Abstract
At later ages, humans have high risk of developing Alzheimer disease (AD) which may afflict up to 50% by 90 years. While prosimians and monkeys show more substantial changes, the great apes brains examined show mild neurodegenerative changes. Compared with rodents, primates develop and reproduce slowly and are long lived. The New World primates contain some of the shortest as well as some of the longest-lived monkey species, while the prosimians develop the most rapidly and are the shortest lived. Great apes have the largest brains, slowest development, and longest lives among the primates. All primates share some level of slowly progressive, age-related neurodegenerative changes. However, no species besides humans has yet shown regular drastic neuron loss or cognitive decline approaching clinical grade AD. Several primates accumulate extensive deposits of diffuse amyloid-beta protein (Aβ) but only a prosimian—the gray mouse lemur—regularly develops a tauopathy approaching the neurofibrillary tangles of AD. Compared with monkeys, nonhuman great apes display even milder brain-aging changes, a deeply puzzling observation. The genetic basis for these major species differences in brain aging remains obscure but does not involve the Aβ coding sequence which is identical in nonhuman primates and humans. While chimpanzees merit more study, we note the value of smaller, shorter-lived species such as marmosets and small lemurs for aging studies. A continuing concern for all aging studies employing primates is that relative to laboratory rodents, primate husbandry is in a relatively primitive state, and better husbandry to control infections and obesity is needed for brain aging research.
Keywords: Brain aging, Neuropathology, Primates, Alzheimer disease, Primate life histories, Animal husbandry
At later ages, the human brain has a high risk of developing Alzheimer disease, which may afflict up to 50% of those reaching advanced years: extensive neuronal loss in forebrain and subcortical regions is accompanied by accumulations of brain amyloid-beta protein (Aβ) in senile plaques and neuronal cytoskeletal abnormalities with hyperphosphorylated tau (tauopathy). In contrast, the few great ape brains examined have shown very mild neurodegenerative changes while prosimians and monkeys show substantial accumulations of brain amyloid and neuronal cytoskeletal abnormalities. Thus, there are profound species differences in outcomes of brain aging. To approach this puzzle, we first discuss the different life spans and life history patterns of primates and then discuss the current evidence on brain aging in detail. We suggest that variations in animal husbandry have some role in these puzzling species differences.
Primates compared with rodents in the mammalian scheme
Primates comprise about 5% of the roughly 5,000 extant mammal species. As a group, they specialize in arboreal locomotion, stereoscopic vision, manual dexterity, and complex social organization and behavior (Vaughan et al. 2000). With several exceptions, primates are distributed in the tropics and subtropics. By contrast, rodents are highly speciose—by far the largest mammalian order, representing more than 40% of living mammal species. Some rodent species are adapted to nearly every habitat type, and they are well represented in nearly every geographic region. Although rodents range in size from 5 g to 50 kg, most species are small (<100 g). Therefore, primates are characterized by large body size relative to rodents. Even the smallest primate species is larger than many rodents. Furthermore, primates are characterized by large brains relative to rodents or any other mammal group (Eisenberg 1981). Finally, the majority of primates are diurnal, relying on vision as a primary sensory mode whereas rodents are for the most part nocturnal and more reliant on olfaction than vision. These latter facts about rodent biology have been largely neglected in widely used experimental paradigms for learning, memory, and complex cognition.
Relative to mammals generally and rodents in particular, primates display slow life histories. That is, they develop and reproduce slowly and are long-lived. For example, typical small rodents such as the house mouse (Mus musculus) can reach sexual maturity in a couple of months, produce a litter of six or more pups every 2 months, and generally live less than a year in the wild. Even exceptionally long-lived rodents like the naked mole rat reproduce copiously (Buffenstein 2005) compared with any primate. By contrast, even primates with relatively “fast” life histories such as the common marmoset require at least 15–18 months to reach sexual maturity, produce no more than two litters of twins per year, and can live 5+ years in the wild and 12–16 years in captivity (Austad and Fischer 2011). More typical primates take years to reach maturity, reproduce at intervals measured in years, and can live decades in the wild. The great apes are characterized by extremely slow maturation (Table 1), only matched by a few much larger mammals such as elephants and whales. Primates are longer lived than most other mammal groups, surviving in captivity about twice as long as rodents, even after correcting for differences in body size (Austad and Fischer 1991).
Table 1.
Selected life history traits of the great apes and common captive primate study species
| Species | Age at first female birth (years) | Interbirth interval (years) | Maximum reported longevity (field) | Maximum reported longevity (captivity) | References |
|---|---|---|---|---|---|
| Human | 19–20 | 2–4 | 80 | 100 | Robson and Wood (2008) |
| Chimpanzee | 12–16 | 5–6 | 55 | 59 | Wich et al. (2004) |
| Gorilla | 9–13 | 4–6 | 43 | 55 | Yamagiwa (1997) |
| Orangutan | 15–16 | 8–9 | 58 | 59 | Wich et al. (2004) |
| Rhesus | 4–5 | 1–2 | 31b | 40 | Hoffman et al. (2010) |
| Marmoset | 1.3–1.5c | 0.5c | 10 | 17 | Grzimek (1990) and Tardif et al. (2011) |
| Lemur | 0.8–1.0 | 0.33–0.5 | 6 | 18 | Lutermann et al. (2006) |
All data are from field studies except where otherwise noted. Maximum field longevity for humans is estimated from modern hunter–gatherers. “Captive” human longevity represents that under modern technological conditions. Because there are vastly more longevity data on modern humans than any other primate species, maximum captive longevity represents the maximum expected in a sample of 10,000 individuals. All other captive longevities are from the AnAge database (http://genomics.senescence.info/species/)
Chimpanzee common chimpanzee (Pan troglodytes), gorilla mountain gorilla (Gorilla beringei), orangutan Sumatran orangutan (Pongo abelii), rhesus rhesus macaque (Macaca mulatta), marmoset common marmoset (Callithrix jacchus), lemur gray mouse lemur (Microcebus murinus)
aThe alleged 75+ age of a former film-star chimpanzee has been falsified (Rosen 2008)
bFrom provisioned, free-living monkeys introduced on a predator-free island
cCaptive population
Primate phylogeny and life histories
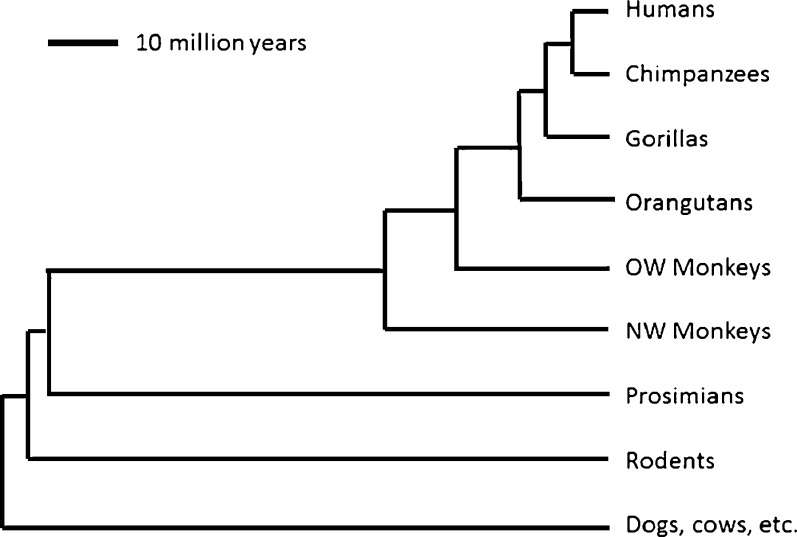
Along with tree shrews (Scandentia) and colugos (Dermoptera), primates form an evolutionary sister group to the rodents and lagomorphs (rabbits, hares) from which they have been separated for about 90 million years (Bininda-Emonds et al. 2007; Murphy et al. 2004) (Fig. 1). Despite this rather close phylogenetic affinity, the more rapid evolutionary pace of chromosomal rearrangements and nucleotide substitution in the mouse genome compared with primates or dogs results in more genomic structural similarity as well as more annotated transcripts shared between dogs and humans than mice and humans (Kirkness et al. 2003; Waterston et al. 2002).
Fig. 1.
Phylogeny of some relevant mammal groups (after Bininda-Emonds et al. 2007). Note that branch lengths are proportional to estimated time of divergence
Primates can be considered in four convenient evolutionary groups: prosimians (e.g., lemurs, lorises, and bushbabies), Old World monkeys (e.g., macaques and baboons), New World monkeys (e.g., marmosets and capuchins), and apes (e.g., humans and gibbons). The apes can be further divided into lesser apes (gibbons and siamang) and greater apes (humans, chimpanzees, gorillas, and orangutans).
Prosimians consist of mostly small, nocturnal species that forage solitarily and eat a mixed diet composed mainly of insects. Exceptions to these generalizations can be found among the 100 or so species of lemurs that diversified into a much wider variety of sizes and ecological niches on the isolated island of Madagascar (Rowe 1996). Compared with other primates, prosimians have low metabolic rates, small brain to body size ratios, and depend less on vision and more on olfaction as a primary sensory modality (Eisenberg 1981). From a life history perspective, compared with other primates, prosimians have a fast life history, developing quickly, having frequent reproductive bouts—often with multiple births, and they are relatively short lived for primates (Austad and Fischer 1992). Only one prosimian has been used in aging-related research, the gray mouse lemur (Microcebus murinus) as discussed below.
Old World monkeys consist of major species radiations in Africa (baboons, guenons, and colobines) and Asia (macaques and langurs). They are typically larger than prosimians, slower to develop, have lower reproductive rates, and are longer lived. Old World monkeys are generally most active during the day and highly reliant on vision, including well-developed color vision. They also have a larger brain to body size ratio, higher metabolic rates, and exhibit considerably more complex social behaviors than prosimians. Some species are entirely arboreal, others such as baboons are primarily terrestrial. Most species are omnivorous although a few are largely folivorous. Old World monkeys are the closest primate relatives of humans outside the apes (Fig. 1). Although several species of macaques and the baboon have been used in aging-related research, by far the most commonly used species for a wide range of biomedical studies is the rhesus macaque (Macaca mulatta).
DNA sequence data indicate that New World monkeys diverged from their African ancestors about 35 million years ago (MYA), presumably immigrating to South America on rafts of floating vegetation at a time when the Atlantic Ocean was considerably narrower than it is today (Chatterjee et al. 2009). New World monkeys are arboreal, with many species having prehensile tails. Except for the genus Aotus, they are most active during the day and have well-developed vision, although fully developed color vision is confined to a few species. New World monkeys exhibit a diversity of life histories. Small species such as marmosets and tamarins develop quickly, reproduce copiously, and are relatively short lived (Table 1). By contrast capuchin monkeys (genus Cebus) display slow life histories and have the longest reported captive lifespan of any monkey, a remarkable 54 years, only slightly exceeded by the great apes (Judge and Carey 2000). Because of its small size, rapid reproduction, and relatively short life, the common marmoset has been increasingly used in aging-related research (Tardif et al. 2011).
The great apes consist of humans, chimpanzees, bonobos, gorillas, and orangutans. All great apes are characterized by large brains, exceptionally slow life histories—particularly with respect to prolonged maternal care—and long lives compared with other primates. Humans are a medium size great ape of African origin—smaller than gorillas or male orangutans, larger than female orangutans or chimpanzees (Nowak 1999) but with a comparatively enormous brain relative to any other species. Diverging from a common ancestor with chimpanzee lineage about 6 MYA, the multiple species in the human lineage began increasing both body size and brain size about 2 MYA, reaching modern levels within the past 500,000 years (Robson and Wood 2008). Anatomically, modern humans appeared roughly 200,000 years ago. The great apes are of exceptional interest because of their close evolutionary affinity with humans. Because of the interest in the genetics of life history evolution in humans, the most informative comparisons will be those assessing life histories in a state-of-nature, the environment that shaped their evolution.
Current information suggests that in nature, gorillas have the most rapid life history of the great apes, followed by chimpanzees, orangutans, and humans (Kaplan et al. 2000; Robbins et al. 2004; Wich et al. 2004). Gorillas reach sexual maturity more rapidly and have shorter interbirth intervals than any of the other great apes except humans; they also have the shortest maximum longevity in nature and in captivity (Table 1). By the same metrics, chimpanzees appear to have a faster life history than orangutans (Bronikowski et al. 2011; Wich et al. 2004).
We consider these conclusions about species difference as provisional because the habitat of all the great apes has been dramatically reduced and degraded in recent decades and all species have been considerably affected by human diseases (Finch 2010). Moreover, field estimates of lifespans are sensitive to numerous confounding factors including the length of time and depth of study. Thus, whether the life histories we observe now, such as the much higher infant mortality rate in chimpanzees than in any of the other great apes (Wich et al. 2004; Bronikowski et al. 2011), represent those experienced prior to modern human encroachment isn’t clear and may never be known. Zoo longevity records only partially support the above conclusions. For instance, in zoo records gorilla life expectancy is slightly longer than that of orangutans, although maximum longevity is a bit shorter (Kohler et al. 2006). Zoo records, however, are strongly affected by husbandry knowledge and implementation (see below).
Although earlier researchers in gerontology frequently assumed that animals in nature did not live long enough to incur aging, this supposition has been overthrown by decades of field research. All primates that have been carefully studied in nature with long-term monitoring of identifiable individuals display the classic demographic signatures of senescence, increasing mortality and decreasing reproduction with age (Bronikowski et al. 2011; Atsalis and Videan 2009; Hoffman et al. 2010). Reproductive decline often occurs relatively late in life compared with humans though. Chimpanzee menopause is about the same age as in humans in captivity and in the wild (Atsalis and Videan 2009; Finch and Holmes 2010; Hawkes and Smith 2010). Female baboons show significant reproductive decline at 18 years of age and undergo menopause in their early 20s in a feral population where adult life expectancy is only 12 years (Altmann et al. 2010; Bronikowski et al. 2002). Ironically, reproductive senescence in these baboons occurs much earlier in males, although it is mediated by male–male dominance interactions and represents reduced access to females rather than reduced male fertility (Altmann et al. 2010). The importance of studying animals in nature to understand life history evolution becomes evident when considering that female reproductive senescence may occur earlier in captive populations than in the wild, possibly due to accelerated maturation combined with captive breeding practices (Atsalis and Videan 2009).
Humans are unique among other ape species in that they develop slowly and are long-lived but paradoxically exhibit more rapid reproduction once sexual maturity has been reached (Table 1). Puberty is followed by a phase of adolescent sterility and high-risk pregnancy until later teen years when the pelvis has fully matured (Bogin 1999; Finch 2007; Moerman 1982; Hawkes and Smith 2010). This more rapid reproduction is enabled by earlier weaning and necessarily with assistance (alloparenting) of other females—is likely responsible for the phenomenal expansion of human populations relative to other great apes (Hawkes and Paine 2006).
For biomedical research, life history variables of study species under captive conditions are more relevant for the design and interpretation of experiments. Because of a more favorable energy balance, primates (and rodents) in captivity grow faster, reach larger adult body size, mature early, and reproduce more often than in the wild. For instance, female common chimpanzees experience menarche at 7–8 years in captivity, 10–12 years in the wild, their first birth at 8–11 years of age in captivity compared with 12–16 years in the wild, and they exhibit interbirth intervals of 2–4 years in captivity compared with 5–6 years in the wild (Atsalis and Videan 2009; Wich et al. 2004). Longevity, as previously noted, is a more complex issue and will be discussed below.
Humans diverged from their closest living relatives, the chimpanzees and bonobos about 6 MYA, from the Old World monkeys such as the macaques and baboons roughly 25 MYA, from the New World monkeys such as marmosets and capuchins about 35 MYA ( Bininda-Emonds et al. 2007; Glazko and Nei 2003; Fabre et al. 2009). The average sequence identity for orthologous genes among these groups is 98–99% between humans and chimpanzees (Chimpanzee Sequencing and Analysis Consortium 2005), 93% between humans and rhesus macaques (Gibbs et al. 2007), and—to give these numbers some perspective—70% between humans and mice (Waterston et al. 2002).
Primate neurobiology
The human brain shows greater vulnerability to age-related Alzheimer disease (AD) neuropathology than other primates studied so far (Table 2; Figs. 2 and 3): All species examined regularly show progressive age-related changes after maturation with dendritic atrophy, damage to white matter, gliosis (activation of astrocytes and microglia), and increased Aβ in brain parenchyma and in cerebral vasculature. Note that all data on aging primates are based on captives in laboratory colonies.
Table 2.
Overview of neurodegenerative changes with age in selected primate species (in order of lifespan)
| Dendritic atrophy | Neuron loss | Diffuse amyloid | Tauopathy | AD | |
|---|---|---|---|---|---|
| Human, clinically normal | ++ | ± | + | ++ after 80 | +++ after 80 |
| Great apea | ? | ? | + | ± | – |
| Baboon | ++ after 20 | ? | |||
| Rhesus monkey | ++ | +? | ++ | – | – |
| Squirrel monkey | ++ | ? | ++ | – | – |
| Marmoset | ? | ? | ++ | – | ? |
| Mouse lemur | ? | ? | ++ | ++ | ? |
aChimpanzee, 41 years (Rosen et al. 2008), 59 years (Gearing et al. 1994, 1996), and 2 “aged,” no age given (Gearing et al. 1994); gorilla, 44 years (Kimura et al. 2001); orangutans—28, 31, and 36 years (Gearing et al. 1997); baboon (Schultz et al. 2000); and lemur (Bons et al. 2006; Kraska et al. 2011; Marchal et al. 2012)
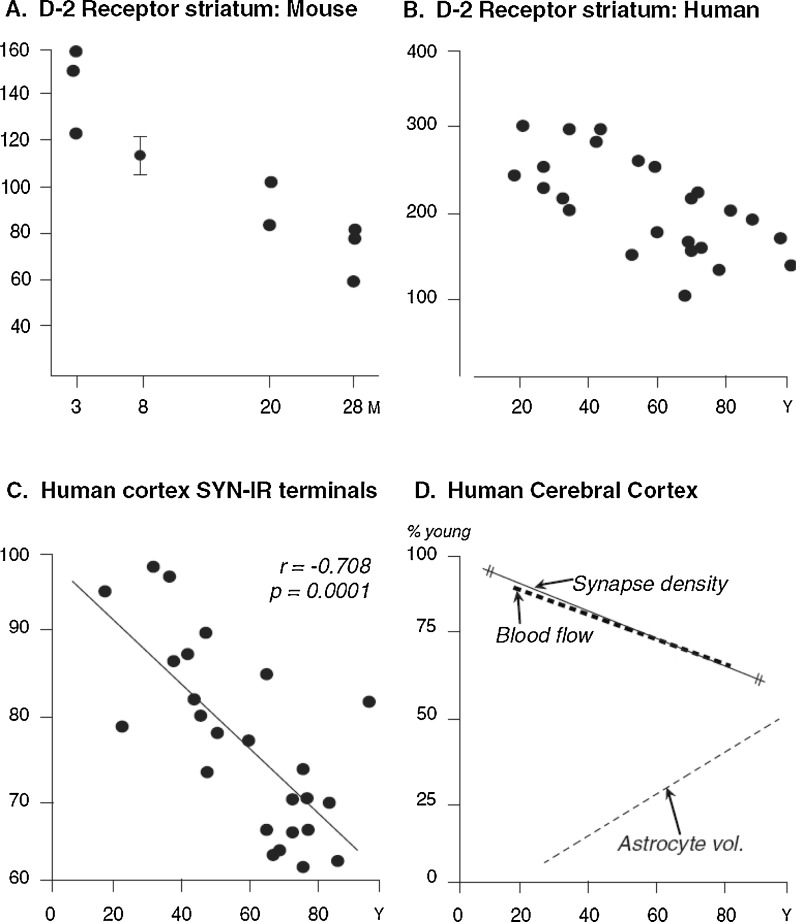
Fig. 2.
Synaptic loss progresses across adult ages in rodents and humans without neuropathology. Adapted from Finch (2007, pp. 24–25 and 98). a Dopamine D2 receptors in postmortem striatum mouse, determined as Bmax by Scatchard analysis (Severson and Finch 1980). b Human D2 sites (Severson et al. 1982). c Synaptophysin immunoreactive presynaptic terminals (SYN-IR) in human cerebral cortex (Masliah et al. 1993). The rate of loss approximates 0.5%/year in humans after age 20 for striatal D2 sites and for SYN-IR cortical synapses. These synaptic losses are independent of β-amyloid accumulation because they arise in aging rodents which do not accumulate Aβ deposits and because losses begin before age 40 in humans when there is negligible Aβ deposition except in rare familial Alzheimer disease. d Declining blood flow parallels the loss of synapses, while astrocyte fibrosis increases progressively (Hansen et al. 1987) in the same brains characterized for SYN-IR terminals. The synapse density line was regraphed from (c). Blood flow was measured by xenon in cortical grey matter of 13 healthy adults (Amano et al. 1982)
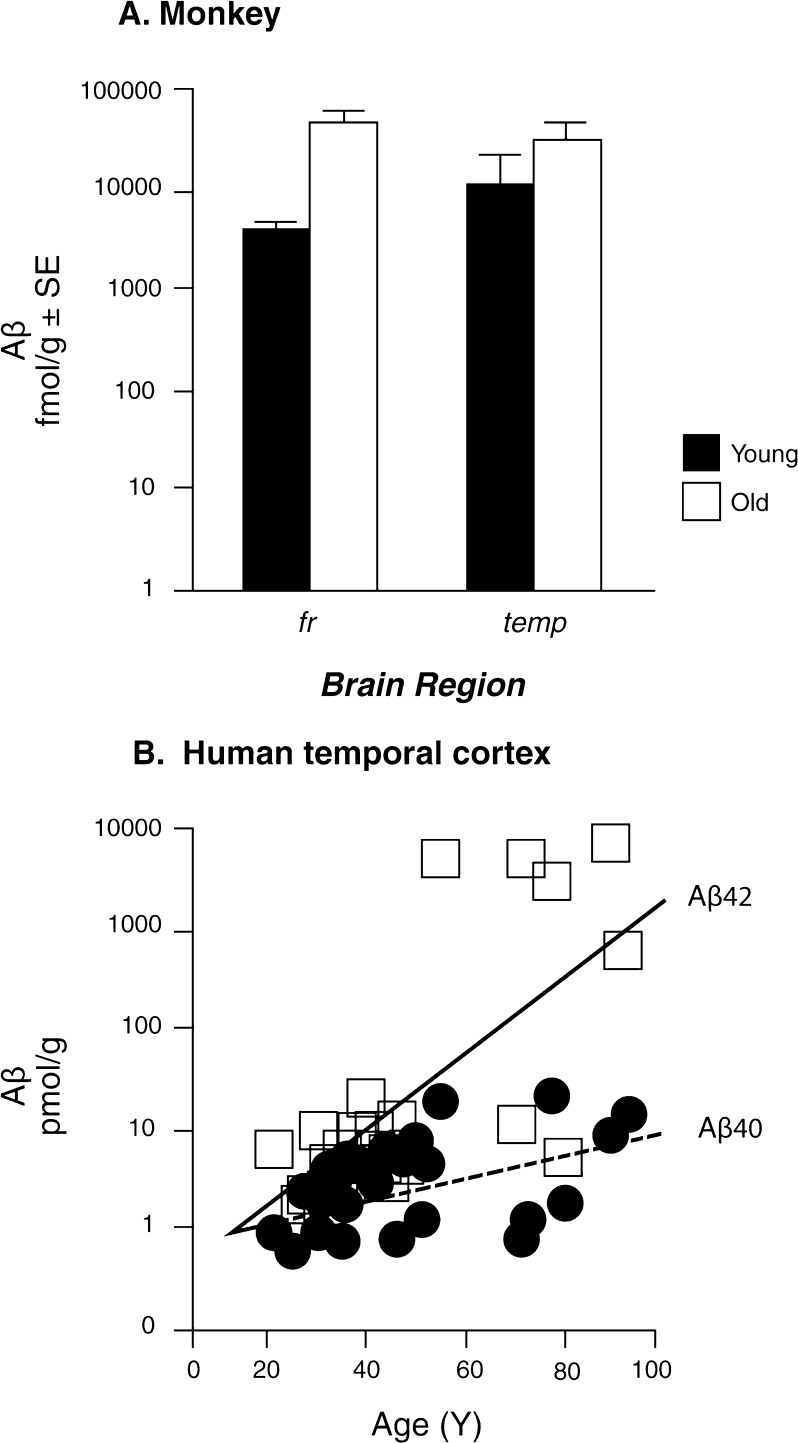
Fig. 3.
Brain amyloid-beta peptide increases progressively during normal aging in (b) rhesus monkey and (b) human cerebral cortex. The peptides analyzed were extracted in cold formic acid. Redrawn from Fukumoto et al. 2004
Against the apparent universals of slow atrophic changes during primate and human brain aging, no species besides humans have shown evidence of regular major neuron loss or drastic cognitive decline that would approach clinical grade AD. It is critical to recognize that humans and all the mammalian lab models examined have progressive mild atrophic changes in brain that are distinct from clinical neurodegenerative diseases of advanced aging, particularly cerebrovascular disease, Alzheimer disease, and Parkinson disease, in rank order of prevalence. The differences between general aging changes and clinical disease may be described as eugeric vs. pathogeric age changes (Finch 1972). For example, in US-wide samples of the NHANES, fluid intelligence scores peak at about 25 years and decline progressively at about 0.5%/year up through age 70 (McArdle 2009; Salthouse 2009). Complex multi-tasking (“walking while memorizing”) also declines progressively across middle age (Li et al. 2001). The rhesus monkey also shows mild progressive cognitive changes from maturation through middle age into old age, in spatial memory (delayed recognition) (Lacreuse et al. 2005) and executive function (Moore et al. 2006); also see chapter by Morrison in this issue. A small sample of great apes (chimpanzees and orangutans) showed similar age trends for other cognitive tests (Anderson et al. 2007, 2005).
Paralleling the subtle cognitive changes during middle age are mild, progressive atrophic changes. In humans, brain atrophy is well characterized from longitudinal and cross-sectional MRI imaging analysis (Fjell et al. 2010; McDonald et al. 2009). Greater cortical atrophy in late middle age is a strong AD risk factor (Dickerson et al. 2011).
Three species of aging nonhuman primates show important differences. Two short-lived species showed progressive atrophy resembling those of humans. An MRI study of aging mouse lemurs showed region-specific atrophy in association with impaired executive function (Picq et al. 2011). Aging rhesus monkeys have been studied in most detail. Regional atrophy of prefrontal cortex by MRI correlated with impaired spatial and recognition memory (Shamy et al. 2011). However, a very recent MRI analysis of chimpanzees aged 10–46 years (N = 69; Yerkes National Primate Center) did not find any indication of atrophy in cerebral cortex, hippocampus, or white matter (Sherwood et al. 2011). These findings challenge earlier postmortem findings of 10% decrease of total brain weight in chimpanzees grouped as 15–30 vs. 30–59 years (Herndon et al. 1998). Because the details of diet, infections, social environment, and other husbandry factors that influence brain development and aging are not known for this study, we consider the findings of Sherwood et al. (2011) to be more definitive, pointing to the conclusion that chimpanzees have milder presentation of gross atrophic changes during aging than humans or shorter- lived primates.
Loss of synapses begins soon after maturation in many brain regions of humans and of monkeys during middle-age and is concurrent with enlargement and activation of astrocytes (Finch 2009) (Fig. 2). Synapse loss progressing across middle-age was first described by Finch and colleagues for dopaminergic D2 binding sites in rodents (Severson and Finch 1980) and in humans (Severson et al. 1982; Morgan et al. 1987), where it approximates a loss of 0.5–1.0%/year in humans and comparable percent for rodents when normalized per unit adult lifespan. The same synapse loss rate was verified for a large set of normal human brains in immunocytochemically identified synaptophysin (SYN) by Robert Terry and Bob Katzman’s group (Masliah et al. 1993). Rhesus monkeys show similar progressive changes in association with complex changes in synaptic markers: SYN and D2 decreased (Harada et al. 2002; Haley et al. 2010; Ingram et al. 2001), but MAP-2 increased (Haley et al. 2010). The progressive age-related loss of caudate D2 receptors in rhesus monkeys is paralleled in caudate volume shrinkage (Ingram et al. 2001).
Astrocyte activation during middle age is identified by increased expression per cell of glial fibrillary acid protein (GFAP), an intermediate filament, and increased fibrous astrocytes in same brains studied for SYN-terminal density (Hansen et al. 1987). GFAP also increases per cell in aging rhesus hippocampus and prefrontal cortex (Haley et al. 2010). However, the numbers of astrocytes (density per area) does not increase with age in these and other studies (Finch et al. 2002). Finch and colleagues have hypothesized that astrocyte activation may be driving the loss of synapses during aging. This hypothesis is based on experimentally evidence that the neurite outgrowth (E18 rat cortical neurons) is slower in co-cultures of astrocytes from old rat cortex (24- vs. 3-month-old donors) (Rozovsky et al. 2005). Thus, aging in astrocytes decreases neurotrophic support. These age changes are reversed by down regulation of GFAP by RNAi, which restored the ability of old astrocytes to support E18 neuron outgrowth. These findings support the role of increased GFAP transcription during middle age in synaptic atrophy of normal aging. Recent experiments extend these findings to the role of the estrogen receptor ERα, which increases during aging in male and female rodent brains (Arimoto et al., in preparation).
Rodent models of brain aging are important in this discussion of species differences because the aging rodent does not accumulate the brain parenchymal β-amyloid deposits (Aβ1-40 and Aβ1-42) (Sullivan et al. 2008) observed in primates and human at later ages to varying degrees. Whereas all primates examined share the same β-amyloid (Aβ1-42) amino acid sequence with humans, the Aβ peptide of lab rodents diverges from primates in three amino acids that reduce its spontaneous aggregation (Boyd-Kimball et al. 2004). The absence of brain Aβ deposits in aging rodents gives an important comparative control in aging mechanisms because Aβ is pro-inflammatory and can activate astrocytes and microglia. Thus, increased GFAP expression during aging and other markers of glial activation during middle age does not require depend on Aβ deposits; nonetheless, Aβ deposits could also contribute to glial activation at later ages.
Other insights come from gene expression comparison of aging rat and monkey hippocampus by the Landfield group: about 35% of the 1,500 changes in different coding RNAs in the hippocampus during middle age in rats (Kadish et al. 2009) are also shared with rhesus monkey (Blalock et al. 2010). Thus, rodent models of aging allow us to begin dissection of complex cellular changes during aging which, in primates appear to involve the additional factor of Aβ amyloid accumulation.
The Aβ peptide is strongly implicated in AD from many lines of evidence (Klein et al. 2001; Walsh and Selkoe 2007; Hardy 2009). Particularly cogent is that AD is universal in the Down syndrome from overexpression of Aβ (trisomy 21): the specific role of the amyloid precursor protein (APP) locus in chromosome 21 is demonstrated by an unusual case of clinically diagnosed Down’s syndrome which proved to have a segmental deletion of the APP locus which produced the equivalent of disomy for the APP locus, thereby normalizing the gene dose. In this case, AD-like changes were absent during a 78 year lifespan, which is much longer than expected for Down’s syndrome (Prasher et al. 1998). Conversely, duplication of the APP locus caused premature AD and cerebral angiopathy (cerebrovascular Aβ amyloid) in five unrelated families (Rovelet-Lecrux et al. 2006). Moreover, the human APP gene shows epigenetic variation, such that some cells express only one allele (50-fold transcription above the silent allele), while others express both parental alleles (Gimelbrant et al. 2007). Somatic cell epigenetic variation in APP expression and possible age changes have not been studied at population levels in humans or primates.
Of major potential significance, humans and rhesus monkey brains both accumulate solubilizable Aβ during middle age (Fig. 3) (Fukumoto et al. 2004); these data do not resolve if solubilizable Aβ (1-40 and 1-42) both increase in aging rhesus, as in humans, or if the increase is exponential as for Aβ (1-42) in humans. At the histological level, fibrillar deposits of Aβ (1-42) arise later and are not prominent in the absence of Alzheimer disease (Arriagada et al. 1992). [It was not possible in this review to address the different sized Aβ peptides in parenchymal and vascular amyloid deposits]. Nonetheless, most individuals after the age of 55 harbor some amyloid deposits (amorphous amyloid and senile plaques) and some neurons with neurofibrillary tangles (NFTs) (Arriagada et al. 1992; Zanjani et al. 2005; Ohm et al. 1995). Importantly, the regional distribution of amyloid deposits and NFTs in nondemented elderly closely approximates the distribution of neurodegeneration in AD. Very recent studies show that scattered neurons with hyperphosphorylated tau arise even before puberty (Braak and Del 2011a; Braak and Del 2011b). Taken together, the evidence suggests some association of pre-clinical Aβ and NFT accumulation with the exponential increase of AD after age 60, but we are far from understanding the chain of causality in these complex and evidently life-long processes. We know even less about the neuroproteomics of aging and neurodegeneration in primates.
By the average lifespan when risk of AD increases sharply in human populations, aging primates present a remarkable diversity of brain changes, which are mostly mild and would not qualify as clinical–grade neuropathology (Table 2). Five species of monkeys and one prosimian accumulate extensive deposits of diffuse Aβ, as do humans during at later ages in the absence of AD. The deposits do not meet the criteria for neuritic plaques in AD by their diffuse characteristics and limited presence of degenerating neurites. The rhesus monkey, which has been studied in most detail (Luebke et al. 2010) (Morrison et al. and Rosene et al., this volume), does not present tauopathic abnormalities or major neuron loss during aging (Duan et al. 2003). The cognitive changes are correlated with synapse loss and white matter degeneration, e.g., in executive functions of the prefrontal cortex area 46 (Rosene et al., this volume). However, the age-associated cognitive changes are mild by comparison with AD. Moreover, some individual monkeys age “successfully” with minimal cognitive decline (Rapp and Amaral 1991; Herndon et al. 1997). Thus, the rhesus monkey may be considered a model for normal (eugeric) brain aging in humans. Although tauopathies have not been reported in aging rhesus, nonetheless aging rhesus show neurocytoskeletal abnormalities which cause axonal bulges (diverticula) that suggest impaired axonal flow (Fiala et al. 2007). In contrast, extensive neurofibrillary degeneration with tau hyperphosphorylation is reported in the mouse lemur (short-lived primate); however, the level of neuron death was not established for comparison with AD brains, which typically show >50% pyramidal neuron loss in the entorhinal cortex and CA1 layer of the hippocampus (Simonian and Hyman 1994).
Chimpanzees and other great apes also accumulate low levels of diffuse Aβ peptides at later ages, with notable amounts of Aβ (1-40) as well as Aβ(1-42) (Gearing et al. 1994, 1996, 1997). The brain aging changes appear to be milder than in the mouse lemur, monkeys, or humans of equivalent demographic age, but very few specimens have been studied (Table 2). Neurofibrillary degeneration has been considered absent in aging great apes (Gearing et al. 1994, 1996, 1997). However, we now have one case of classic Alzheimer tauopathy and neurofibrillary degeneration in a 41-year-old female who was euthanized after a stroke (Rosen et al. 2008); this individual had notable obesity and hypercholesterolemia, which are risk factors in human AD and, which in AD-transgenic mice, accelerate neurodegeneration (Cole et al. 2010; Leduc et al. 2010). Numerous tau-immunoreactive paired helical filaments were found in neocortex and subcortical regions. Aβ deposits were moderate, but neuritic plaques were fewer than in typical AD. The tauopathy was present in both cerebral hemi-spheres and was not restricted to the infarcted region. Notably, the hippocampus was not involved. Cerebrovascular Aβ was also present, consistent with prior findings in aging apes (Gearing et al. 1994, 1996, 1997). The authors’ absence of comment on neuron loss should not be interpreted as its absence. While no great ape or monkey has neurodegenerative changes that approach clinical AD, this case report suggests that obesity and hypercholesterolemia can lower the threshold for tauopathy in the chimpanzee. However, the distribution of lesions differed from classic AD in the lack of neurodegeneration in the hippocampus, which is a major target of AD in humans. Given the small numbers of brains evaluated carefully in great apes of advanced ages (about 10) and the absence of any quantitative analysis of neuron numbers, we cannot yet conclude whether or not AD is unique to humans. The rampant obesity in aging primate colonies may yield additional cases. Cerebrovascular aging merits further attention. In contrast to this diversity of amyloid and tauopathy, humans and all primates examined show increased cerebrovascular amyloid during aging, as noted above. Among the shared risk factors in ischemic heart disease and AD in humans are high LDL cholesterol, obesity, and the apoE4 allele. The individual with a stroke and neurofibrillary degeneration did not have any of the mutations in the MAPT locus which are associated with hereditary tauopathies in humans. Lastly we emphasize that all data on primate aging have come from captive animals. We do not know of any case report on the pathology of aging in wild-living animals.
An intriguing evolutionary question concerns why neurodegenerative changes with age are milder in the great apes than in either lemurs and monkeys or humans. One possibility, of course, is that important changes have been missed due to the small number of carefully evaluated brains of elderly apes. However, two other possibilities deserve mention. First, the limited state of ape husbandry has not yet allowed us to routinely produce really old animals. Notably, the longevity records of apes in captivity are only marginally longer than longevities reported from the wild (Table 1). Thus, it may be that as captive husbandry continues to improve and we see more apes of older ages, more dramatic neurodegenerative changes will appear. An alternative possibility concerns the evolutionary trajectory of lifespans within the ape clade. In the 25 million years since they diverged from African monkey, apes evolved considerably longer lives than monkeys. Elderly apes in the wild live into their 40s and even 50s, whereas monkeys in nature are elderly by their late teens or early 20’s (Bronikowski et al. 2002) (Table 1). Accordingly, we suggest that the evolution of lengthening lifespan could have enhanced protection against neurodegenerative changes with age by an additional decade or two. However, humans have undergone a very rapid increase in longevity much more recently—perhaps as recently as <100,000 years (Caspari and Lee 2004). At the age of 50, human and great ape brain aging changes may be considered as equivalently mild by the modest accumulations of diffuse Aβ and atrophic neurons with cytoskeletal abnormalities. We suggest the working hypothesis that, because the human reproductive lifespan has not been lengthened relative to the apes, there was little selection pressure to evolve additional neuroprotection.
Species differences in relevant genes
The genetic basis for major differences in brain aging between humans and other primates is as obscure as is the basis for differences in life span. Aging monkeys and humans apparently accumulate more deposits of Aβ as diffuse amyloid plaques than do some of the great apes examined. Nonetheless, as noted above, the 42-amino acid sequence of Aβ is identical in human, great ape, and monkey (Johnstone et al. 1991). With remarkably few exceptions (lab rodents), the Aβ sequence is identical in other vertebrates, including zebra fish (Coulson et al. 2000). The absence of diffuse Aβ plaques in old lab rodents may be attributed to three amino acid differences that slow aggregation (Boyd-Kimball et al. 2004). Besides its role in neurotoxicity (Klein et al. 2001; Hardy 2009), the Aβ peptide also has strong anti-microbial activity (Soscia et al. 2010), suggesting a basis for its evolutionary stability. However, the APP from which the Aβ peptide is derived by endoproteases, has undergone considerable evolution, and has many diverse implied functions (Maloney et al. 2004; Jacobsen and Iverfeldt 2009). Primate species differences in neurodegeneration could be sought elsewhere in the APP gene, including the APP promoter (Maloney et al. 2004). The tau peptide (MAPT gene) is also 100% identical in humans, chimpanzees, and in few other primates examined (Holzer et al. 2004) as discussed in Rosen et al. (2008).
The apolipoprotein E (ApoE) gene also merits attention in human-chimp aging differences. In the brain, apoE transports cholesterol to neurons, while blood apoE mediates the clearance of triglycerides. Humans have two main alleles: the most prevalent allele in all populations is ApoE3; ApoE4, the minor allele in all human populations (<1%–45%), is considered ancestral in our genus (Fullerton et al. 2000; Mahley et al. 2009). ApoE4 is associated with higher risk of AD and accelerated brain decline. Although the chimpanzee ApoE shares 2 amino acids with ApoE4, chimp ApoE is predicted to function like the human ApoE3 isoform because of a further coding difference that influences peptide folding (Raffai et al. 2001; Vamathevan et al. 2008). The putative ApoE3-like function could contribute to the low levels of Alzheimer’s and ischemic heart disease in chimpanzees (Finch and Stanford 2004; Finch 2010). Subcellularly, ApoE4 causes more lysosomal leakage than ApoE3, due to greater membrane disruption from peptide chain unfolding at lysosomal pH (“molten globule”) (Ji et al. 2006); this biophysical feature of ApoE4 is unique to humans and is implicated in the greater neurotoxicity of Aβ in ApoE4 transgenic models of AD (Mahley et al. 2009). Moreover, in transgenic mice with targeted replacement (TR) of the native apoE with the human isoforms, the TR-ApoE4 mice had extensive deposits of cerebrovascular amyloid and cerebral hemorrhage which were rarer in the TR-ApoE3 mice, and absent from the background strain (Sullivan et al. 2008). Although chimpanzee ApoE has not shown allelic variations in the few individuals sampled (Mahley et al. 2009), serum cholesterol had considerable heritability in a former breeding colony (Williams-Blangero et al. 1994). The uniquely human apoE3 allele spread about 0.226 MYA, range 0.18–0.58 MYA (Fullerton et al. 2000). These dates precede the emigration of modern Homo sapiens from Africa and overlap with the increased organized hunting of large animals and the use of fire (Finch and Stanford 2004).
Overview of primate research models
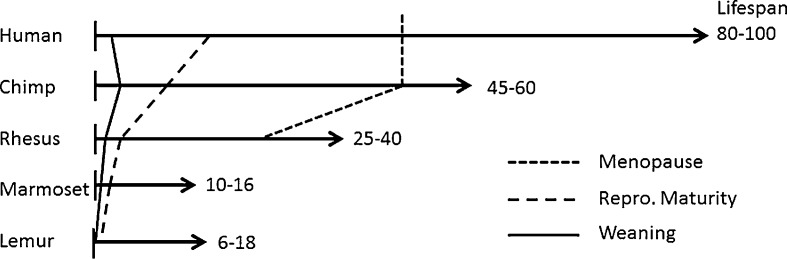
Although numerous primate species have been used for at least some aging research, we will focus on four species—common chimpanzees, rhesus macaques, common marmosets, and gray mouse lemurs—for which there is either the most relevance, the most information, or which exhibit exceptional promise for future aging research. Representative primate life history trajectories are compared with humans in Fig. 4.
Fig. 4.
Comparison of primate life history trajectories. Lifespan ranges represent maximum observed longevities in the wild and in captivity. Maximum human longevity is given as 100 years to compensate for the vast difference in amount of longevity data between humans and other primates. Only about one to two of 10,000 humans live as long as 100 years in modern industrialized populations. Marmoset common marmoset (C. jacchus), lemur gray mouse lemur (M. murinus). Note that marmosets and lemurs do not have menstrual cycles (they have estrous cycles), thus do not have menopause
Because chimpanzees are our closest relatives, both similarities and differences between their patterns of aging and age-related diseases and ours will be particularly enlightening with respect to understanding the genetic basis of aging and disease. The most common causes of older chimpanzee deaths in captivity in current colonies are related to congestive heart failure from myocardial fibrosis (Varki et al. 2009, 2011). Ischemic coronary disease may have been more prevalent in earlier colonies (Finch and Stanford 2004; Finch 2010). Cancers are at much lower prevalence in chimpanzees than in most human populations (Finch 2010; Varki et al. 2011). However, as mentioned above, we know almost nothing about cancer and other conditions of aging in feral populations of chimpanzees. More information on normative aging in chimpanzees would certainly continue to inform our view of the evolution of human aging. However due to the difficulties of research on chimpanzees, it is difficult to imagine that a large corpus of new information will be available in the near future.
Rhesus macaques have been by far the most commonly used primate species in aging research. Ironically given the degree to which they have been used in biomedical research, there is much less information on the life history of wild rhesus than on any of the great apes and what information exists comes mainly from provisioned monkeys on predator-free islands well outside their normal geographical range (Hoffman et al. 2010). In well-maintained captive colonies, mean adult survival is 25–27 years with the oldest animals reaching about 40 years of age (Colman et al. 2009; Bodkin et al. 2003).
The husbandry of rhesus appears to be superior to that of the great apes as shown by their considerably longer lives in captivity than even in provisioned free-living populations. However, on typical captive diets, >40% of older rhesus incur obesity in conjunction with glucoregulatory problems such as hyperinsulinemia and diabetes (Colman et al. 2009; Bodkin et al. 2003). Neoplasia in more than 2600 animals from two well-maintained colonies is reported in about 10% of animals at necropsy. However, among animals older than the median lifespan of 26 years, cancer contributed to approximately 50% of deaths (Simmons and Mattison 2011). Almost 50% of reported cancers affected the gastrointestinal system, possibly suggesting microbial involvement. This is a much higher prevalence of cancer than reported in earlier studies (reviewed in Finch 2010), likely due increasingly adept husbandry over time, such that animals of this species now survive considerably longer than previously. The high incidence of gastrointestinal cancers suggests, however, that environmental conditions could still be improved. Rhesus macaques will continue to make major contributions to our understanding of primate aging.
Two small, shorter-lived and primate species, though somewhat more distantly related to humans than Old World monkeys or apes, may prove exceptionally useful for future research. Advantages of these small primates are related to their relatively rapid life history, multiple births, short life, ease of handling, and reduced maintenance costs compared with larger species (Austad and Fischer 2011). The current and future use of the common marmoset (Callithrix jacchus) in aging research has been recently reviewed in detail (Tardif et al. 2011). A general overview of the use of marmosets in biomedical research may be found in (Mansfield 2003). In brief, the marmosets is among the smallest and shortest lived anthropoid primate with a body mass of 350–400 g, about 4% of the mass of an adult rhesus. Marmoset husbandry is well-enough developed to enable considerably longer lifespans in captivity (5–7 years mean and 17 years maximum) than in the wild (reported maximum of 10 years). Age-specific mortality shows acceleration after age 6 concurrent with declines in lean body mass. The survival curve might become more rectangular if animals were maintained under specific pathogen free (SPF) conditions. SPF conditions are defined by the absence of listed specific infectious pathogens from the colony, as monitored by regular testing. SPF conditions have dramatically improved the health and survival of laboratory rodents since 1970 and made studies of aging among laboratories much more replicable (Finch 2007, pp. 140–142). Although some primate studies employ SPF animals, it is far from the standard of current gerontological rodent colonies. Under current conditions, marmosets are considered to be “old” by the age of 8 years, which is greater than the median adult lifespan and about half of the maximum lifespan (Abbott et al. 2003).
In older marmosets from the New England National Primate Research Center common causes of death were neoplasia (16% of deaths in older animals), amyloidosis (13%), inflammatory bowel disease (13%), chronic renal disease (9%), and diabetes mellitis (7%). The high level of intestinal diseases (the most common malignant neoplasia was adenocarcinoma of the small intestine) suggests that as in virtually all other primate research colonies, intestinal microbiota may play a role in disease. The cerebral cortex showed diffuse Aβ plaques (but no neurofibrillary tangles) (Table 2). However, the frequency and age of onset of this amyloidosis varies widely among colonies in a manner reflective of differences in survival among those colonies (Tardif et al. 2011). Future studies of primate brain aging may access the advantages of the marmoset; their relatively rapid development, multiple births, short life, ease of handling, and reduced maintenance costs compared with larger species (Austad and Fischer 2011).
Lastly, we mention the gray mouse lemur (M. murinus), an even smaller and more rapidly reproducing species than the marmoset. These lemurs weigh 60–110 g, mature in less than 1 year, have multiple births and can reproduce at least twice per year (Picq 2007; Austad and Fischer 2011). Maintained in captivity since the 1950s, large colonies of mouse lemurs have been developed in France since the 1970s and studied for brain aging changes in detail (Bons et al. 2006). Life expectancy in this species is 8–10 years with a reported maximum of 18 years. Signs and symptoms of aging appear within the first decade of life. For instance, 50% of animals have cataracts by age 7 and substantial cognitive decline is evident by age 10 (Beltran et al. 2007; Picq 2007). Older mouse lemurs contract a wide range of tumors (Remick et al. 2009). Although we lack detailed information on the incidence of tumors in this species, about 20% of animal deaths were tumor-associated among captive prosimians generally. Of 123 tumors found in prosimians at the Duke University Lemur Center, none occurred in animals younger than age 5. In addition to multiple cognitive deficits, gray mouse lemurs also develop pathological brain changes including localized atrophy, diffuse and intraneuronal deposits of Aβ, astrocytosis with increased GFAP, and hyperphosphorylated tau in neurons (Dhenain et al. 2000; Bons et al. 2006; Kraska et al. 2011). As discussed below, the distribution of these lesions in the brain is different from humans. Husbandry practices in this species are difficult to assess. While mouse lemurs clearly live longer in research colonies than in the wild (Table 1), we do not know of detailed survival data along with the distribution of causes of death. One drawback of the use of lemurs in biomedical research is that animals cannot be taken from the wild and the international trade of captive individuals is highly regulated (they are listed in Appendix I of the Convention on International Trade in Endangered Species). Consequently, research may proceed at existing colonies but establishing new research colonies presents a challenge.
Husbandry issues
Aging-related biomedical research is most informative when husbandry of the study animals has been optimized to minimize infectious diseases and maximize nutritional and psychological well-being of the animals. Such conditions reveal changes over time specific to aging rather than environmental pathologies. Such detailed attention to husbandry has revolutionized aging research with rodents over the past half century (Finch 2007), making research not only more informative but also more replicable among laboratories.
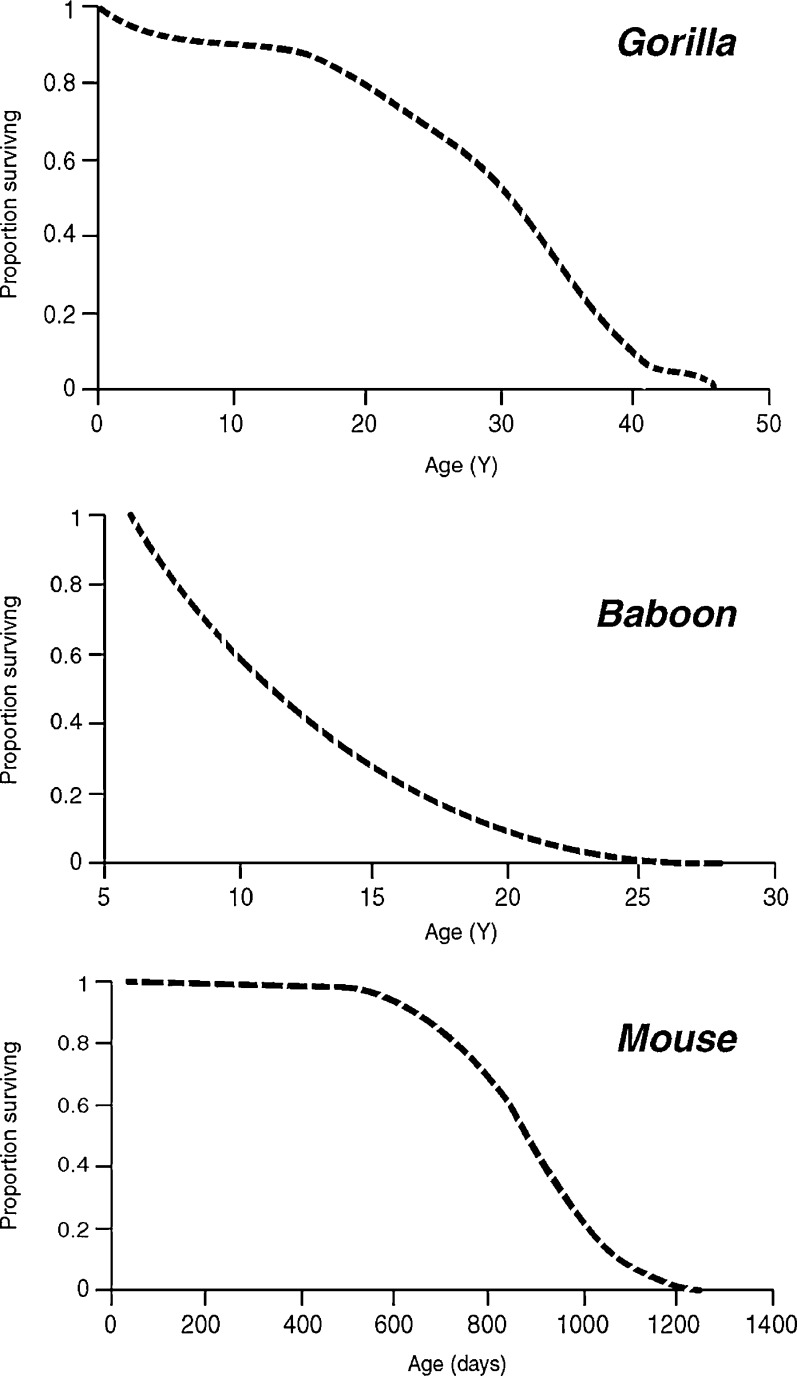
By rodent standards, captive primate husbandry is still at an early stage of development. This is evident either when inspecting captive survival data or when comparing the longevity of captive versus wild populations. For instance, in laboratory mice and rats as well as humans from industrialized countries typically display survival curves with a pronounced right “shoulder” because survival is uniformly high early in adult life but then declines rapidly as senescence progresses (Fig. 5) (Turturro et al. 1999; Austad 2001). Elimination of chronic infections has been a major contributor to producing this survival curve shape (Finch 2007). By contrast, because substantial mortality occurs even early in adulthood and long before evident senescence, captive primate survival curves are typically more linear, even concave (Allman et al. 1998; Smucny et al. 2004). In addition, given optimum husbandry one would expect animals in captivity to survive far longer than animals in the wild. In captivity, food is reliably abundant and predators are absent. Mice and rats clearly survive much longer in the lab than they do in the wild. However, a number of captive primate species live just marginally longer—if that—in captivity compared to the wild (Table 1). For instance, the median longevity of eastern gorillas in a wild Rwandan population is 33 years compared with about 25 years in zoo populations (Kohler et al. 2006). However, maximum zoo longevity for gorillas is substantially longer in zoos than in the wild (Table 1). In orangutans, on the other hand, life expectancy in the wild is considerably longer than in captivity (30 vs. 19 years combining the sexes) and maximum longevity is virtually identical (Wich et al. 2004; Kohler et al. 2006). The causes of reduced captive compared to wild longevity are manifold, no doubt including suboptimal diets, obesity, lack of exercise, inbreeding, poor adaptation to the local climate (for outdoor colonies), and psychological stress. Primate diets in nature are notoriously complex and seasonally variable, as are their social environments. At present, there is little standardization among captive colonies of any species in either diet or social environments. Obesity is rampant in captive mammals of virtually all species, and primates are no exception (Hotta et al. 1996; Klimentidis et al. 2011; Steinetz et al. 1996). A significant challenge is to define healthy (normative) body weight or body mass index for the common research primates. Fortunately, husbandry practices continue to improve. Strikingly, the lifespans of orangutans born in zoos since the middle-1980s approximates that of wild orangutans, for instance (Wich et al. 2009). However, for primates to be maximally informative for aging studies, obesity, stress, and chronic infections will likely need to be controlled to the current biogerontological standards of mouse and rat colonies, or even better. Thus, we cannot yet draw firm conclusions about whether the apparent gross divergence of brain aging patterns between primate species is nature or nurture.
Fig. 5.
Representative survival curves for captive populations of female gorillas (N = 458), baboons (N = 422), and genetically heterogeneous laboratory mice (N = 210). Note the more “rectangular” shape of the mouse curve, particularly when compared with the baboon curve but also in comparison with the gorilla curve. Redrawn from Allman et al. (1998), Martin et al. (2002), and Strong et al. (2008)
Acknowledgments
SA acknowledges the support of NIH grant R24 RR023344. CEF acknowledges the support of NIH grant P01 AG-026572. Molly Austad generously regraphed Figs. 2, 3, and 5.
Footnotes
In, AGE journal special issue “Nonhuman Primate Models of Aging” Ilhem Messaoudi, Ph.D. (Guest Editor) messaoud@ohsu.edu
References
- Abbott DH, Barnett DK, Colman RJ, Yamamoto ME, Schultz-Darken NJ. Aspects of common marmoset basic biology and life history important for biomedical research. Comp Med. 2003;53:339–350. [PubMed] [Google Scholar]
- Allman J, Rosin A, Kumar R, Hasenstaub A. Parenting and survival in anthropoid primates: caretakers live longer. Proc Natl Acad Sci U S A. 1998;95:6866–6869. doi: 10.1073/pnas.95.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann J, Gesquiere L, Galbany J, Onyango PO, Alberts SC. Life history context of reproductive aging in a wild primate model. Ann N Y Acad Sci. 2010;1204:127–138. doi: 10.1111/j.1749-6632.2010.05531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Meyer JS, Okabe T, Shaw T, Mortel KF (1982) Stable xenon CT cerebral blood flow measurements computed by a single compartment–double integration model in normal aging and dementia. J Comput Assist Tomogr 6:923–932 [DOI] [PubMed]
- Anderson US, Stoinski TS, Bloomsmith MA, Maple TL. Relative numerousness judgment and summation in young, middle-aged, and older adult orangutans (Pongo pygmaeus abelii and Pongo pygmaeus pygmaeus) J Comp Psychol. 2007;121:1–11. doi: 10.1037/0735-7036.121.1.1. [DOI] [PubMed] [Google Scholar]
- Anderson US, Stoinski TS, Bloomsmith MA, Marr MJ, Smith AD, Maple TL. Relative numerousness judgment and summation in young and old Western lowland gorillas. J Comp Psychol. 2005;119:285–295. doi: 10.1037/0735-7036.119.3.285. [DOI] [PubMed] [Google Scholar]
- Arriagada PV, Marzloff K, Hyman BT. Distribution of Alzheimer-type pathologic changes in nondemented elderly individuals matches the pattern in Alzheimer’s disease. Neurology. 1992;42:1681–1688. doi: 10.1212/wnl.42.9.1681. [DOI] [PubMed] [Google Scholar]
- Atsalis S, Videan E. Reproductive aging in captive and wild common chimpanzees: factors influencing the rate of follicular depletion. Am J Primatol. 2009;71:271–282. doi: 10.1002/ajp.20650. [DOI] [PubMed] [Google Scholar]
- Austad SN. Concepts and theories of aging. In: Masoro EJ, Austad SN, editors. Handbook of the biology of aging. 5. San Diego: Academic; 2001. pp. 3–22. [Google Scholar]
- Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol. 1991;46:B47–B53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- Austad SN, Fischer KE. Primate longevity: its place in the mammalian scheme. Am J Primatol. 1992;28:251–261. doi: 10.1002/ajp.1350280403. [DOI] [PubMed] [Google Scholar]
- Austad SN, Fischer KE. The development of small primate models for aging research. ILAR J. 2011;52:78–88. doi: 10.1093/ilar.52.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran WA, Vanore M, Ollivet F, Nemoz-Bertholet F, Aujard F, Clerc B, et al. Ocular findings in two colonies of gray mouse lemurs (Microcebus murinus) Vet Ophthalmol. 2007;10:43–49. doi: 10.1111/j.1463-5224.2007.00491.x. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Grondin R, Chen KC, Thibault O, Thibault V, Pandya JD, et al. Aging-related gene expression in hippocampus proper compared with dentate gyrus is selectively associated with metabolic syndrome variables in rhesus monkeys. J Neurosci. 2010;30:6058–6071. doi: 10.1523/JNEUROSCI.3956-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodkin NL, Alexander TM, Ortmeyer HK, Johnson E, Hansen BC. Mortality and morbidity in laboratory-maintained rhesus monkeys and effects of long-term dietary restriction. J Gerontol A Biol Sci Med Sci. 2003;58:212–219. doi: 10.1093/gerona/58.3.b212. [DOI] [PubMed] [Google Scholar]
- Bogin B. Evolutionary perspective on human growth. Ann Rev Anthropol. 1999;28:109–156. doi: 10.1146/annurev.anthro.28.1.109. [DOI] [PubMed] [Google Scholar]
- Bons N, Rieger F, Prudhomme D, Fisher A, Krause KH. Microcebus murinus: a useful primate model for human cerebral aging and Alzheimer’s disease? Genes Brain Behav. 2006;5:120–130. doi: 10.1111/j.1601-183X.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- Boyd-Kimball D, Sultana R, Mohmmad-Abdul H, Butterfield DA. Rodent Abeta(1-42) exhibits oxidative stress properties similar to those of human Abeta(1-42): implications for proposed mechanisms of toxicity. J Alzheimers Dis. 2004;6:515–525. doi: 10.3233/jad-2004-6509. [DOI] [PubMed] [Google Scholar]
- Braak H, Del TK. Alzheimer’s pathogenesis: is there neuron-to-neuron propagation? Acta Neuropathol. 2011;121:589–595. doi: 10.1007/s00401-011-0825-z. [DOI] [PubMed] [Google Scholar]
- Braak H, Del TK. The pathological process underlying Alzheimer’s disease in individuals under thirty. Acta Neuropathol. 2011;121:171–181. doi: 10.1007/s00401-010-0789-4. [DOI] [PubMed] [Google Scholar]
- Bronikowski AM, Alberts SC, Altmann J, Packer C, Carey KD, Tatar M. The aging baboon: comparative demography in a non-human primate. Proc Natl Acad Sci U S A. 2002;99:9591–9595. doi: 10.1073/pnas.142675599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronikowski AM, Altmann J, Brockman DK, Cords M, Fedigan LM, Pusey A, et al. Aging in the natural world: comparative data reveal similar mortality patterns across primates. Science. 2011;331:1325–1328. doi: 10.1126/science.1201571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R. The naked mole-rat: a new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- Caspari R, Lee SH. Older age becomes common late in human evolution. Proc Natl Acad Sci U S A. 2004;101:10895–10900. doi: 10.1073/pnas.0402857101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee HJ, Ho SY, Barnes I, Groves C. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol Biol. 2009;9:259. doi: 10.1186/1471-2148-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing & Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cole GM, Ma QL, Frautschy SA. Dietary fatty acids and the aging brain. Nutr Rev. 2010;68(Suppl 2):S102–S111. doi: 10.1111/j.1753-4887.2010.00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson EJ, Paliga K, Beyreuther K, Masters CL. What the evolution of the amyloid protein precursor supergene family tells us about its function. Neurochem Int. 2000;36:175–184. doi: 10.1016/s0197-0186(99)00125-4. [DOI] [PubMed] [Google Scholar]
- Dhenain M, Michot JL, Privat N, Picq JL, Boller F, Duyckaerts C, et al. MRI description of cerebral atrophy in mouse lemur primates. Neurobiol Aging. 2000;21:81–88. doi: 10.1016/s0197-4580(00)00098-1. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Stoub TR, Shah RC, Sperling RA, Killiany RJ, Albert MS, et al. Alzheimer-signature MRI biomarker predicts AD dementia in cognitively normal adults. Neurology. 2011;76:1395–1402. doi: 10.1212/WNL.0b013e3182166e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Wearne SL, Rocher AB, Macedo A, Morrison JH, Hof PR. Age-related dendritic and spine changes in corticocortically projecting neurons in macaque monkeys. Cereb Cortex. 2003;13:950–961. doi: 10.1093/cercor/13.9.950. [DOI] [PubMed] [Google Scholar]
- Eisenberg JF. The mammalian radiations. Chicago: University of Chicago Press; 1981. [Google Scholar]
- Fabre PH, Rodrigues A, Douzery EJ. Patterns of macroevolution among primates inferred from a supermatrix of mitochondrial and nuclear DNA. Mol Phylogenet Evol. 2009;53:808–825. doi: 10.1016/j.ympev.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Peters A, Barbas H. Mitochondrial degeneration in dystrophic neurites of senile plaques may lead to extracellular deposition of fine filaments. Brain Struct Funct. 2007;212:195–207. doi: 10.1007/s00429-007-0153-1. [DOI] [PubMed] [Google Scholar]
- Finch CE. Enzyme activities, gene function and ageing in mammals. (Review) Exp Gerontol. 1972;7:53–67. doi: 10.1016/0531-5565(72)90035-6. [DOI] [PubMed] [Google Scholar]
- Finch CE. The biology of human longevity: inflammation, nutrition, and aging in the evolution of lifespans. San Diego: Academic; 2007. [Google Scholar]
- Finch CE. The neurobiology of middle-age has arrived. Neurobiol Aging. 2009;30:515–520. doi: 10.1016/j.neurobiolaging.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Finch CE. Evolution in health and medicine Sackler colloquium: evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE, Holmes DJ. Ovarian aging in developmental and evolutionary contexts. Ann N Y Acad Sci. 2010;1204:82–94. doi: 10.1111/j.1749-6632.2010.05610.x. [DOI] [PubMed] [Google Scholar]
- Finch CE, Morgan TE, Rozovsky I, Xie Z, Weindruch R, Prolla T. Microglia and aging in the brain. In: Streit WJ, editor. Microglia in the regenerating and degenerating central nervous system. New York: Springer; 2002. pp. 275–305. [Google Scholar]
- Finch CE, Stanford CB. Meat-adaptive genes and the evolution of slower aging in humans. Q Rev Biol. 2004;79:3–50. doi: 10.1086/381662. [DOI] [PubMed] [Google Scholar]
- Fjell AM, Westlye LT, Espeseth T, Reinvang I, Dale AM, Holland D, et al. Cortical gray matter atrophy in healthy aging cannot be explained by undetected incipient cognitive disorders: a comment on Burgmans et al. 2009. Neuropsychology. 2010;24:258–263. doi: 10.1037/a0018827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto H, Rosene DL, Moss MB, Raju S, Hyman BT, Irizarry MC. Beta-secretase activity increases with aging in human, monkey, and mouse brain. Am J Pathol. 2004;164:719–725. doi: 10.1016/s0002-9440(10)63159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengard JH, et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing M, Rebeck GW, Hyman BT, Tigges J, Mirra SS. Neuropathology and apolipoprotein E profile of aged chimpanzees: implications for Alzheimer disease. Proc Natl Acad Sci USA. 1994;91:9382–93826. doi: 10.1073/pnas.91.20.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing M, Tigges J, Mori H, Mirra SS. A beta40 is a major form of beta-amyloid in nonhuman primates. Neurobiol Aging. 1996;17:903–908. doi: 10.1016/s0197-4580(96)00164-9. [DOI] [PubMed] [Google Scholar]
- Gearing M, Tigges J, Mori H, Mirra SS. A beta-amyloid (A beta) deposition in the brains of aged orangutans. Neurobiol Aging. 1997;18:139–146. doi: 10.1016/s0197-4580(97)00012-2. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Glazko GV, Nei M. Estimation of divergence times for major lineages of primate species. Mol Biol Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- Grzimek B (1990) Grzimek’s Encyclopedia of Mammals, vol 2, Parker SP (ed), McGraw-Hill, New York
- Haley GE, Kohama SG, Urbanski HF, Raber J. Age-related decreases in SYN levels associated with increases in MAP-2, apoE, and GFAP levels in the rhesus macaque prefrontal cortex and hippocampus. Age (Dordr) 2010;32:283–296. doi: 10.1007/s11357-010-9137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LA, Armstrong DM, Terry RD. An immunohistochemical quantification of fibrous astrocytes in the aging human cerebral cortex. Neurobiol Aging. 1987;8:1–6. doi: 10.1016/0197-4580(87)90051-0. [DOI] [PubMed] [Google Scholar]
- Harada N, Nishiyama S, Satoh K, Fukumoto D, Kakiuchi T, Tsukada H. Age-related changes in the striatal dopaminergic system in the living brain: a multiparametric PET study in conscious monkeys. Synapse. 2002;45:38–45. doi: 10.1002/syn.10082. [DOI] [PubMed] [Google Scholar]
- Hardy J. The amyloid hypothesis for Alzheimer’s disease: a critical reappraisal. J Neurochem. 2009;110:1129–1134. doi: 10.1111/j.1471-4159.2009.06181.x. [DOI] [PubMed] [Google Scholar]
- Hawkes K, Paine RR. The evolution of human life history. Santa Fe: School of American Research Press; 2006. [Google Scholar]
- Hawkes K, Smith KR. Do women stop early? Similarities in fertility decline in humans and chimpanzees. Ann N Y Acad Sci. 2010;1204:43–53. doi: 10.1111/j.1749-6632.2010.05527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon JG, Moss MB, Rosene DL, Killiany RJ. Patterns of cognitive decline in aged rhesus monkeys. Behav Brain Res. 1997;87:25–34. doi: 10.1016/s0166-4328(96)02256-5. [DOI] [PubMed] [Google Scholar]
- Herndon JG, Tigges J, Klumpp SA, Anderson DC. Brain weight does not decrease with age in adult rhesus monkeys. Neurobiol Aging. 1998;19:267–272. doi: 10.1016/s0197-4580(98)00054-2. [DOI] [PubMed] [Google Scholar]
- Hoffman CL, Higham JP, Mas-Rivera A, Ayala JE, Maestripieri D. Terminal investment and senescence in rhesus macaques (Macaca mulatta) on Cayo Santiago. Behav Ecol. 2010;21:972–978. doi: 10.1093/beheco/arq098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer M, Craxton M, Jakes R, Arendt T, Goedert M. Tau gene (MAPT) sequence variation among primates. Gene. 2004;341:313–322. doi: 10.1016/j.gene.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Hotta K, Gustafson TA, Ortmeyer HK, Bodkin NL, Nicolson MA, Hansen BC. Regulation of obese (ob) mRNA and plasma leptin levels in rhesus monkeys. Effects of insulin, body weight, and non-insulin-dependent diabetes mellitus. J Biol Chem. 1996;271:25327–25331. doi: 10.1074/jbc.271.41.25327. [DOI] [PubMed] [Google Scholar]
- Ingram DK, Chefer S, Matochik J, Moscrip TD, Weed J, Roth GS, et al. Aging and caloric restriction in nonhuman primates: behavioral and in vivo brain imaging studies. Ann N Y Acad Sci. 2001;928:316–326. doi: 10.1111/j.1749-6632.2001.tb05661.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen KT, Iverfeldt K. Amyloid precursor protein and its homologues: a family of proteolysis-dependent receptors. Cell Mol Life Sci. 2009;66:2299–2318. doi: 10.1007/s00018-009-0020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji ZS, Mullendorff K, Cheng IH, Miranda RD, Huang Y, Mahley RW. Reactivity of apolipoprotein E4 and amyloid beta peptide: lysosomal stability and neurodegeneration. J Biol Chem. 2006;281:2683–2692. doi: 10.1074/jbc.M506646200. [DOI] [PubMed] [Google Scholar]
- Johnstone EM, Chaney MO, Norris FH, Pascual R, Little SP. Conservation of the sequence of the Alzheimer’s disease amyloid peptide in dog, polar bear and five other mammals by cross-species polymerase chain reaction analysis. Brain Res Mol Brain Res. 1991;10:299–305. doi: 10.1016/0169-328x(91)90088-f. [DOI] [PubMed] [Google Scholar]
- Judge DS, Carey JR. Postreproductive life predicted by primate patterns. J Gerontol A Biol Sci Med Sci. 2000;55:B201–B209. doi: 10.1093/gerona/55.4.b201. [DOI] [PubMed] [Google Scholar]
- Kadish I, Thibault O, Blalock EM, Chen KC, Gant JC, Porter NM, et al. Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J Neurosci. 2009;29:1805–1816. doi: 10.1523/JNEUROSCI.4599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan H, Hill K, Lancaster JA, Hurtado AM. A theory of human life history: diet, intelligence, and longevity. Evolutionary Anthropology. 2000;9:156–185. [Google Scholar]
- Kimura N, Nakamura S, Goto N, Narushima E, Hara I, Shichiri S, Saitou K, Nose M, Hayashi T, Kawamura S, Yoshikawa Y (2001) Senile plaques in an aged western lowland gorilla. Exp Anim 50:77–81 [DOI] [PubMed]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Klein WL, Krafft GA, Finch CE. Targeting small Abeta oligomers: the solution to an Alzheimer’s disease conundrum? Trends Neurosci. 2001;24:219–224. doi: 10.1016/s0166-2236(00)01749-5. [DOI] [PubMed] [Google Scholar]
- Klimentidis YC, Beasley TM, Lin HY, Murati G, Glass GE, Guyton M, Newton W, Jorgensen M, Heymsfield SB, Kemnitz J, Fairbanks L, Allison DB. Canaries in the coal mine: a cross-species analysis of the plurality of obesity epidemics. Proc Biol Sci. 2011;278:1626–1632. doi: 10.1098/rspb.2010.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler IV, Preston SH, Lackey LB. Comparative mortality levels among selected species of captive animals. Demogr Res. 2006;15:413–434. [Google Scholar]
- Kraska A, Dorieux O, Picq JL, Petit F, Bourrin E, Chenu E, Volk A, Perret M, Hantraye P, Mestre-Frances N, Aujard F, Dhenain M. Age-associated cerebral atrophy in mouse lemur primates. Neurobiol Aging. 2011;32:894–906. doi: 10.1016/j.neurobiolaging.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacreuse A, Kim CB, Rosene DL, Killiany RJ, Moss MB, Moore TL, et al. Sex, age, and training modulate spatial memory in the rhesus monkey (Macaca mulatta) Behav Neurosci. 2005;119:118–126. doi: 10.1037/0735-7044.119.1.118. [DOI] [PubMed] [Google Scholar]
- Leduc V, Jasmin-Belanger S, Poirier J. APOE and cholesterol homeostasis in Alzheimer’s disease. Trends Mol Med. 2010;16:469–477. doi: 10.1016/j.molmed.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Li KZ, Lindenberger U, Freund AM, Baltes PB. Walking while memorizing: age-related differences in compensatory behavior. Psychol Sci. 2001;12:230–237. doi: 10.1111/1467-9280.00341. [DOI] [PubMed] [Google Scholar]
- Luebke J, Barbas H, Peters A. Effects of normal aging on prefrontal area 46 in the rhesus monkey. Brain Res Rev. 2010;62:212–232. doi: 10.1016/j.brainresrev.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutermann H, Schmelting B, Radespiel U, Ehresmann P, Zimmermann E (2006) The role of survival for the evolution of female philopatry in a solitary forager, the grey mouse lemur (Microcebus murinus). Proc R Soc B 273:2527–2533 [DOI] [PMC free article] [PubMed]
- Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J Lipid Res. 2009;50(Suppl):S183–S188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney B, Ge YW, Greig N, Lahiri DK. Presence of a “CAGA box” in the APP gene unique to amyloid plaque-forming species and absent in all APLP-1/2 genes: implications in Alzheimer’s disease. FASEB J. 2004;18:1288–1290. doi: 10.1096/fj.03-1703fje. [DOI] [PubMed] [Google Scholar]
- Mansfield K. Marmoset models commonly used in biomedical research. Comp Med. 2003;53:383–392. [PubMed] [Google Scholar]
- Marchal S, Givalois L, Verdier JM, Mestre-Francés N (2012) Distribution of lithostathine in the mouse lemur brain with aging and Alzeheimer’s-like pathology. Neurobiol Aging 33:431.e15–25 [DOI] [PubMed]
- Martin LJ, Mahaney MC, Bronikowski AM, Carey KD, Dyke B, Comuzzie AG (2002) Lifespan in captive baboons is heritable. Mech Age Devel 123:1461–1467 [DOI] [PubMed]
- Masliah E, Mallory M, Hansen L, DeTeresa R, Terry RD. Quantitative synaptic alterations in the human neocortex during normal aging. Neurology. 1993;43:192–197. doi: 10.1212/wnl.43.1_part_1.192. [DOI] [PubMed] [Google Scholar]
- McArdle JJ. Latent variable modeling of differences and changes with longitudinal data. Annu Rev Psychol. 2009;60:577–605. doi: 10.1146/annurev.psych.60.110707.163612. [DOI] [PubMed] [Google Scholar]
- McDonald CR, McEvoy LK, Gharapetian L, Fennema-Notestine C, Hagler DJ, Jr, Holland D, et al. Regional rates of neocortical atrophy from normal aging to early Alzheimer disease. Neurology. 2009;73:457–465. doi: 10.1212/WNL.0b013e3181b16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman ML. Growth of the birth canal in adolescent girls. Am J Obstet Gynecol. 1982;143:528–532. doi: 10.1016/0002-9378(82)90542-7. [DOI] [PubMed] [Google Scholar]
- Moore TL, Killiany RJ, Herndon JG, Rosene DL, Moss MB. Executive system dysfunction occurs as early as middle-age in the rhesus monkey. Neurobiol Aging. 2006;27:1484–1493. doi: 10.1016/j.neurobiolaging.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Morgan DG, Marcusson JO, Nyberg P, Wester P, Winblad B, Gordon MN, et al. Divergent changes in D-1 and D-2 dopamine binding sites in human brain during aging. Neurobiol Aging. 1987;8:195–201. doi: 10.1016/0197-4580(87)90002-9. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Pevzner PA, O’Brien SJ. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Nowak RM. Walker’s mammals of the world. 6. Baltimore: Johns Hopkins University Press; 1999. [Google Scholar]
- Ohm TG, Muller H, Braak H, Bohl J. Close-meshed prevalence rates of different stages as a tool to uncover the rate of Alzheimer’s disease-related neurofibrillary changes. Neuroscience. 1995;64:209–217. doi: 10.1016/0306-4522(95)90397-p. [DOI] [PubMed] [Google Scholar]
- Picq JL. Aging affects executive functions and memory in mouse lemur primates. Exp Gerontol. 2007;42:223–232. doi: 10.1016/j.exger.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Picq JL, Aujard F, Volk A, Dhenain M (2011) Age-related cerebral atrophy in nonhuman primates predicts cognitive impairments. Neurobiol Aging (in press) [DOI] [PMC free article] [PubMed]
- Prasher VP, Farrer MJ, Kessling AM, Fisher EM, West RJ, Barber PC, et al. Molecular mapping of Alzheimer-type dementia in Down’s syndrome. Ann Neurol. 1998;43:380–383. doi: 10.1002/ana.410430316. [DOI] [PubMed] [Google Scholar]
- Raffai RL, Dong LM, Farese RV, Jr, Weisgraber KH. Introduction of human apolipoprotein E4 “domain interaction” into mouse apolipoprotein E. Proc Natl Acad Sci U S A. 2001;98:11587–11591. doi: 10.1073/pnas.201279298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp PR, Amaral DG. Recognition memory deficits in a subpopulation of aged monkeys resemble the effects of medial temporal lobe damage. Neurobiol Aging. 1991;12:481–486. doi: 10.1016/0197-4580(91)90077-w. [DOI] [PubMed] [Google Scholar]
- Remick AK, Wettere AJ, Williams CV. Neoplasia in prosimians: case series from a captive prosimian population and literature review. Vet Pathol. 2009;46:746–772. doi: 10.1354/vp.08-VP-0154-R-FL. [DOI] [PubMed] [Google Scholar]
- Robbins MM, Bermejo M, Cipolletta C, Magliocca F, Parnell RJ, Stokes E. Social structure and life-history patterns in western gorillas (Gorilla gorilla gorilla) Am J Primatol. 2004;64:145–159. doi: 10.1002/ajp.20069. [DOI] [PubMed] [Google Scholar]
- Robson SL, Wood B. Hominin life history: reconstruction and evolution. J Anat. 2008;212:394–425. doi: 10.1111/j.1469-7580.2008.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen RD (2008) The lie of the jungle. Washington Post, 7 December 2008, p. W14
- Rosen RF, Farberg AS, Gearing M, Dooyema J, Long PM, Anderson DC, et al. Tauopathy with paired helical filaments in an aged chimpanzee. J Comp Neurol. 2008;509:259–270. doi: 10.1002/cne.21744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le MN, Laquerriere A, Vital A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- Rowe N. Pictorial guide to the living primates. East Hampton: Pogonias Press; 1996. [Google Scholar]
- Rozovsky I, Wei M, Morgan TE, Finch CE. Reversible age impairments in neurite outgrowth by manipulations of astrocytic GFAP. Neurobiol Aging. 2005;26:705–715. doi: 10.1016/j.neurobiolaging.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Salthouse TA. When does age-related cognitive decline begin? Neurobiol Aging. 2009;30:507–514. doi: 10.1016/j.neurobiolaging.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C, Hubbard GB, Rüb U, Braak E, Braak H (2000) Age-related progression of tau pathology in brains of baboons. Neurobiol Aging 21:905–912 [DOI] [PubMed]
- Severson JA, Finch CE. Reduced dopaminergic binding during aging in the rodent striatum. Brain Res. 1980;192:147–162. doi: 10.1016/0006-8993(80)91015-x. [DOI] [PubMed] [Google Scholar]
- Severson JA, Marcusson J, Winblad B, Finch CE. Age-correlated loss of dopaminergic binding sites in human basal ganglia. J Neurochem. 1982;39:1623–1631. doi: 10.1111/j.1471-4159.1982.tb07996.x. [DOI] [PubMed] [Google Scholar]
- Shamy JL, Habeck C, Hof PR, Amaral DG, Fong SG, Buonocore MH, et al. Volumetric correlates of spatiotemporal working and recognition memory impairment in aged rhesus monkeys. Cereb Cortex. 2011;21:1559–1573. doi: 10.1093/cercor/bhq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood CC, Gordon AD, Allen JS, Phillips KE, Erwin JW, Hof PW, et al. (2011) Aging of the cerebral cortex differs between humans and chimpanzees. Proc Natl Acad Sci U S A (in press) [DOI] [PMC free article] [PubMed]
- Simmons HA, Mattison JA. The incidence of spontaneous neoplasia in two populations of captive rhesus macaques (Macaca mulatta) Antioxid Redox Signal. 2011;14:221–227. doi: 10.1089/ars.2010.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonian NA, Hyman BT. Functional alterations in Alzheimer’s disease: selective loss of mitochondrial-encoded cytochrome oxidase mRNA in the hippocampal formation. J Neuropathol Exp Neurol. 1994;53:508–512. doi: 10.1097/00005072-199409000-00010. [DOI] [PubMed] [Google Scholar]
- Smucny DA, Abbott DH, Mansfield KG, Schultz-Darken NJ, Yamamoto ME, Alencar AI, et al. Reproductive output, maternal age, and survivorship in captive common marmoset females (Callithrix jacchus) Am J Primatol. 2004;64:107–121. doi: 10.1002/ajp.20065. [DOI] [PubMed] [Google Scholar]
- Soscia SJ, Kirby JE, Washicosky KJ, Tucker SM, Ingelsson M, Hyman B, et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One. 2010;5:e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinetz BG, Randolph C, Cohn D, Mahoney CJ. Lipoprotein profiles and glucose tolerance in lean and obese chimpanzees. J Med Primatol. 1996;25:17–25. doi: 10.1111/j.1600-0684.1996.tb00188.x. [DOI] [PubMed] [Google Scholar]
- Strong R, Miller RA, Astle CM, Floyd RA, Flurkey K, Hensley KL, Javors MA, Leeuwenburgh C, Nelson JF, Ongini E, Nadon NL, Warner HR, Harrison DE (2008) Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell 7:641–650 [DOI] [PMC free article] [PubMed]
- Sullivan PM, Mace BE, Estrada JC, Schmechel DE, Alberts MJ. Human apolipoprotein E4 targeted replacement mice show increased prevalence of intracerebral hemorrhage associated with vascular amyloid deposition. J Stroke Cerebrovasc Dis. 2008;17:303–311. doi: 10.1016/j.jstrokecerebrovasdis.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Tardif SD, Mansfield KG, Ratnam R, Ross CN, Ziegler TE. The marmoset as a model of aging and age-related diseases. ILAR J. 2011;52:54–65. doi: 10.1093/ilar.52.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–B501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- Vamathevan JJ, Hasan S, Emes RD, Amrine-Madsen H, Rajagopalan D, Topp SD, et al. The role of positive selection in determining the molecular cause of species differences in disease. BMC Evol Biol. 2008;8:273. doi: 10.1186/1471-2148-8-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki N, Anderson D, Herndon JG, Pham T, Gregg CJ, Cheriyan M, et al. Heart disease is common in humans and chimpanzees, but is caused by different pathological processes. Evol Appl. 2009;2:101–112. doi: 10.1111/j.1752-4571.2008.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki NM, Strobert E, Dick EJ, Jr, Benirschke K, Varki A. Biomedical differences between human and nonhuman hominids: potential roles for uniquely human aspects of sialic acid biology. Annu Rev Pathol. 2011;6:365–393. doi: 10.1146/annurev-pathol-011110-130315. [DOI] [PubMed] [Google Scholar]
- Vaughan TA, Ryan JM, Czaplewski NJ. Mammalogy. 4. Orlando: Harcourt; 2000. [Google Scholar]
- Walsh DM, Selkoe DJ. A beta oligomers—a decade of discovery. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wich SA, Shumaker RW, Perkins L, Vries H. Captive and wild orangutan (Pongo sp.) survivorship: a comparison and the influence of management. Am J Primatol. 2009;71:680–686. doi: 10.1002/ajp.20704. [DOI] [PubMed] [Google Scholar]
- Wich SA, Utami-Atmoko SS, Setia TM, Rijksen HD, Schurmann C, Hooff JA, et al. Life history of wild Sumatran orangutans (Pongo abelii) J Hum Evol. 2004;47:385–398. doi: 10.1016/j.jhevol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Williams-Blangero S, Butler T, Brasky K, Murthy KK. Heritabilities of clinical chemical traits in chimpanzees. Lab Anim Sci. 1994;44:141–143. [PubMed] [Google Scholar]
- Yamagiwa J (1997) Mushamuka’s story: the largest group and the longest tenure. Gorilla J 15:7–9
- Zanjani H, Finch CE, Kemper C, Atkinson J, McKeel D, Morris JC, et al. Complement activation in very early Alzheimer disease. Alzheimer Dis Assoc Disord. 2005;19:55–66. doi: 10.1097/01.wad.0000165506.60370.94. [DOI] [PubMed] [Google Scholar]