the sidechain guanidinium group of Arg57; this arrangement positions the hydrophobic portion of Glu54 against BMP-9 Phe43, consistent with the greater loss of affinity (17-fold) upon substitution with alanine than phenylalanine (no loss). Arg53 faces the solvent in the model, consistent with the modest reduction in affinity upon substitution with alanine (3-fold); the larger reduction in affinity upon replacement with aspartic acid (15-fold) is probably a consequence of repulsion with the sidechain carboxylate of BMP-9 Asp23. Two of the other three residues evaluated, His52, and Arg59, also engage in hydrophobic interactions in the wrist region, consistent with the 9- to 10-fold loss in affinity upon substitution with alanine. The third residue, Asn50, is buried within the interface and led to a 9-fold reduction in affinity when substituted with alanine. The underlying basis for the reduction is not obvious, but might be due to interactions with nearby residues on BMP-9, such as Thr50 or Thr52. Though not tested by mutagenesis, it is notable that Leu51 of Alk1 is positioned between Trp22 and Trp25 of BMP-9; this closely mimics the interaction between Phe85 of Alk3 and Trp28 and Trp31 of BMP-2 and may therefore play a similar stabilizing role.
Discussion
The type I receptors of the superfamily that have been structurally characterized, Alk3 (19, 20, 52, 53), Alk5 (39, 40, 54), and Alk6 (41), share the same three finger toxin fold and bind using the solvent-exposed loop bridging β-strands 4 and 5. There are nevertheless important differences in both the flexibility of the β4-β5 loop and the conformation it adopts in the bound state. The loop is flexible in unbound Alk3 and undergoes a disorder-to-order transition upon binding (20). The bound conformation, which includes a 1-1/2 turn helix near the center of the loop, is similar to that of Alk6, consistent with the similar though not identical manner of ligand binding exhibited by the these two otherwise closely related type I receptors (19, 41). The N-terminal portion of the binding region of the TGF-β type I receptor Alk5 in contrast includes a five-residue PRDRP extension. The extension is structurally ordered in the unbound form (40) and assumes a conformation similar to the bound form (39, 54). This has been suggested to be important for Alk5’s restricted binding to the TGF-βs, but not other superfamily ligands, such as BMPs and activins (40). The type I receptor Alk1, which was earlier considered as an orphan receptor, has recently been shown to bind BMP-9 and BMP-10 with high specificity and affinity (14), yet the underlying mechanism responsible for its restricted binding is not known.
The results presented here have shown that the Alk1 extracellular domain binds with high affinity to BMP-9, but not to BMP-2 (Fig. 3 and Table 1). The precise affinity differential could not be determined due to difficulty quantifying Alk1’s weak binding to BMP-2, but is estimated to be 1000-fold or higher based on the Kd of 29 ± 5 nM for binding BMP-9 and an estimated Kd of 50 μM or higher for binding BMP-2. This is the opposite of Alk3, which binds BMP-2 with an affinity estimated to be at least 500-fold greater than that of BMP-9. The analysis of the Alk1 structure showed that the overall fold and secondary structure elements are highly similar to Alk3, yet there are significant differences in the conformations of the β1-β2 and β4-β5 loops (Fig. 7A, B). The differences of the β4-β5 loop are very pronounced for the unbound form of Alk3 where the loop was shown by NMR to be flexible on the ns-ps timescale and disordered in the ensemble of calculated structures (20), but is also evident for the bound form of Alk3, where this region is ordered and assumes a longer, though similarly positioned central helix (19) (Fig. 7A, B). The β1-β2 and β4-β5 loops have been shown previously as the specificity determining elements in Alk3 and Alk6 (21, 41) and therefore could be significant for Alk1 binding specificity as well.
This was tested by swapping the Arg53-Glu54-Leu55 tripeptide from Alk1 with the corresponding tripeptide from Alk3, Asp84-Phe85-Gln86. This reduced the binding affinity of both receptors for their respective ligands several hundred fold, but did not engender the receptors with high affinity binding to their non-cognate ligands. This indicates that other residues and the conformation of the β1-β2 and β4-β5 loops might also be important for determining the affinity and specificity. This is supported by the mutational analysis that showed substitution of residues outside of the tripeptide motif but within the β4-β5 loop of Alk1 reduced its affinity for binding BMP-9 (Table 1). This is also supported by the model of the Alk1:BMP-9 complex which showed that Alk1 binds to the wrist in a manner similar to Alk3, but is rotated by roughly 40 ° due to steric clashes caused by differences in three different loops, including the β1-β2 and β4-β5 loops in Alk1 which were shown to be largely rigid and adopt a distinct conformation relative to the corresponding loops of Alk3 (Fig. 7A, B) and the finger 1–2 loop of the ligands (Fig. 7C). The alternate positioning allows Alk1 to bind BMP-9 through a large interface dominated by hydrophobic interactions. These interactions are consistent with the mutational analysis and place the aliphatic portion of Glu54 and the sidechains of His52 and Leu55 in the hydrophobic pocket on the wrist region of BMP-9 and the sidechain of Leu51 between the indole rings of Trp22 and Trp25. The aliphatic portion of Glu54 is constrained by a hydrogen-bonded ion pair formed between its sidechain carboxylate group and the guandinium group of Arg57 and Arg53 protrudes into the solvent. The importance of Leu51, His52, Glu54 and Leu55 for binding is further supported by the finding that these residues are highly conserved among mammalian Alk1 sequences, whereas Arg53 is not (Supplementary Material, Figure 7). The bound form of Alk3 is similarly incompatible with the Alk1 manner of binding - this is a result of the longer length of the β4-β5 loop in Alk3 as well as a displacement of the finger 3–4 loop of BMP-2 toward the wrist (Supplementary Material, Figure 8). This positioning also places the sidechain carboxylate of Glu81 between the indole rings of Trp28 and Trp31, which is expected to further disfavor this manner of binding.
Taken together, these results suggest that the specificity of Alk1 and Alk3 for their respective ligands arise as a consequence of relatively minor, but important structural and residue changes in the binding regions of the receptors and ligands that reposition the receptor relative to the ligand. This provides distinct interfaces and stabilizing interactions and enables the high degree of specificity for these otherwise similar ligands and receptors. This type of repositioning and corresponding expansion of specificity has been previously observed for the TGF-β type I receptor, Alk5. This receptor includes a five residue extension in the N-terminal portion of the β4-β5 loop that is both rigid and forms a tight turn with an N-terminal cis proline (40). The extension precludes Alk5 from binding in an Alk3 manner due to clashes with underlying hydrophobic residues in the wrist – this causes the receptor to reposition so that its shifted toward the fingertips where it interacts extensively with both TGF-β and TβR-II (39, 54). The extension binds in the cleft between TGF-β and TβR-II and orchestrates a number of interactions that are specifically required for assembly of the TGF-β signaling complex (39, 54). Thus, changes in the length and composition of the N-terminal portion of the β4-β5 loop - anticipated to be relatively common mutational event – provides a mechanism for shifting the binding interface and in turn expanding the range of specificity between ligands and receptors that otherwise share the same structural scaffold.
The alternative manner of Alk1 binding identified through the docking calculations is dependent on the underlying assumption that the backbone conformation of the unbound form of Alk1 and unbound form of BMP-9 are representative of the bound forms. This is not known with certainty, though the fact that the structural elements involved – the β1-β2 loop and the N-terminal portion of the β4-β5 on the receptor are both rigid and reinforced by disulfide bonds - suggests that they might well be. The other important point is that the docking calculations yielded a consistent docking solution – this would not occur unless the backbone conformations are already close to those of the bound form as the backbone conformation of neither the receptor nor the ligand was adjusted in the docking calculations. The final point is that even if the β1-β2 and β4-β5 loops were able to adapt to bind BMP-9 in an Alk-3 manner – something that is not anticipated due to the demonstrated rigidity of the β1-β2 and β4-β5 loops - the interactions at the interface would be far from optimal. This includes not only the unfavorable interaction between Arg57 and Trp22 noted earlier, but as well between Alk1 Arg 59 and BMP-9 Leu63 and between Alk1 Lys 21 and BMP-9 Gly5. Thus, it appears likely that Alk1 binds in an alternative manner relative to Alk3 and that this underlies the high specificity of Alk1 and Alk3/6 for their respective ligands, though to be certain, this will have to await the determination of the structure of Alk1:BMP-9 complex using crystallography, which is ideally suited for this system owing to the high affinity of Alk1 for BMP-9 and the relatively high molecular weight of the complex (50 kDa).
The prior binding studies have shown that endoglin and Alk1 non-competitively bind BMP-9, while endoglin and ActRIIb competitively bind BMP-9 (16, 17). This suggests that endoglin binds BMP-9 at a site that overlaps with ActRIIb – this almost certainly includes the knuckle epitope as all BMP type II receptors studied to date have been shown to bind to the knuckle (52, 53, 55). The fact that endoglin non-competitively binds with Alk1, together with our finding that Alk1 binds on the opposite side of the ligand at the wrist, indicates that the endoglin binding site does not extend into the wrist. The fact that both endoglin and Alk1 non-competively bind BMP-9 suggests that endoglin and Alk1 function together to bind and capture BMP-9. The type II receptor BMPRII might then function to displace the bound endoglin to form a type I:type II receptor signaling complex. This is suggested by the fact that BMPRII has been shown to bind BMP-9 with the highest affinity of the type II receptors that bind BMP-9, ActRII, ActRIIb, and BMPRII (18). These biochemical observations fit nicely with several lines of evidence that like Alk1 and endoglin, BMPRII also plays a role in normal vasculogenesis (56).
The mutations in Alk1 that have been linked to HHT, a disease caused by the fragility of the vasculature, are found throughout the protein (9). The most prominent ectodomain mutations involve either the addition or elimination of a cysteine residue. The cysteines are strictly conserved in all known type I receptors of the superfamily – addition or elimination of a cysteine would be expected to disrupt the formation of the disulfide bonds and thus interfere with the function of the receptor. There are eleven additional non-cysteine HHT mutations in the ectodomain (R26P, G27R, G27E, A28P, T31A, H45P, R46W, R46Q, G58R, N75D and N77S). The majority of these mutations are nonconservative – one falls in the predicted binding interface and would be expected to effect BMP-9 binding (G58R). The others fall outside the interface and may lead to disease by disrupting the folding of the receptor. This is suggested by the fact that many mutated residues lie at positions in the structure that would be expected to disrupt the fold - the R26P mutation for instance replaces an arginine residue located directly adjacent to one of the conserved cysteines with a constrained proline. This is further suggested by the finding that Alk1 ectodomain HHT mutants are either not expressed on the cell surface or are expressed poorly (57).
Supplementary Material
Acknowledgments
The authors would like to acknowledge Dr. Eileen Lafer who directs the SPR Core and who helped troubleshoot some of the SPR experiments and Dr. Susan Weintraub and Sammy Pardo who assisted with analysis of the Alk1-ED and Alk3-ED using ESI-MS.
This research was supported by grants from the NIH (GM58670 and RR13879 awarded to A.P.H. and CA54174 awarded to the U. Texas HSC San Antonio Cancer Therapy and Research Center) and the Robert A. Welch Foundation (AQ-1431 awarded to A.P.H.).
Abbreviations
- Alk
Activin-like kinase
- BMP
bone morphogenetic protein
- BMPR-Ia
BMP type I receptor a
- GDF
growth and differentiation factor
- TβR-II
TGF-β type II receptor
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser enhancement
- RDC
residual dipolar coupling
- TGF-β
transforming growth factor β
- SPR
surface plasmon resonance
- HHT
hereditary hemorrhagic telangiectasia
Footnotes
Supporting Information Available
Supplementary Figures 1–8. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes & development. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 3.Reddi AH. BMPs: from bone morphogenetic proteins to body morphogenetic proteins. Cytokine & growth factor reviews. 2005;16:249–250. doi: 10.1016/j.cytogfr.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 5.Derynck R. TGF-beta-receptor-mediated signaling. Trends Biochem Sci. 1994;19:548–553. doi: 10.1016/0968-0004(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 6.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 8.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson DW, Berg JN, Baldwin MA, Gallione CJ, Marondel I, Yoon SJ, Stenzel TT, Speer M, Pericak-Vance MA, Diamond A, Guttmacher AE, Jackson CE, Attisano L, Kucherlapati R, Porteous ME, Marchuk DA. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet. 1996;13:189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- 10.Oh SP, Seki T, Goss KA, Imamura T, Yi Y, Donahoe PK, Li L, Miyazono K, ten Dijke P, Kim S, Li E. Activin receptor-like kinase 1 modulates transforming growth factor-beta 1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci U S A. 2000;97:2626–2631. doi: 10.1073/pnas.97.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthur HM, Ure J, Smith AJ, Renforth G, Wilson DI, Torsney E, Charlton R, Parums DV, Jowett T, Marchuk DA, Burn J, Diamond AG. Endoglin, an ancillary TGFbeta receptor, is required for extraembryonic angiogenesis and plays a key role in heart development. Dev Biol. 2000;217:42–53. doi: 10.1006/dbio.1999.9534. [DOI] [PubMed] [Google Scholar]
- 12.Perlingeiro RC. Endoglin is required for hemangioblast and early hematopoietic development. Development. 2007;134:3041–3048. doi: 10.1242/dev.002907. [DOI] [PubMed] [Google Scholar]
- 13.David L, Feige JJ, Bailly S. Emerging role of bone morphogenetic proteins in angiogenesis. Cytokine Growth Factor Rev. 2009;20:203–212. doi: 10.1016/j.cytogfr.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 14.David L, Mallet C, Mazerbourg S, Feige JJ, Bailly S. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood. 2007;109:1953–1961. doi: 10.1182/blood-2006-07-034124. [DOI] [PubMed] [Google Scholar]
- 15.Scharpfenecker M, van Dinther M, Liu Z, van Bezooijen RL, Zhao Q, Pukac L, Lowik CW, ten Dijke P. BMP-9 signals via ALK1 and inhibits bFGF-induced endothelial cell proliferation and VEGF-stimulated angiogenesis. J Cell Sci. 2007;120:964–972. doi: 10.1242/jcs.002949. [DOI] [PubMed] [Google Scholar]
- 16.Alt A, Miguel-Romero L, Donderis J, Aristorena M, Blanco FJ, Round A, Rubio V, Bernabeu C, Marina A. Structural and functional insights into endoglin ligand recognition and binding. PLoS One. 2012;7:e29948. doi: 10.1371/journal.pone.0029948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castonguay R, Werner ED, Matthews RG, Presman E, Mulivor AW, Solban N, Sako D, Pearsall RS, Underwood KW, Seehra J, Kumar R, Grinberg AV. Soluble endoglin specifically binds bone morphogenetic proteins 9 and 10 via its orphan domain, inhibits blood vessel formation, and suppresses tumor growth. The Journal of biological chemistry. 2011;286:30034–30046. doi: 10.1074/jbc.M111.260133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown MA, Zhao Q, Baker KA, Naik C, Chen C, Pukac L, Singh M, Tsareva T, Parice Y, Mahoney A, Roschke V, Sanyal I, Choe S. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J Biol Chem. 2005;280:25111–25118. doi: 10.1074/jbc.M503328200. [DOI] [PubMed] [Google Scholar]
- 19.Kirsch T, Sebald W, Dreyer MK. Crystal structure of the BMP-2-BRIA ectodomain complex. Nat Struct Biol. 2000;7:492–496. doi: 10.1038/75903. [DOI] [PubMed] [Google Scholar]
- 20.Klages J, Kotzsch A, Coles M, Sebald W, Nickel J, Muller T, Kessler H. The solution structure of BMPR-IA reveals a local disorder-to-order transition upon BMP-2 binding. Biochemistry. 2008;47:11930–11939. doi: 10.1021/bi801059j. [DOI] [PubMed] [Google Scholar]
- 21.Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD. Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol. 2004;11:481–488. doi: 10.1038/nsmb756. [DOI] [PubMed] [Google Scholar]
- 22.Franzen P, ten Dijke P, Ichijo H, Yamashita H, Schulz P, Heldin CH, Miyazono K. Cloning of a TGF beta type I receptor that forms a heteromeric complex with the TGF beta type II receptor. Cell. 1993;75:681–692. doi: 10.1016/0092-8674(93)90489-d. [DOI] [PubMed] [Google Scholar]
- 23.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3-D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J Magn Reson Ser B. 1993;101 [Google Scholar]
- 24.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 25.Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic side-chain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J Magn Reson Ser B. 1993;101:114–119. [Google Scholar]
- 26.Kay LE, Ikura M, Tschudin R, Bax A. 3-Dimensional Triple-Resonance Nmr-Spectroscopy of Isotopically Enriched Proteins. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Clubb RT, Thanabal V, Wagner G. A Constant-Time 3-Dimensional Triple-Resonance Pulse Scheme to Correlate Intraresidue H-1(N), N-15, and C-13(‘) Chemical-Shifts in N-15-C-13-Labeled Proteins. J Magn Reson. 1992;97:213–217. [Google Scholar]
- 28.Kay LE, Xu GY, Singer AU, Muhandiram DR, Forman-Kay JD. A graidient-enhanced HCCH-TOCSY experiment for recording side-chain H-1 and C-13 correlations in H2O samples of proteins. J Magn Reson Ser B. 1993;101:333–337. [Google Scholar]
- 29.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 30.Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 32.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple 2-parameter model-free approach to the interpretation of N-15 nulcear magnetic -relaxation of protein. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- 33.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 34.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis and experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 35.Mandel AM, Akke M, Palmer AG., 3rd Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 36.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. Journal of molecular biology. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 37.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, Schueler-Furman O, Baker D. Improved side-chain modeling for protein-protein docking. Protein science : a publication of the Protein Society. 2005;14:1328–1339. doi: 10.1110/ps.041222905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 40.Zuniga JE, Ilangovan U, Mahlawat P, Hinck CS, Huang T, Groppe JC, McEwen DG, Hinck AP. The TbetaR-I Pre-Helix Extension Is Structurally Ordered in the Unbound Form and Its Flanking Prolines Are Essential for Binding. J Mol Biol. 2011;412:601–618. doi: 10.1016/j.jmb.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotzsch A, Nickel J, Seher A, Sebald W, Muller TD. Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity. EMBO J. 2009;28:937–947. doi: 10.1038/emboj.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell D, Pobre EG, Mulivor AW, Grinberg AV, Castonguay R, Monnell TE, Solban N, Ucran JA, Pearsall RS, Underwood KW, Seehra J, Kumar R. ALK1-Fc inhibits multiple mediators of angiogenesis and suppresses tumor growth. Mol Cancer Ther. 2010;9:379–388. doi: 10.1158/1535-7163.MCT-09-0650. [DOI] [PubMed] [Google Scholar]
- 43.Zuniga JE, Groppe JC, Cui Y, Hinck CS, Contreras-Shannon V, Pakhomova ON, Yang J, Tang Y, Mendoza V, Lopez-Casillas F, Sun L, Hinck AP. Assembly of TbetaRI:TbetaRII:TGFbeta ternary complex in vitro with receptor extracellular domains is cooperative and isoform-dependent. J Mol Biol. 2005;354:1052–1068. doi: 10.1016/j.jmb.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 44.Kirsch T, Nickel J, Sebald W. Isolation of recombinant BMP receptor IA ectodomain and its 2:1 complex with BMP-2. FEBS letters. 2000;468:215–219. doi: 10.1016/s0014-5793(00)01214-x. [DOI] [PubMed] [Google Scholar]
- 45.Komesli S, Vivien D, Dutartre P. Chimeric extracellular domain type II transforming growth factor (TGF)-beta receptor fused to the Fc region of human immunoglobulin as a TGF-beta antagonist. Eur J Biochem. 1998;254:505–513. doi: 10.1046/j.1432-1327.1998.2540505.x. [DOI] [PubMed] [Google Scholar]
- 46.Heinecke K, Seher A, Schmitz W, Mueller TD, Sebald W, Nickel J. Receptor oligomerization and beyond: a case study in bone morphogenetic proteins. BMC Biol. 2009;7:59. doi: 10.1186/1741-7007-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eghbalnia HR, Wang L, Bahrami A, Assadi A, Markley JL. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J Biomol NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- 48.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 49.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 50.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of biomolecular NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 51.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 52.Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci U S A. 2006;103:7643–7648. doi: 10.1073/pnas.0602558103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber D, Kotzsch A, Nickel J, Harth S, Seher A, Mueller U, Sebald W, Mueller TD. A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct Biol. 2007;7:6. doi: 10.1186/1472-6807-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, Sun PD. Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J Biol Chem. 2010;285:14806–14814. doi: 10.1074/jbc.M109.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 56.David L, Feige JJ, Bailly S. Emerging role of bone morphogenetic proteins in angiogenesis. Cytokine & growth factor reviews. 2009;20:203–212. doi: 10.1016/j.cytogfr.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 57.Ricard N, Bidart M, Mallet C, Lesca G, Giraud S, Prudent R, Feige JJ, Bailly S. Functional analysis of the BMP9 response of ALK1 mutants from HHT2 patients: a diagnostic tool for novel ACVRL1 mutations. Blood. 2010;116:1604–1612. doi: 10.1182/blood-2010-03-276881. [DOI] [PubMed] [Google Scholar]
- 58.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.