Abstract
Accumulation of DNA lesions compromises replication and transcription and is thus toxic to cells. DNA repair deficiencies are generally associated with cellular replicative senescence and premature aging syndromes, suggesting that efficient DNA repair is required for normal longevity. It follows that the evolution of increasing lifespan amongst animal species should be associated with enhanced DNA repair capacities. Although UV damage repair has been shown to correlate positively with mammalian species lifespan, we lack similar insight into many other DNA repair pathways, including base excision repair (BER). DNA is continuously exposed to reactive oxygen species produced during aerobic metabolism, resulting in the occurrence of oxidative damage within DNA. Short-patch BER plays an important role in repairing the resultant oxidative lesions. We therefore tested whether an enhancement of BER enzyme activities has occurred concomitantly with the evolution of increased maximum lifespan (MLSP). We collected brain and liver tissue from 15 vertebrate endotherm species ranging in MLSP over an order of magnitude. We measured apurinic/apyrimidinic (AP) endonuclease activity, as well as the rates of nucleotide incorporation into an oligonucleotide containing a single nucleotide gap (catalyzed by BER polymerase β) and subsequent ligation of the oligonucleotide. None of these activities correlated positively with species MLSP. Rather, nucleotide incorporation and oligonucleotide ligation activities appeared to be primarily (and negatively) correlated with species body mass.
Keywords: Base excision repair (BER), DNA repair, Maximum lifespan, Body mass, Polymerase β, AP endonuclease, APE1
Introduction
When the rate of ROS production exceeds that of neutralization, oxidative damage can accumulate within proteins, lipids, and DNA. The accumulation of DNA damage, however, may be the most detrimental to cell viability due to the risk that replication and transcription of the genome will be permanently compromised. The exposure of mammalian DNA to endogenous genotoxic agents has been estimated to account for on the order of 10,000 DNA lesions per cell per day (Lindahl 1993), including base loss or modification and strand breaks (Barnes and Lindahl 2004). The failure to properly remove and repair DNA lesions can result in genomic instability, mutagenesis, transcriptional blockage, and/or apoptotic cell death.
DNA repair deficiencies are associated with replicative senescence and premature aging syndromes, suggesting that competent DNA repair pathways are essential to achieving normal lifespan. A corollary of this is that longer-lived animal species, or long-lived strains of a particular species, should presumably have greater DNA repair capacities, thus allowing the maintenance of genomic integrity over longer lifespans. Several reports support this hypothesis. Hart and Setlow (1974), Francis et al. (1981), Hall et al. (1984), and Licastro and Walford (1985) have all demonstrated positive correlations between species maximum lifespan (MLSP) and UV-induced DNA damage repair in fibroblasts or lymphocytes. The pyrimidine dimers and photoproducts associated with UV radiation are removed by global nucleotide excision repair (NER). Although the accumulation of pyrimidine dimers and other lesions repaired by NER leads to significant genomic instability, a perhaps more common and ubiquitous source of DNA lesions is reactive oxygen species (ROS) which are produced by mitochondria as normal by-products of aerobic metabolism (reviewed in Lambert and Brand 2009). Therefore, removal of lesions arising from endogenous ROS production may be particularly important in the highly oxidative cells comprising organs like brain and liver.
Many forms of oxidative damage, and also alkylative damage and spontaneous base loss, are repaired by enzymes of the base excision repair (BER) pathway. BER is subdivided into short- and long-patch pathways, catalyzed by a number of enzymes, but the “core” BER pathway in mammals (Robertson et al. 2009) includes a number of DNA glycosylases, an apurinic/apyrimidinic (AP) endonuclease (APE), polymerase β (polβ), and DNA ligase. Two steps in BER are potentially “rate limiting” to the overall processing capacity. APE possesses a 3'-phosphoesterase activity, which is believed to be rate limiting specifically following oxidation-induced DNA base loss (Izumi et al. 2000). Polβ possesses a putatively rate-limiting deoxyribophosphatase activity (Srivastava et al. 1998). Cabelof et al. (2003a) have demonstrated the dependence of BER activity on polβ in polβ haploinsufficient mice. Therefore, measurements of APE and polβ activities should provide insight into overall BER capacity.
While there is evidence that DNA BER is upregulated concomitantly with increased longevity in mice (e.g., Cabelof et al. 2003b; Stuart et al. 2004; Kisby et al. 2010), other results question the relationship between BER and longevity. For example, ∼50% reduction in either APE or polβ activity is not associated with reduced lifespan in unstressed mice (Meira et al. 2001; Cabelof et al. 2006). Similarly, shorter-lived mammalian species, which tend also to be smaller, do not show elevated levels of oxidative damage in nuclear DNA in all tissues (Barja and Herrero 2000), despite apparently higher rates of respiratory ROS production owing to greater mitochondrial abundance (e.g., Porter and Brand 1995) and enhanced ROS production per mitochondrion (e.g., Lambert et al. 2007). An important observation is that these smaller and shorter-lived species also excrete higher levels of stable DNA repair end products in their urine (Foksinski et al. 2004), which presumably indicates higher rates of repair. Taken together, these observations are consistent with the hypothesis that smaller, shorter-lived species with higher mass-specific metabolic rates sustain more DNA oxidative damage in their highly oxidative tissues but also repair it rapidly, thus leading to greater rates of excretion of DNA repair end products. Where the rates of damage incidence and repair are balanced, steady-state levels of oxidative DNA damage show no relationship to body mass (e.g., brain; Barja and Herrero 2000). Where they are unbalanced, oxidative DNA damage may accumulate (e.g., heart; Barja and Herrero 2000), with negative results for genomic stability.
Here, we have tested the hypothesis that DNA BER capacity of highly oxidative tissues is actually higher in smaller, shorter-lived species, by measuring BER activities in brain and liver tissues of 15 vertebrate endotherm species (Table 1; Fig. 1) with naturally disparate lifespans. We measured the rate of oligonucleotide incision at an AP site (primarily indicating APE activity) and the rate at which an oligonucleotide containing a processed single nucleotide gap was filled and ligated (polβ and DNA ligase activities). We found that none of these activities positively correlated with species MLSP. Rather, nucleotide incorporation and incorporation + ligation activities in both brain and liver tissues were actually negatively correlated with species body mass, an observation consistent with our hypothesis.
Table 1.
Sex, age, mass, and maximal lifespan (MLSP) of the mammalian and avian species
| Common name | Scientific name | Number and sex | Age | Mass (kg) | MLSP (years)* | Source |
|---|---|---|---|---|---|---|
| Mammalia | ||||||
| C57BL/6Ncr1BR mouse | Mus musculus | 4 F, 4 M | 4 months | 0.028 | 4 | Charles River (Wilmington, MA, USA) |
| Syrian hamster | Mesocricetus auratus | 4; sex unknown | 4–8 months | 0.130 | 3.9 | Equitech-Bio, Inc. (Kerrville, TX, USA) |
| Norway rat | Rattus norvegicus | 6 M | 3 months | 0.550 | 4 | Charles River (Wilmington, MA, USA) |
| Mongolian gerbil | Meriones unguiculatus | 4; sex unknown | 4–8 months | 0.070 | 6.3 | Equitech-Bio, Inc. (Kerrville, TX, USA) |
| 13-lined ground squirrel | Spermophilus tridecemlineatus | 8 F, 3 M | Unknown | 0.205 | 7 | University of Manitoba Field Research Station (Carmen MB, Canada) |
| Rabbit | Oryctolagus cuniculus | 4 F, 1 M | 3.5 months | 2.60 | 9 | Local abattoir (Fort Erie, ON, Canada) |
| Guinea pig | Cavia porcellus | 4; sex unknown | Unknown | 1.05 | 12 | Rockland (Gilbertsville, PA, USA) |
| Big brown bat | Eptesicus fuscus | 3 F, 2 M | 6–12 months | 0.025 | 19 | McMaster University Bat Laboratory (Hamilton, ON, Canada) |
| Sussex sheep | Ovis aries | 5 F | 6–12 months | 29.5 | 22.8 | Local abattoir (Fort Erie, ON, Canada) |
| White-tailed deer | Odocoileus virginianus | 3 M | 2.5–3.5 years | 110 | 21.6 | Local hunters (Cobden, ON, Canada) |
| Domestic dog | Canis familiaris | 3 M | 2–8 years | 11.0 | 24 | Equitech-Bio, Inc. (Kerrville, TX, USA) |
| Yorkshire/Hampshire pig | Sus scrofa | 2 F, 3 M | 6 months | 99.9 | 27 | Local abattoir (Fort Erie, ON, Canada) |
| Black angus/Charlet cow | Bos taurus | 4 F, 1 M | 1.5–2.5 years | 379.5 | 20 | Local abattoir (Fort Erie, ON, Canada) |
| Aves | ||||||
| Japanese quail | Coturnix japonica | 2 F, 2 M | 4–5 months | 0.264 | 6 | Cro Quail Farms Inc (St. Anne's, ON, Canada) |
| Zebra finch | Taeniopygia guttata | 3 F, 3 M | 1 year | 0.017 | 14.5 | Trent University Animal Care Facility (Peterborough, ON, Canada) |
aMLSP data are from AnAge (de Magalhaes et al. 2005)
Mass data are either measured directly or estimated based on age, sex, and visual assessment (see “Materials and methods” section)
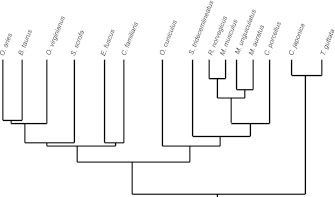
Fig. 1.
Phylogeny of the 15 species used in this study. The tree is a compilation of four phylogenies (Stuart et al. 2002; Springer and Murphy 2007; Hackett et al. 2008; Prasad et al. 2009). Branch lengths are best estimates from the cited phylogenies
Materials and methods
Materials
Chemicals were purchased from Sigma-Aldrich (Oakville, Ontario, Canada), Fisher Scientific (Mississauga, Ontario, Canada) and Bioshop (Burlington, Ontario, Canada). BioRad protein dye was purchased from BioRad laboratories (Hercules, California, USA). Oligonucleotides were purchased from Midland Certified Reagent Co. (Midland, Texas, USA). [γ-32P]-Adenosine 5'-triphosphate and [α-32P]-dCTP were purchased from Perkin Elmer (Waltham, Massachusetts, USA).
Animals
Between three and eight individuals of each of thirteen mammalian and two avian species were included in this study (Table 1). Species were selected on the bases of: (1) phylogenetic position; (2) MLSP; (3) availability. We used domestic and, where possible, wild-caught animals. In most cases, age and sex were known; however, this was not always possible to determine depending on source. All animals were healthy young adults. We attempted to sample from both male and female individuals of each species and, where sex differences in adult body mass occur, our values represent averages that incorporate both.
Tissue collection
Normal C57BL mice were euthanized by cervical dislocation following which brain, heart, and liver tissues were immediately excised, flash frozen in liquid nitrogen, and stored at −80°C. Norway laboratory rats were euthanized with pentabarbital sodium injection (Euthanyl®; 2 μg/g body mass). Brain, heart, and liver tissues were removed and flash frozen in liquid nitrogen and stored at −80°C. Tissues from 13-lined ground squirrels were collected at the University of Western Ontario (London, ON, Canada) as previously described in Page et al. (2009). Active (i.e., non-hibernating) animals were euthanized by Euthanyl overdose (270 mg/ml, 0.2 ml/100 g), brain, heart, and liver tissue were rapidly removed, flash frozen in liquid nitrogen, and stored at −80°C. Big brown bats were from a colony housed at McMaster University (Hamilton, ON, Canada). Bats were euthanized using 0.6 mg/g body wt. sodium pentabarbital followed by decapitation. Brain, heart, and liver tissues were excised and flash frozen in liquid nitrogen and stored at −80°C. Zebra finch and Japanese quail tissues were collected at Trent University (Peterborough, ON, Canada). Birds were euthanized with 2 (finch) or 0.6 (quail) mg/g body wt. sodium pentabarbital (Euthasol®), followed by decapitation. Brain, heart, and liver tissues from both bird species were removed and flash frozen in liquid nitrogen and stored at −80°C. Dogs, gerbils, and hamsters were euthanized by Euthanyl overdose, and the tissues were frozen in liquid nitrogen then shipped to Brock University (St. Catharines, ON, Canada) on dry ice. Guinea pigs were euthanized in a manner unknown to the authors; however, the tissue was rinsed in 1× PBS and frozen in liquid nitrogen and shipped to Brock University (St. Catharines, ON, Canada) on dry ice. Domesticated livestock were collected during normal processing at a local abattoir, with the exception of rabbits which were collected during processing from a local farmer. Brain and liver tissues were collected from these animals within approximately 30 min post mortem. Due to the organ size of the domesticated livestock and rabbits, tissue samples were isolated from the following locations; for brain, from the cortex and striatum; and for the liver, from the tip of the leftmost lobe. The sample sites for the brain and liver were similar in all domesticated livestock, including the rabbit. Enzyme activities were measured separately for cortex and striatum tissue; however, values were similar between both sites and therefore the data was compiled. Tissues were frozen on dry ice and shipped to Brock University (St. Catharines, ON, Canada), where they were stored at −80°C. White-tailed deer were killed by hunters in Cobden (ON, Canada) following which brain, heart, and liver tissue was collected within 1 h post mortem and frozen at −20°C for 1 month. The tissues were transferred to Brock University where they were stored at −80°C. Tissue sampling sites were unknown for the white-tailed deer.
Isolation of nuclear fractions
Frozen brain and liver tissue were used to isolate nuclear fractions as in Cabelof et al. (2002a). Tissue was powdered using a mortar and pestle and rinsed twice in 1× PBS. Subsequent steps in nuclear isolation were essentially as described in Stuart et al. (2004). Powdered tissue was further minced in 20 volumes of MSHE buffer (0.21 M mannitol, 70 mM sucrose, 10 mM HEPES pH 7.4, 1 mM EGTA, 2 mM EDTA, with 5 mM DTT and 1 μg/ml aproptinin added immediately prior to use) and homogenized with ten strokes of a Potter-Elvejhem homogenizer. Homogenization was carried out on ice. Samples were centrifuged for 10 min at 500×g (4°C). The nuclear pellet was resuspended in lysis buffer A (20 mM HEPES, pH 7.5, 1 mM EDTA, 5% glycerol, 0.5% Triton X-100, 300 mM KCl), 5 mM DTT, 1 μg/ml aproptinin, 1 mM PMSF were added immediately prior to use. Resuspended samples were incubated in ice with intermittent vortexing for 30 min and then centrifuged for 1 h at 20,000×g. The concentration of the supernatants was measured using the Bradford technique with a BioRad protein kit, and nuclear protein homogenates were stored at −80°C.
Base excision repair assays
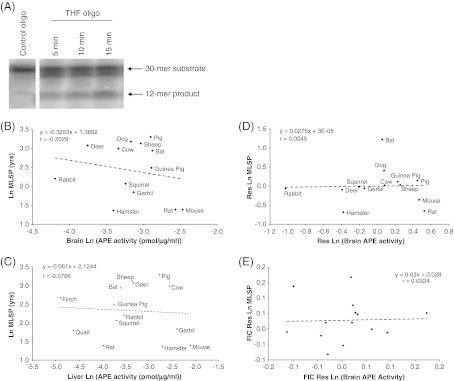
APE activity was measured by incubating 100 ng protein with 1 pmol of [γ-32P]-tetrahydrofuran-containing 30-mer oligonucleotide (THF; Table 2). The percent incision of the THF oligonucleotide was determined at 5, 10, 15 min at 37°C in a 10 μl reaction buffer (50 mM HEPES (pH 7.5), 50 mM KCl, 100 μg/ml BSA, 100 mM MgCl2, 10% glycerol, and 0.05% Triton X-100). Reactions were terminated with the addition of 10 μl formamide loading dye (80% formamide, 10 mM EDTA, 1 mg/ml xylene cyanol, and 1 mg/ml bromophenol blue) followed by incubation at 90°C for 10 min. Two controls were run in tandem with each APE activity assay: an oligonucleotide without the abasic site analog (THF control; Table 2) and the THF oligonucleotide incubated in the absence of cell lysate for the full duration of the assay (data not shown). Reaction products were resolved by electrophoresis at 15 W for 1 h 30 min in 20% polyacrylamide gels containing 7 M urea. Gels were visualized with PhosphoImager (Fujifilm FLA-3000) and analyzed with ImageGuageTM software. APE activity was quantified as the intensity of the product bands relative to the combined intensity of the product and substrate bands, essentially as in Stuart et al. (2004).
Table 2.
Oligonucleotides used to measure BER enzyme activity
| Name | Sequence | |
|---|---|---|
| AP endonuclease | THF control | 5′ ATATACCGCGGCCGGCCGATCAAGCTTATT 3′ |
| THF | 5′ ATATACCGCGG(THF)CGGCCGATCATCAAGCTTATT 3′ | |
| THF Comp | 5′ AATAAGCTTGATGATCGGCCGGCCGCGGTATAT 3′ | |
| Polymerase β | Polβ control | 5′ CTGCAGCTGATGCGCCGTACGGATCCCCGGGTTAC 3′ |
| GAP | 15mer—5′ CTGCAGCTGATGCGC-OH 3′ | |
| 18mer—5′ P-TACGGATCCCCGGGTTAC 3′ | ||
| GAP Comp | 5′ GTAACCCGGGGATCCGTACGGCGCATCAGCTGCAG 3′ |
THF tetrahydrofuran, Comp complementary sequence, GAP double-stranded oligonucleotide containing single nucleotide gap
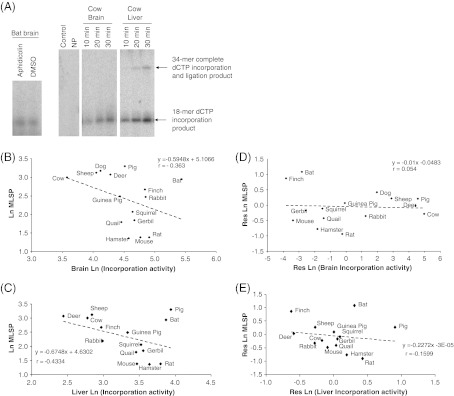
Polβ-mediated nucleotide incorporation was measured by incubating 1 μg nuclear extract protein with 1 pmol of an oligonucleotide with a single nucleotide gap (GAP; Table 2) for 10, 20, 30 min at 37°C, in a total volume of 10 μl reaction buffer (40 mM HEPES, 0.1 mM EDTA, 5 mM MgCl2, 0.2 mg/ml BSA, 50 mM KCl, 1 mM DTT, 40 mM phosphocreatine, 100 μg/ml phosphocreatine kinase, 3% glycerol, 2 mM ATP, 40 μM dCTP, and 4 μCi [α-32P]dCTP). Reactions were terminated with the addition of 5 μg proteinase K, 1 μl 10% SDS and incubation at 55°C for 30 min. DNA was precipitated with 1 μg glycogen, 4 μl 11 M ammonium acetate, and 90% ethanol overnight (−20°C). Samples were washed with 75% ethanol, centrifuged (16,000×g and 4°C) and speed vacuumed. Dried DNA was then resuspended in 10 μl formamide loading buffer and incubated at 90°C for 10 min. Two controls were run in parallel with each polβ activity assay: the identical oligonucleotide with no gap (GAP-control; Table 2); and the GAP oligonucleotide without cell lysate, incubated for the full duration of the assay (Fig. 3a). In addition, on each gel, an internal standard (4 μg) was included to control for inter-gel variation. Gels were visualized in a similar manner as above; however, polβ activity was quantified as the increase in signal intensity due to incorporation of [α-32P]dCTP. Complete incorporation and ligation activity was measured using the conditions as above for polβ activity. A higher shifted band (30-mer) represented complete repair of the gapped oligonucleotide. Gels were visualized as above and DNA ligase activity was quantified as the increase in signal intensity due to the incorporation of [32P]dCTP.
Fig. 3.
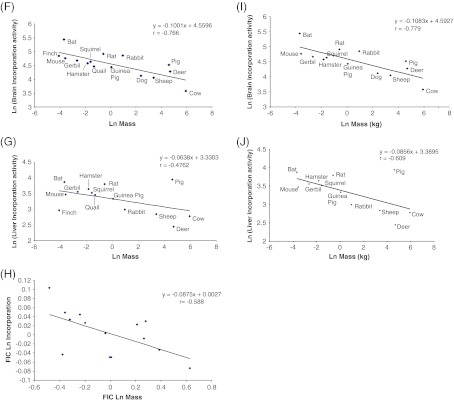
Polβ nucleotide incorporation activity is negatively correlated with body mass, but not MLSP. a Representative gel showing products of dCTP incorporation assay. The leftmost panel shows aphidicolin insensitivity of nucleotide incorporation in nuclear extracts from big brown bat brain tissue. The rightmost panels show incorporation/ligation products in cow brain and liver. Control = polβ control (no gap; see Table 1) and NP = no nuclear protein extract. Polβ activity in b brain and c liver appears to be negatively correlated negatively with MLSP, though significance is not reached in either tissue. The negative correlations between Polβ nucleotide incorporation activity and MLSP in d brain and e liver are lost following analysis of residuals, which removes the effect of body mass. Polβ nucleotide incorporation activity is significantly (negatively) correlated with body mass in both brain (f; p = 0.0004) and liver (g; p = 0.043). h The negative correlation with body mass remains following FIC analysis. i–j show only the mammalian data in f and g, respectively, without bird data. Values are natural log transformations of data from Tables 1 and 4
Statistical analyses
Correlational analyses of DNA BER activity data with either species' MLSP or body mass were performed. MLSP was chosen over other lifespan parameters (e.g., mean lifespan) because it most closely reflects the lifespan potential of a species removed from external stressors. All raw data were natural log-transformed prior to correlation analyses because the relationships between MLSP or body mass and the variables of interest appeared to be power functions which can be estimated well using log transformation on both axes. Amongst vertebrates, body mass and MLSP are highly correlated, and for this reason, it is necessary to determine whether apparent correlations with MLSP are actually driven by body mass. Residuals were calculated from simple linear regression of the dependent variable of interest on body mass to remove this confounding effect of body mass. Significance of the coefficient of correlation was used to determine if the line was different from horizontal, and significance was based on the t value. A p value of <0.05 was considered significant. The correlation analysis assumes that individual data points (species) are independent of each other. However, shared evolutionary history violates this assumption. To address this limitation, Felsenstein's phylogenetically independent contrasts (Felsenstein 1985) were calculated using PDAP (Garland et al. 1992; Garland et al. 1993). The phylogenetic tree was constructed from four phylogenies from which branch length estimates were taken (Stuart et al. 2002; Springer and Murphy 2007; Hackett et al. 2008; Prasad et al. 2008) (Fig. 1). The analysis was repeated with all branch lengths set to one, as described for speciation models of character change (Martins and Garland 1991 and Price 1997). This latter approach avoids errors associated with the estimation of branch lengths.
Results
AP site processing is the first of two putative rate-limiting steps in short-patch DNA BER, and therefore between-species differences in this activity can provide insight into differences in BER capacity. The rate of incision by nuclear protein extracts of an oligonucleotide containing an abasic site analog was measured for brain and liver tissues of 13 mammalian and 2 avian species (Table 3; Fig. 2). Two experimental controls were included for all species: a THF control oligonucleotide (see Table 2) without the abasic site analog to quantify non-specific degradation of the oligonucleotide (Fig. 2a), and the THF oligonucleotide incubated in assay buffer with all constituents but nuclear protein extract (not shown). Non-specific degradation of the control oligonucleotide was either absent or minor and in the latter case was subtracted from the measured incision activity. There was no evidence that the rate of incision of the THF oligonucleotide in either brain or liver correlated with species MLSP (Fig. 2b, c). This result persisted following analysis of residuals to account for effects of body mass (Table 5 reports liver; Fig. 2d shows brain). Furthermore, because all extant vertebrate species share an evolutionary history with a common ancestor, they cannot be treated as independent data (see Speakman 2005 for an excellent review of this issue). Therefore, to account for potential effects of phylogenetic relatedness on the correlational analysis, data were re-analyzed using Felsenstein's phylogenetic independent contrasts (FIC; Felsenstein, 1985), which again revealed no significant correlations between MLSP and THF incision activity (Table 5; Fig. 2e). There were similarly no significant correlations of this activity with species body mass, when data were analyzed with or without FIC transformation (Table 6).
Table 3.
Rates of THF incision (APE activity) in brain and liver tissue homogenates from 15 mammalian and avian species
| Species | Brain APE mean ± SEM | Liver APE mean ± SEM |
|---|---|---|
| Mammalian | ||
| Bat | 0.0563 ± 0.006 | 0.0273 ± 0.002 |
| Mouse | 0.0845 ± 0.011 | 0.1219 ± 0.007 |
| Gerbil | 0.0437 ± 0.005 | 0.0915 ± 0.038 |
| Hamster | 0.0331 ± 0.003 | 0.0668 ± 0.018 |
| Squirrel | 0.0390 ± 0.005 | 0.0251 ± 0.003 |
| Rat | 0.0770 ± 0.019 | 0.0188 ± 0.003 |
| Guinea pig | 0.0554 ± 0.014 | 0.0236 ± 0.003 |
| Rabbit | 0.0150 ± 0.003 | 0.0292 ± 0.005 |
| Dog | 0.0421 ± 0.005 | N/A |
| Sheep | 0.0480 ± 0.018 | 0.0348 ± 0.003 |
| Pig | 0.0548 ± 0.002 | 0.0630 ± 0.005 |
| Deer | 0.0022 ± 0.010 | 0.0358 ± 0.007 |
| Cow | 0.0355 ± 0.005 | 0.0779 ± 0.013 |
| Avian | ||
| Finch | 0.0106 ± 0.001 (0.0140) | 0.0074 ± 0.001 (0.010) |
| Quail | 0.0094 ± 0.001 (0.0125) | 0.0098 ± 0.002 (0.012) |
Data are in presented in the units pmol/μg/min. Sample size (n) for each species is as in Table 1. Each sample (from a given individual) was assayed twice to provide an average for that individual. Numbers in parentheses represent calculated corrections for higher avian body temperatures using an estimated Q10 = 2 and a temperature differential (avian versus mammalian body temperature) of 4°C
Fig. 2.
Abasic site incision activity does not correlate with MLSP. a Representative gel showing incision of an oligonucleotide containing a THF abasic site analog (see Table 2) by a cow liver nuclear extract. Control oligo is THF control (Table 2). b–c No correlation between MLSP and abasic site incision activity of b brain and c liver nuclear extracts. d Residual analysis of brain APE activity and animal MLSP showing no significant correlation. e FIC analysis (see “Materials and methods”) of brain APE activity and MLSP showing no significant correlation. Values are natural log transformations of data from Tables 1 and 4
Table 5.
Results of correlational analysis of enzyme activities as a function of MLSP
| BER enzyme activity | Correlation coefficient | Slope | p value |
|---|---|---|---|
| Liver APE | |||
| Residual | −0.292 | −0.206 | 0.16 |
| FIC analysis of residuals | −0.230 | −0.134 | 0.22 |
| Brain nucleotide incorporation | |||
| FIC analysis of residuals | −0.442 | −0.101 | 0.06 |
| Liver nucleotide incorporation | |||
| FIC analysis of residuals | −0.335 | −0.482 | 0.13 |
| Liver incorporation + ligation | |||
| FIC analysis of residuals | −0.226 | −0.143 | 0.22 |
Table 6.
Results of correlational analysis of enzyme activities as a function of body mass
| BER enzyme activity | Correlation coefficient | Slope | p value |
|---|---|---|---|
| Brain APE | |||
| Versus mass | 0.041 | 0.0083 | 0.44 |
| FIC analysis | −0.103 | −0.0229 | 0.36 |
| Liver APE | |||
| Versus mass | 0.251 | 0.0602 | 0.19 |
| FIC analysis | −0.229 | −0.0776 | 0.23 |
| Liver nucleotide incorporation | |||
| FIC analysis | −0.047 | −0.3554 | 0.44 |
| Liver incorporation + ligation | |||
| FIC analysis | 0.053 | 0.1759 | 0.43 |
Polβ activity was measured as the rate of aphidicolin-insenstive [32P]dCTP incorporation into the GAP oligonucleotide (see Table 2) by nuclear protein extracts (Table 4). In this assay, either one or two products were visible on PAGE gels: the band representing incorporation of [32P]dCTP into the oligonucleotide and, in some instances, an additional band representing subsequent ligation to complete the repair (Fig. 3a). Quantification of [32P]dCTP incorporation alone (smaller product) revealed a weak but statistically non-significant correlation with MLSP in either brain (Fig. 3b; p = 0.091) or liver tissue (Fig. 3c; p = 0.061). For both the brain and liver (non-significant) correlations with MLSP, analysis of the residuals of MLSP versus body mass and [32P]dCTP incorporation versus body mass indicated that the weak correlation with MLSP was in fact due to the correlation between MLSP and body mass (Fig. 3d, e). Indeed, statistically significant correlations of [32P]dCTP incorporation activities with body mass were evident in both brain (p = 0.0004) and liver (p = 0.043) (Fig. 3f, g). This correlation remained for brain following FIC analysis to account for the phylogenetic signal (Fig. 3h; p = 0.012), but became non-significant for liver (Table 5). In addition, the significant correlations of both brain and liver [32P]dCTP incorporation activities with body mass remained even if only data from the 13 mammalian species was included in the analysis (Fig. 3i, j for brain (p = 0.00085) and liver (p = 0.017), respectively.
Table 4.
Rates of [32P]dCTP incorporation or incorporation followed by ligation in brain and liver tissue homogenates from 15 mammalian and avian species
| Species | Brain incorporation mean ± SEM | Liver incorporation mean ± SEM | Liver incorporation + ligation mean ± SEM |
|---|---|---|---|
| Mammalian | |||
| Bat | 229.96 ± 31.78 | 47.98 ± 7.17 | 7.21 ± 2.13 |
| Mouse | 117.44 ± 7.80 | 32.13 ± 5.66 | 8.51 ± 2.48 |
| Gerbil | 106.78 ± 19.41 | 35.00 ± 2.80 | 34.73 ± 15.85 |
| Hamster | 97.41 ± 13.98 | 38.18 ± 1.50 | 39.25 ± 8.84 |
| Squirrel | 103.37 ± 9.32 | 33.86 ± 3.40 | 11.53 ± 2.91 |
| Rat | 135.54 ± 16.72 | 44.51 ± 3.91 | 24.02 ± 6.08 |
| Guinea pig | 84.29 ± 17.23 | 28.05 ± 3.31 | 10.26 ± 2.45 |
| Rabbit | 128.04 ± 13.10 | 19.91 ± 1.68 | 4.94 ± 0.95 |
| Dog | 61.61 ± 9.73 | N/A | N/A |
| Sheep | 57.46 ± 11.28 | 17.11 ± 1.56 | 1.73 ± 0.22 |
| Pig | 91.40 ± 7.66 | 51.55 ± 7.29 | 13.74 ± 3.50 |
| Deer | 72.12 ± 15.72 | 11.51 ± 0.89 | 3.09 ± 0.26 |
| Cow | 35.73 ± 5.66 | 16.10 ± 1.09 | 3.10 ± 0.58 |
| Avian | |||
| Finch | 126.50 ± 4.90 (168) | 19.43 ± 2.19 (25) | 13.21 ± 3.08 (17) |
| Quail | 86.35 ± 7.94 (114) | 31.59 ± 4.22 (41) | 14.53 ± 3.33 (18) |
Data are presented in the units Δ signal strength/μg/min. Sample size (n) for each species is as in Table 1. Each sample (from a given individual) was assayed twice to provide an average for that individual. Numbers in parentheses represent calculated corrections for higher avian body temperatures using an estimated Q10 = 2 and a temperature differential (avian versus mammalian body temperature) of 4°C
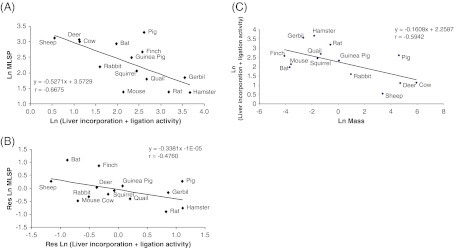
The product representing full repair of the GAP oligonucleotide following [32P]dCTP incorporation and subsequent ligation was sufficient to be accurately quantified in liver tissue, but not brain, of all species. Analysis of the rate at which this product was formed by liver nuclear protein extracts revealed a strongly negative correlation with species MLSP (p = 0.004; Fig. 4a). This correlation remained following analysis of residuals to account for effects of body mass, though it was highly dependent upon inclusion of the bat and finch data points (Fig. 4b; p = 0.042) and was absent when these were excluded (not shown). Additionally, there was again no correlation of incorporation + ligation activity with MLSP following FIC analysis (Table 5). A strong negative correlation with species' body mass was also found (Fig. 4c; p = 0.012), though this was lost following FIC analysis (Table 6).
Fig. 4.
Incorporation plus ligation activity of liver nuclear extracts is negatively correlated with species MLSP and body mass. Incorporation plus ligation is significantly and negatively correlated with species MLSP (a) and remains following residual analysis to account for effects of body mass (b). The correlation with body mass is also significant (c). Values are natural log transformations of data from Tables 1 and 4
Discussion
Although it has long been hypothesized that enhanced DNA repair capacity drives increases in longevity, tests of the hypothesis in naturally long-lived species have typically focused on highly mitotic cell types (e.g., fibroblasts) and upon the repair of UV-induced lesions by NER (e.g., Hart and Setlow 1974; Francis et al. 1981; Hall et al. 1984; Licastro and Walford 1985; Cortopassi and Wang 1996). There have been no comparative studies of DNA repair activities in the highly oxidative somatic cells that comprise many critical tissues such as brain, heart, kidney, and liver. These somatic cells are continuously exposed to the endogenous oxidants produced during normal mitochondrial respiration, which is exacerbated in highly oxidative tissues. Although NER and double-strand break repair pathways play roles in the repair of oxidative DNA damage (reviewed by Slupphaug et al. 2003), the BER pathway is an important means of repairing oxidized bases and abasic sites that occur commonly (Croteau and Bohr 1997). BER should therefore be particularly important in highly oxidative cells, such as neurons and hepatocytes that produce significant amounts of ROS during respiration. In these cells, which are generally either post-mitotic or slowly mitotic, the primary contribution of BER to cellular homeostasis is presumably the preservation of transcriptional fidelity and prevention of transcriptional stalling.
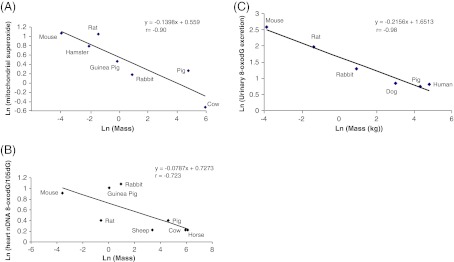
Our data represent the first multi-species measurements of BER in highly oxidative tissues, and the lack of correlation with MLSP is perhaps a surprising result. Interestingly, some BER activities, such as nucleotide incorporation, were actually negatively correlated with species body mass, though this appeared to be associated with phylogenetic predispositions toward higher activities in the Rodentia and lower activities in the ungulates based on the loss of significance following FIC analysis in many instances. Nonetheless, this negative correlation is consistent with four related observations from multi-species comparisons: (1) cellular respiration rates (e.g., liver hepatocytes, Porter and Brand 1995) and the corresponding proportion of cell volume occupied by mitochondria (multiple cell types, Else and Hulbert 1985; Porter and Brand 1995) are greater in smaller species; (2) ROS production per mitochondrion (multiple tissues; Sohal et al. 1989; Sohal et al. 1990; Ku et al. 1993; Barja 1998; Wright et al. 2004; Lambert et al 2007) is generally greater in smaller, shorter-lived species (Fig. 5a); (3) steady-state levels of nuclear DNA oxidative damage are greater in some tissues of smaller species (see Fig. 5b; data re-plotted from Barja and Herrero 2000); (4) urinary excretion of multiple DNA repair end products is significantly greater in smaller species, and inversely correlated with body mass (Fig. 5c; data re-plotted from Foksinski et al 2004). These disparate results are well reconciled by the observation here that some BER activities are also elevated in brain and liver of smaller species and correlate negatively with body mass. Taken together, the data from all of these studies support a scenario where smaller species maintain higher cellular respiratory activities with more mitochondria and concomitantly higher rates of intracellular ROS production leading to a higher incidence of DNA damage that is countered by enhanced BER and perhaps also other DNA repair pathways. Cabelof et al. (2002b) demonstrated that polβ activity in vivo is upregulated by intracellular oxidative stress, which supports the idea that chronically higher rates of endogenous respiratory ROS production in smaller species might be sufficient to stimulate BER. Given the kinetics outlined above, it seems likely that smaller species with higher metabolic rates could be particularly susceptible to perturbations that upset what appears to be a highly dynamic balance between genomic damage and repair.
Fig. 5.
Species' body mass is negatively correlated with a mitochondrial ROS production (heart data from Ku et al (1993) re-plotted against body mass); b urinary 8-oxodG excretion (data from Foksinski et al (2004) re-plotted against body mass); and c nuclear DNA 8-oxodG levels (heart data from Barja and Herrero (2000) re-plotted against body mass)
It is interesting to note that the trends observed here for BER-related activities are similar to those reported for other cellular macromolecular maintenance activities, such as those involved in protein repair and degradative recycling. Salway et al. (2011) showed that the 20S/26S proteasome, as well as the activities of enzymes capable of maintaining the redox state of proteins, generally correlate negatively with species body mass but show no relationship with MLSP. This suggests that the rate at which macromolecular oxidative damage (to both proteins and DNA) occurs probably decreases with increasing body mass, and therefore the rates of recycling/repair required to maintain cellular homeostasis and viability are concomitantly reduced. An important caveat, however, is that both protein (Salway et al. 2011) and DNA homeostasis activities were measured under non-stressed conditions, so we cannot detect any differences in the inducibility of these activities that might exist between species and be related to MLSP.
Another concept critical to the interpretation of the data in this study is that relationships between BER (or other DNA repair activities) and lifespan may be highly cell-type specific. Previously, various authors have described positive correlations between species' MLSP and poly(ADP-ribose) polymerase activity (Grube and Bürkle 1992), NER (Cortopassi and Wang 1996), double-strand break recognition (Lorenzini et al 2009), and telomere length maintenance (Haussmann et al 2003) in fibroblasts and blood cells. These cell types will have very different characteristics than those in brain and liver. Our measurements in these tissues will disproportionately reflect the activities of highly differentiated post-mitotic cells (particularly in brain) that account for the majority of cells in these highly oxidative tissues. The functions and regulation of BER and other DNA repair pathways will certainly differ between these types of cells and those with greater mitotic potential such as fibroblasts or perhaps tissue resident stem and progenitor cells. Narciso et al. (2007) demonstrated that a significant reduction in BER capacity occurs during differentiation of actively mitotic skeletal muscle progenitors (myoblasts) to post-mitotic myotubes (myofiber-like). Other authors have shown that BER enzyme activities are upregulated at specific times during the cell cycle (Chaudhry 2007) and are generally elevated in tissues containing higher proportions of actively mitotic cells (see Karahalil et al. 2002). Perhaps the expected correlations of DNA repair activities with MLSP manifest in actively mitotic cells. However, it is also possible that the proportion of actively mitotic cells in brain and liver (and presumably other tissues) is itself a function of species' body mass. Thus, if larger and/or longer-lived species express lower levels of mitogenic signals, then perhaps they possess a lower proportion of actively cycling mitotic cells. In this context, it is interesting to note that larger mammalian species have lower circulating levels of the mitogen insulin-like growth factor-1 (IGF-1) (Stuart and Page 2010). Similarly, the activity of telomerase (in various tissues), which is associated with cell replication, also correlates negatively with body mass (Seluanov et al. 2007). Both observations are consistent with a general reduction in the proportion of actively mitotic cells within tissues as body mass increases, though we are not aware of data that could confirm this hypothesis.
Despite the observed negative correlations between species' body mass and nucleotide incorporation + oligonucleotide ligation activities, there was no evidence for a similar relationship with abasic site processing, which would be expected based on the arguments made above. However APE1, the main enzyme catalyzing this step in mammals, is a dual function protein (also Ref-1) involved in redox-sensitive transcriptional regulation. It may therefore be affected by additional regulatory controls that do not affect polβ or DNA ligases. In any event, the absence of positive correlations of AP endonuclease and polβ activities in both brain and liver indicate that BER activity in these tissues does not increase concomitantly with MLSP.
Finally, multiple two-species comparisons can be made within the dataset, but this approach also fails to provide unequivocal support for the hypothesis that BER activities are greater in longer-lived species. For example, AP site incision activity in either brain or liver was not higher in the bat than in shorter-lived rodent species. Nor was this activity greater in the finch compared to the mouse, even after the finch activity was corrected for the effect of this bird's higher body temperature (using an estimated Q10 = 2, and body temperatures of 42°C for birds versus ∼38°C for mammalian species). Similarly, we could identify no consistent pattern of greater nucleotide incorporation/ligation activities in the long-lived species. While bat tissues did have higher [32P]dCTP incorporation activities than in rodent species (Table 4), there was no similar elevation in the long-lived zebra finch compared to the short-lived quail. Taken together, these multiple two-species comparisons do not consistently support the hypothesis that BER activities are elevated in longer-lived species.
In conclusion, we make two key observations based on this unique dataset. Firstly, there is no evidence that increases in BER enzyme activities in brain or liver tissues have co-evolved with longevity in vertebrates. Secondly, our data indicate that some important steps in the BER pathway are actually negatively correlated with species body mass. Taken together, these observations are consistent with the interpretation that BER capacity of highly differentiated and oxidative tissues under non-stressed conditions may be related to the metabolic activity of cells comprising those tissues rather than to longevity of the host species. Our data do not, however, allow us to detect potential differences in the timing or robustness of BER induction that may relate to MLSP.
Acknowledgements
MMP was supported by an Ontario Graduate Scholarship. Work at Brock University was supported by the Natural Sciences and Engineering Research Council, the Canada Foundation for Innovation and an Early Researcher Award from the Ontario Ministry of Research and Innovation to JAS. We thank Gary Burness, Esther Tiedke, and Paul Faure for providing bird, livestock, and bat samples.
References
- Barja G. Mitochondrial free radical production and aging in mammals and birds. Ann NY Acad Sci. 1998;854:224–238. doi: 10.1111/j.1749-6632.1998.tb09905.x. [DOI] [PubMed] [Google Scholar]
- Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Yanamadala S, Ganir C, Guo Z, Heydari AR. Attenuation of DNA polymerase β-dependent base excision repair and increased DMS-induced mutagenicity in aged mice. Mutat Res. 2002;500:135–145. doi: 10.1016/S0027-5107(02)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Yanamadala S, Guo Z, Heydari AR. Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis. 2002;23:1419–1425. doi: 10.1093/carcin/23.9.1419. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Guo Z, Raffoul JJ, Sobol RW, Wilson SH, Richardson A, Heydari AR. Base excision repair deficiency caused by polymerase beta haploinsufficiency: accelerated DNA damage and increased mutational response to carcinogens. Cancer Res. 2003;63:5799–5807. [PubMed] [Google Scholar]
- Cabelof DC, Yanamadala S, Raffoul JJ, Guo Z, Soofi A, Heydari AR. Caloric restriction promotes genomic stability by induction of base excision repair and reversal of its age-related decline. DNA Repair (Amst) 2003;2:295–307. doi: 10.1016/S1568-7864(02)00219-7. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Ikeno Y, Nyska A, Busuttil RA, Anyangwe N, Vijg J, Matherly LH, Tucker JD, Wilson SH, Richardson A, Heydari AR. Haploinsufficiency in DNA polymerase beta increases cancer risk with age and alters mortality rate. Cancer Res. 2006;66:7460–7465. doi: 10.1158/0008-5472.CAN-06-1177. [DOI] [PubMed] [Google Scholar]
- Chaudhry MA. Base excision repair of ionizing radiation-induced DNA damage in G1 and G2 cell cycle phases. Cancer Cell Int. 2007;7:15. doi: 10.1186/1475-2867-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi GA, Wang E. There is substantial agreement among interspecies estimates of DNA repair activity. Mech Ageing Dev. 1996;91:211–218. doi: 10.1016/S0047-6374(96)01788-5. [DOI] [PubMed] [Google Scholar]
- Croteau DL, Bohr VA. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J Biol Chem. 1997;272:25409–25412. doi: 10.1074/jbc.272.41.25409. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Costa J, Toussaint O (2005) HAGR: the human ageing genomic resources. Nucleic Acids Res 33:D537–D543 [DOI] [PMC free article] [PubMed]
- Else PL, Hulbert AJ. Mammals: an allometric study of metabolism at tissue and mitochondrial level. Am J Physiol. 1985;248:R415–R421. doi: 10.1152/ajpregu.1985.248.4.R415. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. doi: 10.1086/284325. [DOI] [Google Scholar]
- Foksinski M, Rozalski R, Guz J, Ruszkowska B, Sztukowska P, Piwowarski M, Klungland A, Olinski R. Urinary excretion of DNA repair products correlates with metabolic rates as well as maximum life spans of different mammalian species. Free Radic Biol Med. 2004;37:1449–1454. doi: 10.1016/j.freeradbiomed.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Francis AA, Lee WH, Regan JD. The relationship of DNA excision repair of ultraviolet-induced lesions to the maximum life span of mammals. Mech Ageing Dev. 1981;16:181–189. doi: 10.1016/0047-6374(81)90094-4. [DOI] [PubMed] [Google Scholar]
- Garland T, Jr, Harvey PH, Ives AR. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst Biol. 1992;41:18–32. [Google Scholar]
- Garland T, Jr, Dickerman AW, Janis CM, Jones JA. Phylogenetic analysis of covariance by computer simulation. Syst Biol. 1993;42:265–292. [Google Scholar]
- Grube K, Bürkle A. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc Natl Acad Sci USA. 1992;89:11759–11763. doi: 10.1073/pnas.89.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han K-L, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Hall KY, Hart RW, Benirschke AK, Walford RL. Correlation between ultraviolet-induced DNA repair in primate lymphocytes and fibroblast and species maximum achievable life span. Mech Ageing Dev. 1984;24:163–173. doi: 10.1016/0047-6374(84)90068-X. [DOI] [PubMed] [Google Scholar]
- Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc Nat Acad Sci USA. 1974;71:2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann MF, Winkler DW, O'Reilly KM, Huntington CE, Nisbet ICT, Vleck CM. Telomeres shorten more slowly in long-lived birds and mammals than in short-lived ones. Proc R Soc Lond B. 2003;270:1387–1392. doi: 10.1098/rspb.2003.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Hazra TK, Boldogh I, Tomkinson AE, Park MS, Ikeda S, Mitra S. Requirement for human AP endonuclease 1 for repair of 3'-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000;21:1329–1334. doi: 10.1093/carcin/21.7.1329. [DOI] [PubMed] [Google Scholar]
- Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- Kisby GE, Kohama SG, Olivas A, Churchwell M, Doerge D, Spangler E, Cabo R, Ingram DK, Imhof B, Bao G, Kow YW. Effect of caloric restriction on base-excision repair (BER) in the aging rat brain. Exp Gerontol. 2010;45:208–216. doi: 10.1016/j.exger.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku H-H, Brunk UT, Sohal RS. Relationship between mitochondrial superoxide and hydrogen peroxide production and longevity of mammalian species. Free Radic Biol Med. 1993;15:621–627. doi: 10.1016/0891-5849(93)90165-Q. [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Brand MD. Reactive oxygen species production by mitochondria. Methods Mol Biol. 2009;554:165–181. doi: 10.1007/978-1-59745-521-3_11. [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Boysen HM, Buckingham JA, Yang T, Podlutsky A, Austad SN, Kunz TH, Buffenstein R, Brand MD. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6:607–618. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- Licastro F, Walford RL. Proliferative potential and DNA repair in lymphocytes from short-lived and long-lived strains of mice, relation to aging. Mech Ageing Dev. 1985;31:171–186. doi: 10.1016/S0047-6374(85)80028-2. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lorenzini A, Johnson FB, Oliver A, Tresini M, Smith JS, Hdeib M, Sell C, Cristofalo VJ, Stamato TD. Significant correlation of species longevity with DNA double strand break recognition but not with telomere length. Mech Ageing Dev. 2009;130:784–792. doi: 10.1016/j.mad.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins EP, Garland T., Jr Phylogenetic analyses of correlated evolution of continuous characters: a simulation study. Evolution. 1991;45:534–557. doi: 10.2307/2409910. [DOI] [PubMed] [Google Scholar]
- Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, Friedberg EC. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–5557. [PubMed] [Google Scholar]
- Narciso L, Fortini P, Pajalunga D, Franchitto A, Liu P, Degan P, Frechet M, Demple B, Crescenzi M, Dogliotti E. Terminally differentiated muscle cells are defective in base excision DNA repair and hypersensitive to oxygen injury. Proc Natl Acad Sci USA. 2007;104:17010–17015. doi: 10.1073/pnas.0701743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MM, Peters CW, Staples JF, Stuart JA. Intracellular antioxidant enzymes are not globally upregulated during hibernation in the major oxidative tissues of the 13-lined ground squirrel Spermophilus tridecemlineatus. Comp Biochem Physiol A Mol Integr Physiol. 2009;152:115–122. doi: 10.1016/j.cbpa.2008.09.032. [DOI] [PubMed] [Google Scholar]
- Porter RK, Brand MD. Causes of differences in respiration rate of hepatocytes from mammals of different body mass. Am J Physiol Regul Integr Comp Physiol. 1995;269:1213–1224. doi: 10.1152/ajpregu.1995.269.5.R1213. [DOI] [PubMed] [Google Scholar]
- Prasad AB, Allard MW, Comparative Sequencing Program NISC, Green ED. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T. Correlated evolution and independent contrasts. Phil Trans R Soc Lond B. 1997;352:519–529. doi: 10.1098/rstb.1997.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salway KD, Page MM, Faure PA, Burness G, Stuart JA. Enhanced protein repair and recycling are not correlated with longevity in 15 vertebrate endotherm species. Age. 2011;33:33–47. doi: 10.1007/s11357-010-9157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Chen Z, Hine C, Sasahara TH, Ribeiro AA, Catania KC, Presgraves DC, Gorbunova V. Telomerase activity coevolves with body mass not lifespan. Aging Cell. 2007;6:45–52. doi: 10.1111/j.1474-9726.2006.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Svensson I, Sohal BH, Brunk UT. Superoxide anion radical production in different animal species. Mech Ageing Dev. 1989;49:129–135. doi: 10.1016/0047-6374(89)90096-1. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Svensson I, Brunk UT. Hydrogen peroxide production by liver mitochondria in different species. Mech Ageing Dev. 1990;53:209–215. doi: 10.1016/0047-6374(90)90039-I. [DOI] [PubMed] [Google Scholar]
- Speakman JR. Correlations between physiology and lifespan—two widely ignored problems with comparative studies. Aging Cell. 2005;4:167–175. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ. Mammalian evolution and biomedicine: new views from phylogeny. Biol Rev. 2007;82:375–392. doi: 10.1111/j.1469-185X.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Page MM. Plasma IGF-1 is negatively correlated with body mass in a comparison of 36 mammalian species. Mech Ageing Dev. 2010;131:591–598. doi: 10.1016/j.mad.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Moffett K, Leader JJ. A comprehensive vertebrate phylogeny using vector representations of protein sequences from whole genomes. Mol Biol Evol. 2002;19:554–562. doi: 10.1093/oxfordjournals.molbev.a004111. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA base excision repair are affected differently by caloric restriction. FASEB J. 2004;18:595–597. doi: 10.1096/fj.03-0890fje. [DOI] [PubMed] [Google Scholar]
- Wright AF, Jacobson SG, Cideciyan AV, Roman AJ, Shu X, Vlachantoni D, McInnes RR, Riemersma RA. Lifespan and mitochondrial control over neurodegeneration. Nat Genet. 2004;36:1153–1158. doi: 10.1038/ng1448. [DOI] [PubMed] [Google Scholar]