Abstract
The RNA-binding protein RNPC1 is a target of the p53 family and forms a feedback regulatory loop with the p53 family proteins. The MDM2 oncogene, a key negative regulator of p53, plays a critical role in a variety of fundamental cellular processes. MDM2 expression is found to be regulated via gene amplification, transcription, protein translation, and protein stability. In the current study, we reported a novel regulation of MDM2 by RNPC1 via mRNA stability. Specifically, we found that over-expression of RNPC1 decreases, whereas knockdown or knockout of RNPC1 increases, the level of MDM2 transcript and protein independent of p53. To uncover the underlying mechanism, we found that RNPC1 is able to destabilize the MDM2 transcript via binding to multiple AU-/U-rich elements in MDM2 3′UTR. Consistent with this, we showed that RNPC1 inhibits expression of exogenous MDM2 from a expression vector as long as the vector contains an AU-/U-rich element from MDM2 3′UTR. Finally, we showed that the RNA-binding activity of RNPC1 is required for binding to MDM2 transcript and consequently, for inhibiting MDM2 expression. Together, we uncover a novel regulation of MDM2 by the RNA-binding protein RNPC1 via mRNA stability.
Keywords: RNA-binding proteins, RNPC1, MDM2, mRNA stability
Introduction
The murine double minute-2 (MDM2) gene was originally identified as one of three genes (MDM1, MDM2, and MDM3), which were located on double minute chromosomes (acentromeric extrachromosomal nuclear bodies) in a spontaneously transformed mouse cell line, 3T3-DM [1]. The MDM2 gene was subsequently found to be amplified in 3T3-DM cells and to induce tumorigenicity when overexpressed [2]. Soon after its discovery, MDM2 was found to be a negative regulator of tumor suppressor p53. This occurs through two main mechanisms. First, MDM2 directly binds to the N-terminus of p53 and inhibits its transcriptional activities [3, 4]. Second, MDM2 possesses E3 ubiquitin ligase activity that targets p53 for proteasomal degradation [5–7]. The importance of Mdm2 as a negative regulator of p53 is demonstrated by elegant mouse models. Homozygous deletion of the MDM2 gene leads to embryonic lethality due to inappropriate apoptosis. This lethal phenotype can be rescued by concomitant deletion of the p53 gene [8, 9]. Additionally, MDM2 interacts with an array of proteins, such as RB, Numb, and p21, and modulates with their expression and/or activities [10, 11].
Due to its critical role in growth control and tumourigenesis, much work has been done to elucidate the underlying mechanisms by which MDM2 expression is regulated. For example, MDM2 transcription is induced by p53 through its P2 promoter, and thus forms a feedback regulatory loop with p53 [12, 13]. Moreover, MDM2 protein translation is enhanced by RNA-binding protein La, which binds to a 27-nt segment in the MDM2 5′ untranslated region (5′ UTR) [14]. Similarly, MDM2 translation is found to be regulated by the hepatocyte growth factor (HGF) receptor (Met) and the insulin-like growth factor 1 (IGF-1) receptor (IGF-1R) [15, 16]. Furthermore, MDM2 functions as an E3 ubiquitin ligase, targeting itself for proteasomal degradation [17, 18]. Recent studies showed that MDM2 is regulated by a number of ribosomal proteins, including RPL5, RPS7, RPL11, RPL23, and RPS27L [19–25], all of which play a critical role in ribosomal stress-induced p53 activation. Nevertheless, it is not clear whether MDM2 can be regulated by other posttranscriptional mechanisms, such as mRNA stability.
The RNPC1 gene, also called RBM38, encodes a RNA-binding protein (RBP) and is expressed as two isoforms, RNPC1a with 239 aa and RNPC1b with 121 aa. Structure analysis reveals that both RNPC1a and RNPC1b contain a putative RNA recognition motif (RRM) (aa 36–82) and belong to the RRM-containing RBP family, which includes Musashi, HuR, and nucleolin. Recently, we identified that RNPC1 is a target of the p53 tumor suppressor family, including p53, p63, and p73 [26]. Interestingly, RNPC1 can in turn posttranscriptionally regulate the p53 family proteins, including p53 and p63 [27, 28]. Thus, RNPC1 forms a feedback regulatory loop with the p53 family proteins. In addition, RNPC1a, the large isoform of RNPC1, was found to regulate p21 mRNA stability via interacting with the AU-rich elements (ARE) in p21 3′ untranslated region (3′UTR) [26, 29–31]. Since MDM2 is a target of p53 and its 3′UTR contains several AREs, we thus asked whether RNPC1 can posttranscriptionally regulate MDM2 expression. Indeed, we found that RNPC1 is able to bind to and destabilize the MDM2 transcript, leading to decreased expression of MDM2 transcript and protein. Furthermore, we found that the RNA-binding activity of RNPC1 is required for binding to the MDM2 transcript and for inhibiting MDM2 expression.
Results
Ectopic expression of RNPC1 inhibits MDM2 expression independent of p53
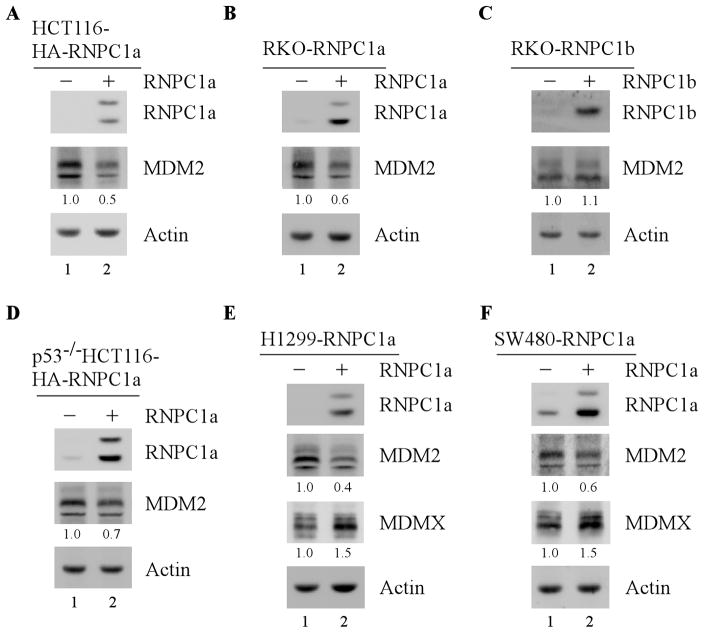
To investigate whether RNPC1 regulates MDM2 expression, wild-type p53-containing HCT116 and RKO cells that can inducibly express RNPC1a under the control of the tetracycline-regulated promoter were utilized. Specifically, cells were uninduced or induced with tetracycline to express RNPC1a for 24 h, and the level of MDM2 protein was determined by Western blot analysis. We found that upon RNPC1a expression, MDM2 level was markedly decreased (Fig. 1A–B, MDM2 panel, compare lane 1 with 2), consistent with previous report [28]. By contrast, ectopic expression of RNPC1b did not have any, if little, effect on MDM2 expression (Fig. 1C and supplemental Fig. 1A, compare lane 1 with 2). As RNPC1b does not affect MDM2 expression, this study will focus on RNPC1a. To simplify, RNPC1 and RNPC1a are interchangeably used here.
Figure 1. Ectopic expression of RNPC1 inhibits MDM2 expression independent of p53.
(A–B) The level of MDM2 protein is decreased by RNPC1 in HCT116 (A) and RKO (B) cells. HCT116 (A) and RKO (B) cells were uninduced or induced with 0.25 μg/ml tetracycline to express RNPC1 for 24 h. Cell lysates were collected and the level of RNPC1, MDM2, and actin was analyzed by Western blot analysis. The level of MDM2 protein was normalized to that of actin control and the fold change was shown below each lane. Data are representative from three independent experiments. (C) Ectopic expression of RNPC1b does not affect MDM2 expression. The level of RNPC1b, MDM2, and actin was determined in RKO cells uninduced or induced to express RNPC1b for 24 h. The level of MDM2 protein was normalized to that of actin control and the fold change was shown below each lane. (D–E) Over-expression of RNPC1 inhibits MDM2 expression in p53-null cells. p53−/− HCT116 (D) and H1299 (E) cells was uninduced or induced to express RNPC1 for 24 h, followed by Western blot analysis to determine the level of RNPC1, MDM2, MDMX, and actin. The level of MDM2 and MDMX protein was normalized to that of actin control, respectively, and the fold change was shown below each lane. Data are representative from three independent experiments. (F) The experiment was performed as in (E) except that SW480 cells were uninduced or induced to express RNPC1 for 24 h. The relative fold change was shown below each lane. Data are representative from three independent experiments.
Next, to determine whether RNPC1 regulates MDM2 expression in the absence of p53, p53-null H1299 and p53−/−HCT116 cells were used to determine the level of MDM2. We showed that the level of MDM2 was greatly decreased by RNPC1 in H1299 and p53−/−HCT116 cells (Fig. 1D–E, compare lane 1 with 2). Consistent with this, ectopic expression of RNPC1 repressed the level of MDM2 in mutant p53-containing SW480 cells (Fig. 1F, compare lane 1 with 2). Interestingly, we also found that in H1299 and SW480 cells, the level of MDMX, the MDM2 homolog, was increased upon expression of RNPC1, which is likely due to decreased expression of MDM2, an E3 ligase of MDMX [32, 33]. Taken together, these data suggest that RNPC1 inhibits MDM2 expression independent of p53.
Knockdown or knockout of RNPC1 increases expression of MDM2
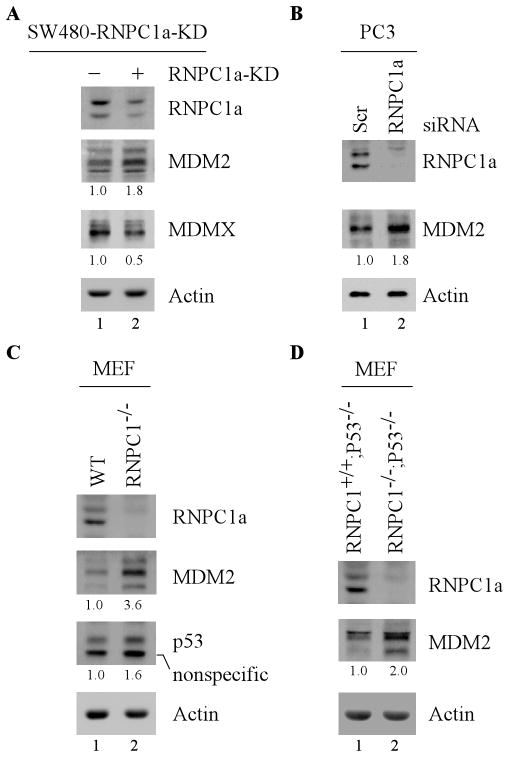
To determine whether endogenous RNPC1 has an effect on MDM2 expression, SW480 cells that can inducibly express an shRNA against RNPC1 were utilized. Specifically, SW480 cells were uninduced or induced to knock down RNPC1 for 3 days, followed by Western blot analysis. We found that upon RNPC1 knockdown, the level of MDM2 protein was greatly increased, concomitantly with decreased expression of MDMX (Fig. 2A, compare lane1 with 2). To rule out potential off-target effects of RNPC1 shRNA, a second siRNA, which targets another region of RNPC1 mRNA, was introduced into p53-null PC3 cells along with a scrambled control siRNA. We found that upon knockdown of RNPC1, MDM2 expression was increased in PC3 cells (Fig. 2B, compare lane 1 with 2). To further verify this, the level of MDM2 was determined in primary mouse embryonic fibroblasts (MEFs) isolated from wild-type and RNPC1−/− embryos. We found that RNPC1 deficiency markedly increased expression of MDM2 in RNPC1−/− MEFs as compared to that in wild-type MEFs (Fig. 2C, compare lane 1 with 2). Consistent with previous report [28], the level of p53 was accumulated in RNPC1−/−MEFs (Fig. 2C, compare lane1 with 2). Furthermore, to ensure that RNPC1 knockout increases MDM2 expression in the absence of p53, the level of MDM2 was examined in p53−/−;RNPC1+/+ and p53−/−;RNPC1−/− MEFs. We found that the level of MDM2 was still increased in p53−/−;RNPC1−/−MEFs as compare to that in p53−/−;RNPC1+/+ MEFs (Fig. 2D, compare lane 1 with 2). Together, these data suggest that knockdown or knockout of RNPC1 increases MDM2 expression independent of p53.
Figure 2. Knockdown or knockout of RNPC1 increases expression of MDM2.
(A) Knockdown of RNPC1 results in an increased expression of MDM2 in SW480 cells. SW480 cells were mock-treated or treated with 0.5 μg/ml tetracycline to induce RNPC1 shRNA for 3 days, and the level of RNPC1, MDM2, MDMX, and actin was determined by Western blot analysis. The level of MDM2 and MDMX protein was normalized to that of actin control, respectively, and the fold change was shown below each lane. Data are representative from three independent experiments. (B) PC3 cells were transiently transfected with scrambled or RNPC1 siRNA for 3 days, followed by Western blot analysis to determine the level of RNPC1, MDM2, and actin. The level of MDM2 was normalized to that of actin control and the fold change was shown below each lane. Data are representative from three independent experiments. (C) RNPC1 deficiency in primary mouse embryonic fibroblasts (MEFs) increases MDM2 expression. The level of RNPC1, MDM2, p53, and actin was analyzed in MEFs isolated from wild-type and RNPC1-null embryos. The level of MDM2 and p53 protein was normalized to that of actin control, respectively, and the fold change was shown below each lane. Data are representative from three independent experiments. (D) The level of RNPC1, MDM2, and actin was analyzed in MEFs isolated from p53−/−;RNPC1+/+and p53−/−;RNPC1−/−embryos. The level of MDM2 was normalized to that of actin control and the fold change was shown below each lane. Data are representative from three independent experiments.
RNPC1 destabilizes the MDM2 transcript
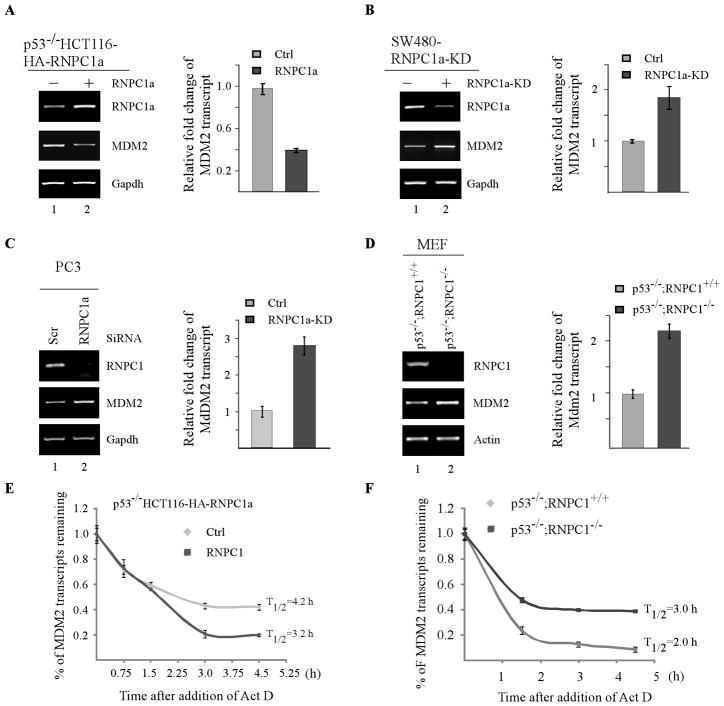
To gain further insight into how RNPC1 regulates MDM2 expression, the level of precursor MDM2 mRNA, the nuclear MDM2 RNA prior to splicing, was measured and found not to be altered by ectopic expression of RNPC1 in SW480 and p53−/− HCT116 cells (Supplemental Fig. 2A–B). Similarly, knockdown of RNPC1 in PC3 and SW480 cells or knockout of RNPC1 in p53−/− MEFs had no effect on the level of MDM2 precursor mRNA (Supplemental Fig. 2C–E). These data suggest that RNPC1 regulation of MDM2 occurs mainly through a posttranscriptional mechanism. In this regard, semi-quantitative and quantitative RT-PCR analysis were performed to determine the level of mature MDM2 mRNA by using p53−/−HCT116 cells uninduced or induced to express RNPC1. We found that ectopic expression of RNPC1 reduced the level of mature MDM2 transcript by 60% (Fig. 3A). To determine whether endogenous RNPC1 has an effect on the mature MDM2 transcript, SW480 and PC3 cells with or without endogenous RNPC1 knockdown were used to perform the same experiment. We found that RNPC1 knockdown increased the level of mature MDM2 mRNA by 1.9- and 2.9-fold in SW480 and PC3 cells, respectively (Fig. 3B–C). To further verify this, the level of mature MDM2 mRNA was measured in p53−/−;RNPC1+/+ and p53−/−;RNPC1−/− MEFs. We found that RNPC1 knockout led to a 2.2-fold increase in the mature MDM2 mRNA in RNPC1−/−;p53−/−MEFs as compared to that in p53−/−;RNPC1+/+ MEFs (Fig. 3D).
Figure 3. RNPC1 destabilizes the MDM2 transcript.
(A) The level of MDM2 transcript is decreased by RNPC1 in p53−/− HCT116 cells. Total RNAs were isolated from p53−/− HCT116 cells uninduced or induced to express RNPC1 for 48 h, followed by cDNA synthesis. Left panel: Semi-quantitative RT-PCR analysis was performed to determine the level of RNPC1, MDM2, and actin. Right panel: Quantitative RT-PCR analysis was performed in triplicates and the level of MDM2 transcript was normalized to that of GAPDH control. The relative fold change of MDM2 transcript was a ratio by dividing the level of MDM2 transcript in RNPC1-expressing cell to that in control cells. (B) Inducible knockdown of RNPC1 increases the level of MDM2 transcript in SW480 cells. Total RNAs were isolated from SW480 cells uninduced or induced to express RNPC1 shRNA for 3 days, followed by cDNA synthesis. Left panel: The level of RNPC1, MDM2, and actin transcripts was determined by semi-quantitative RT-PCR analysis. Right panel: Quantitative RT-PCR analysis was performed in triplicates and the level of MDM2 transcript was normalized to that of GAPDH control. The relative fold change of MDM2 transcript was a ratio by dividing the level of MDM2 transcript in RNPC1-knockdown cells to that in control cells. (C) The level of MDM2 transcript is increased in PC3 cells upon RNPC1 knockdown. The experiment was performed as in (B) except that PC3 cells were transiently transfected with scrambled or RNPC1 siRNA for 3 days. (D) RNPC1 deletion in primary MEFs increases the level of MDM2 transcript independent of p53. The experiments were performed as in (B) except that p53−/−;RNPC1+/+and p53−/−;RNPC1−/− MEFs were used. (E) Ectopic expression of RNPC1 shortens the half-life of MDM2 transcript. p53−/−HCT116 cells were uninduced or induced to express RNPC1 for 24 h, followed by treatment with 5 μg/ml actinomycin D for various times. Total RNAs were isolated and then subjected to quantitative RT-PCR analysis. The level of MDM2 transcript was normalized to that of GAPDH control and the relative half-life of MDM2 was calculated. Data were presented as Mean±S.D. from triplicate samples. (F) Knockout of RNPC1 prolongs the half-life of MDM2 transcript in p53-null MEFs. The experiment was performed as in (E) except that p53−/−;RNPC1+/+ andp53−/−;RNPC1−/− MEFs were used.
To further explore the underlying mechanism by which RNPC1 inhibits the level of MDM2 transcript, we determined whether RNPC1 has an effect on the half-life of MDM2 transcript. To approach this, p53−/− HCT116 cells were uninduced or induced to express RNPC1 for 24 h, followed by treatment with actinomycin D for various times to inhibit the de novo RNA synthesis. The level of MDM2 transcripts was determined by qRT-PCR analysis and subsequently normalized to that of GAPDH control. We found that ectopic expression of RNPC1 significantly decreased the half-life of MDM2 transcript, from 4.2 h in control cells to 3.2 h in RNPC1-expressing cells. To further verify this, the same experiment was carried out with p53−/−;RNPC1+/+ and p53−/−;RNPC1−/− MEFs. As shown in Fig. 3F, the half-life of MDM2 transcript was significantly prolonged from 2 h in p53−/−;RNPC1+/+ MEFs to 3.0 h in p53−/−;RNPC1−/− MEFs. We would like to note that the half-life of human MDM2 transcript (4.2 h) was longer than that of mouse MDM2 transcript (2.0 h), likely due to differential posttranscriptional regulations via their 3′UTRs. Taken together, these data suggest that RNPC1 is able to destabilize the MDM2 transcript.
RNPC1 binds to the 3′UTR of MDM2 transcript in vivo and in vitro
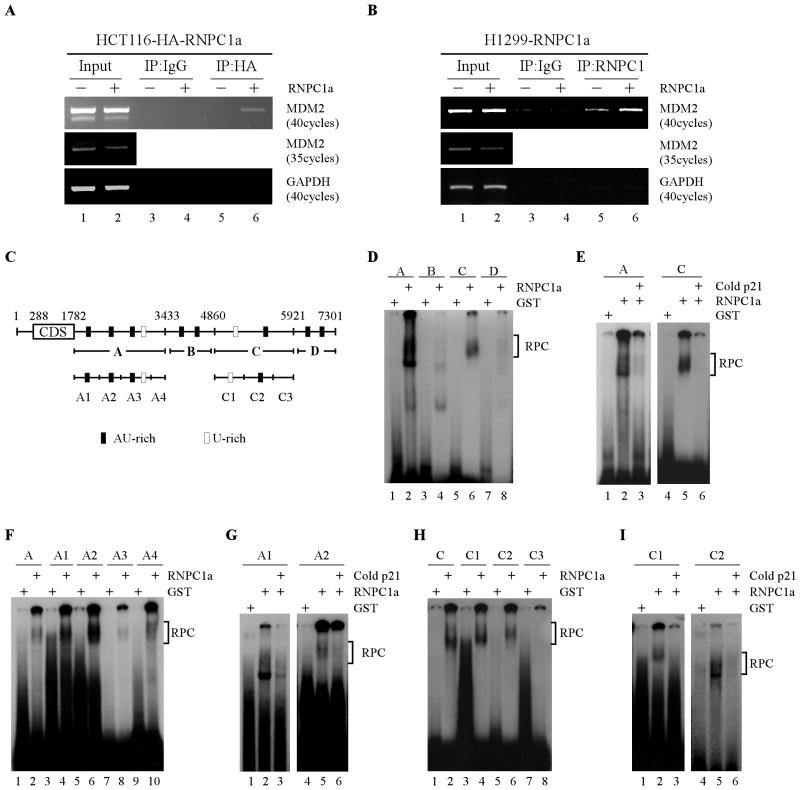
We next evaluated whether RNPC1 can bind to the MDM2 transcript in vivo using RNA-chip analysis. To do so, HCT116 cells were uninduced or induced to express HA-tagged RNPC1 for 36 h, and an anti-HA antibody, along with an isotype control IgG, was used to immunoprecipitate potential RNPC1-RNA complexes, followed by RT-PCR analysis to determine the association of RNPC1 with MDM2 transcript. Five percent of cell extracts were used as an input control. We found that upon RNPC1 induction, MDM2 mRNA was detectable in RNPC1 (anti-HA antibody), but not control IgG, immunoprecipitates (Fig. 4A, compare lanes 3 and 5 with 4 and 6, respectively). As a control, GAPDH transcripts were not found to interact with RNPC1 (Fig. 4A). To further verify this, H1299 cells that inducibly express RNPC1 (no tag) were used to perform the same experiment except that anti-RNPC1 antibody was used. As revealed in Fig. 4B, MDM2 mRNA was present in RNPC1, but not control IgG, immunoprecipitates (Fig. 4B, compare lanes 3 and 5 with 4 and 6, respectively). Interestingly, we showed that endogenous RNPC1 was able to associate with MDM2 mRNA although to a less extent (Fig. 4B, lane 5). Finally, we would like to mention that the level of MDM2 mRNA was decreased by RNPC1 when the input controls were amplified with fewer PCR cycles (Fig. 4A–B).
Figure 4. RNPC1 binds to the 3′UTR of MDM2 transcript in vivo and in vitro.
(A–B) RNPC1 associates with MDM2 transcript in vivo. HCT116 (A) or H1299 (B) cells were uninduced or induced to express HA-tagged RNPC1 (A) or RNPC1 (B) for 48 h, followed by immunoprecipitation with anti-HA (A), anti-RNPC1 (B), or an isotype control IgG (A–B). RT-PCR analysis was performed to measure the level of MDM2 and GAPDH transcript in the control and RNPC1-RNA immunocomplexes. Five percent of cell lysate was used as input. (C) Schematic presentation of MDM2 transcript and the location of probes used for REMSA. The AU- or U-rich elements were shown in shaded boxes. (D) Probes A and C, but not probes B and D, associate with RNPC1. REMSA was performed by mixing 32P-labeled probe A, B, C, or D with recombinant GST or GST-RNPC1 protein. The bracket indicates RNA-protein complexes (RPC). (E) Competition assay was performed by addition of unlabeled p21 probe, derived from p21 3′UTR, to the reaction mix prior to incubation with the 32P-labeled probe A or C. The bracket indicates RNA-protein complexes. (F) REMSA was performed by mixing recombinant GST or GST-RNPC1 protein with 32P-labeled probe A, or A1–4. The bracket indicates RNA-protein complexes. (G) Competition assay was performed by adding unlabeled p21 probe to the reaction mix prior to incubation with the 32P-labeled probe A1 or A2. The bracket indicates RNA-protein complexes. (H) REMSA was performed by mixing recombinant GST or GST-RNPC1 protein with 32P-labeled probe C, or C1–3. The bracket indicates RNA–protein complexes. (I) Competition assay was performed by adding unlabeled p21 probe to the reaction mix prior to incubation with the 32P-labeled probe C1 or C2. The bracket indicates RNA-protein complexes.
Next, to delineate RNPC1-binding site(s) in MDM2 transcript, RNA electrophoretic mobility assay (REMSA) was performed by using radiolabeled probes (probes A, B, C, and D), spanning the entire MDM2 3′ UTR (Fig. 4C). We found that recombinant GST-fused RNPC1 but not GST alone was able to form a complex with probes A and C (Fig. 4D, compare lanes 1 and 5 with 2 and 6, respectively). In addition, the formation of the complexes was significantly inhibited by cold p21 probe (Fig. 4E, lanes 3 and 6). The p21 probe is derived from p21 3′UTR and known to bind with RNPC1 [26, 29]. By contrast, probes B and D were unable to associate with recombinant RNPC1 (Fig. 4D, lanes 4 and 8). Next, to further delineate the RNPC1-binding region(s) within probe A, several subfragments within probe A (probes A1, A2, A3, and A4) (Fig. 4C) were made for REMSA. We found that like probe A, probes A1 and A2 showed a strong binding to RNPC1 (Fig. 4F, compare lanes 3 and 5 with 4 and 6, respectively) and this binding was significantly inhibited by adding cold p21 probe (Fig. 4G, lanes 3 and 6). However, probes A3 and A4 showed very weak binding with RNPC1 (Fig. 4F, lanes 8 and 10). To map the RNPC1-binding region(s) within probe C, probes C1, C2, and C3 were generated for REMSA (Fig. 4C). We showed that probes C1 and C2, but not probe C3, formed a complex with RNPC1 (Fig. 4H, compare lanes 3, 5, and 7 with 4, 6 and 8, respectively), which was also inhibited by cold p21 probe (Fig. 4I, lanes 3 and 6).
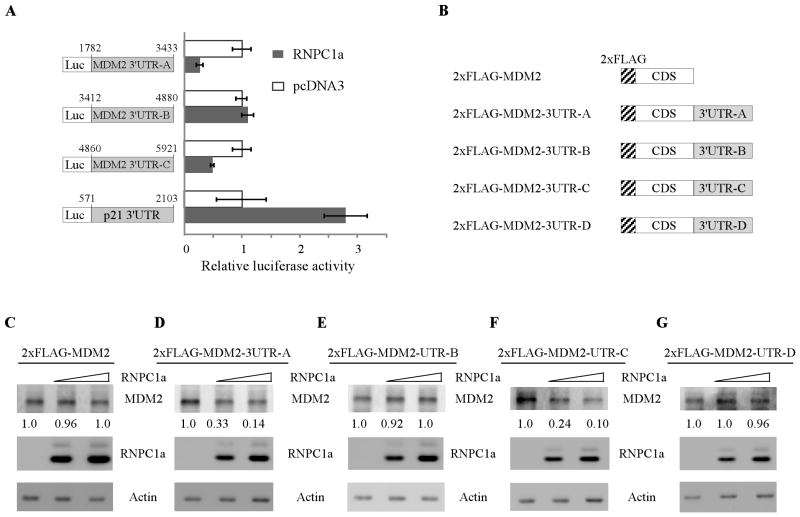
The 3′UTR in MDM2 transcript is required for RNPC1 to inhibit MDM2 expression
To verify that the 3′UTR in MDM2 transcript is indeed required for RNPC1 to inhibit MDM2 expression, two approaches were employed. Firstly, a dual-luciferase assay was performed using pGL3 reporters that carry various regions of MDM2 3′UTR, including 3′UTR-A, 3′UTR-B, and 3′UTR-C, whose sequences are identical to probes A, B, and C, respectively (Fig. 5A, left panel). We found that the luciferase activity for a reporter carrying MDM2 3′UTR-A and -C was significantly repressed by RNPC1 (Fig. 5A, right panel). By contrast, the MDM2 3′UTR-B was not responsive to RNPC1 (Fig. 5A, right panel). As a control, RNPC1 was able to increase the luciferase activity for a reporter carrying p21 3′UTR (Fig. 5A, right panel), consistent with previous report [26]. Secondly, we generated various expression vectors that contain the MDM2 coding region alone or in combination with various regions of the MDM2 3′UTR (Fig. 5B). We showed that RNPC1 markedly inhibited ectopic expression of MDM2 in a dose-dependent manner when MDM2 3′UTR-A or -C was inserted into downstream of the MDM2 open reading frame (Fig. 5D and 5F). By contrast, RNPC1 had no effect on ectopic MDM2 expression from expression vectors that contain MDM2 coding region alone (Fig. 5C) or in combination with 3′UTR-B (Fig. 5E) or 3′UTR-D (Fig. 5G). Taken together, these data suggest that MDM2 3′UTR-A from nt 1782 to 3443 and MDM2 3′UTR-C from nt 4860 to 5921 are responsive to RNPC1 and that each region is sufficient for RNPC1 to inhibit MDM2 expression.
Figure 5. The 3′UTR in MDM2 transcript is required for RNPC1 to inhibit MDM2 expression.
(A) The luciferase activity for the reporter carrying MDM2 3′UTR-A or -C is repressed by RNPC1. H1299 cells were cotransfected with pGL3 reporter carrying various regions of MDM2 3′UTR or p21 3′UTR along with pcDNA3 or pcDNA3 expressing HA-tagged RNPC1 for 48 h. Cells were then harvested for luciferase assay as described in “Materials and Methods”. The fold increase in relative luciferase activity is a product of the luciferase activity induced by RNPC1 divided by that induced by an empty pcDNA3 vector. (B) Schematic representation of the constructs containing the 2xFLAG-tagged MDM2 alone or in combination with various region of MDM2 3′UTR. (C–G) MDM2 expression is inhibited by RNPC1 for MDM2-expressing vectors, which contain MDM2 3′UTR-A and -C. Various amounts of RNPC1 expression vectors were transfected into H1299 cells along with a fixed amount of MDM2 expression vectors that contain the coding region alone (C) or in combination with MDM2 3′UTR-A (D), 3′UTR-B (E), 3′UTR-C (F), or 3′UTR-D (G). Twenty-four hours posttransfection, cell lysates were collected and the level of RNPC1, MDM2, and actin was determined by Western blot analysis. The level of MDM2 was normalized to that of actin and the fold change was shown below each lane.
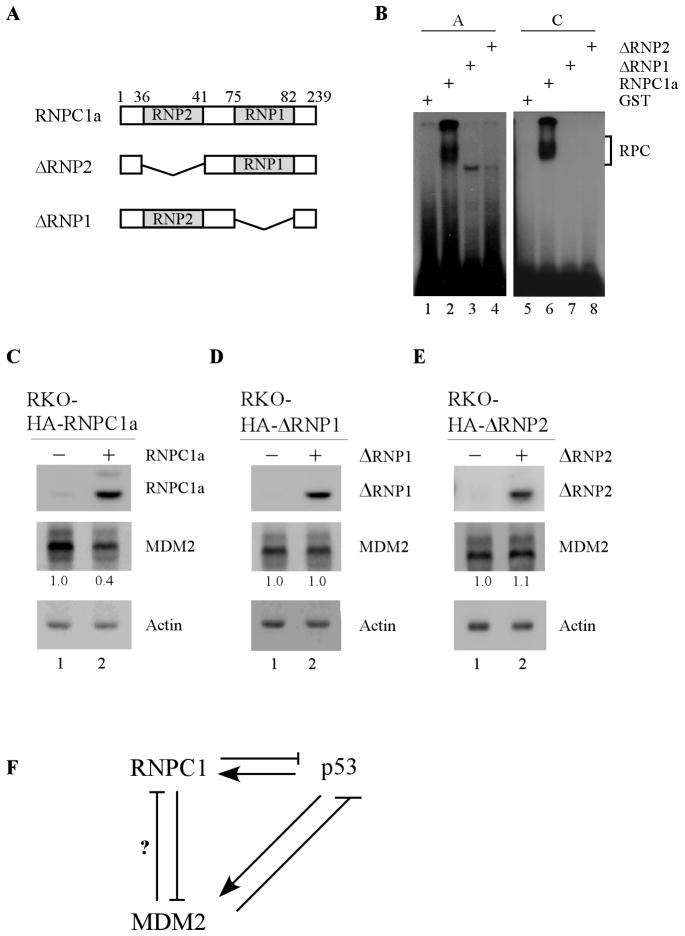
The RNA-binding activity of RNPC1 is required for inhibiting MDM2 expression
The RNA binding domain in RNPC1 is composed of two submotifs, RNP1 and RNP2 (Fig. 6A). Thus, to determine whether the RNA-binding domain in RNPC1 is required for its binding to the MDM2 transcript, two RNPC1 mutants deficient in RNP1 or RNP2 (ΔRNP1 and ΔRNP2) (Fig. 6A) were generated for REMSA. We found that unlike the full-length RNPC1, both ΔRNP1- and ΔRNP2-deletion mutants failed to associate with probe A or C (Fig. 5B, compare lane 2 and 6 with 3–4 and 7–8, respectively). To further verify this, the level of MDM2 was examined in RKO cells that can inducibly express RNPC1, ΔRNP1, or ΔRNP2. As shown in Fig. 6C, the level of MDM2 was repressed upon expression of RNPC1 (Fig. 6C, compare lane 1 with 2). By contrast, both ΔRNP1- and ΔRNP2-deletion mutants had no effect on MDM2 expression (Fig. 6D–E, compare lane 1 with 2). Taken together, these data suggest that the RNA-binding activity of RNPC1 is required for binding to the MDM2 transcript and consequently, for inhibiting MDM2 expression.
Figure 6. The RNA-binding activity of RNPC1 is required for inhibiting MDM2 expression.
(A) Schematic representation of RNPC1 mutants deficient in RNP1- or RNP2-submotif. (B) The RNA-binding domain in RNPC1 is required for binding to MDM2 3′UTR. REMSA was performed by incubating 32P-labeled probe A or C with recombinant GST, GST-RNPC1, GST-ΔRNP1, and GST-ΔRNP2. The bracket indicates the RNA-protein complexes. (C–E) The RNA-binding activity of RNPC1 is required for inhibiting MDM2 expression. RKO cells were uninduced or induced to express HA-tagged RNPC1 (C), ΔRNP1 (D), and ΔRNP2 (E) for 24 h, followed by Western blot analysis. The level of MDM2 was normalized to that of actin and the fold change was shown below each lane. Data are representative from three independent experiments. (F) A model for the regulation of MDM2 by RNPC1 and the interplay among RNPC1, MDM2, and p53.
Discussion
The MDM2 oncogene is frequently overexpressed in a subset of human cancers [34]. Thus, understanding how MDM2 expression is controlled will provide insight into the mechanisms of tumorigenesis and ultimately, how to target MDM2 for cancer therapeutics. In the current study, we reported a novel regulation of MDM2 by the RNA binding protein RNPC1. Specifically, we found that overexpression of RNPC1 shortened, whereas knockdown or knockout of RNPC1 prolonged, the half-life of MDM2 transcript, resulting in altered expression of MDM2 transcript and protein. Moreover, we showed that RNPC1 was able to bind to multiple sites in the 3′UTR of MDM2 transcript, and these sites were required for RNPC1 to inhibit ectopic MDM2 expression. Furthermore, we showed that the RNA binding activity of RNPC1 was required for binding to and destabilizing MDM2 transcript. Based on these findings, we propose a model for the regulation of MDM2 by RNPC1 and the interplay among RNPC1, MDM2, and p53 (Fig. 6F).
Our results indicated that RNPC1 was able to bind multiple sites in the MDM2 3′UTR, which were located in two regions, 3′UTR-A from nt 1782 to 3433 and 3′UTR-C from nt 4860 to 5621 (Fig. 4D–I). Moreover, we showed that the luciferase activity for a reporter carrying each region can be significantly repressed by RNPC1 (Fig. 5A). Consistent with this, RNPC1 was able to inhibit ectopic MDM2 expression only when MDM2 3′UTR-A or -C was cloned downstream of the MDM2 open reading frame (Fig. 5D and 5F). These data suggest that RNPC1 is able to bind multiple sites in MDM2 3′UTR and subsequently inhibits MDM2 expression. The presence of multiple binding sites may be critical to ensure RNPC1 binding to MDM2 transcript when some sites are not available. As a matter of fact, the MDM2 3′UTR contains several cis-regulatory elements, such as AREs, which can be bound by microRNAs and/or other RNA-binding proteins. For example, several microRNAs, including miR-143, miR-145, and miR-605, were found to inhibit MDM2 expression at a posttranscriptional level [35, 36]. Moreover, HuR, an ARE-interacting RNA-binding protein, was recently found to bind to and stabilize the MDM2 transcript via its 3′UTR although the binding site is not mapped [37]. Thus, future studies are warranted to examine whether RNPC1 associates or competes with microRNAs and/or other RNA-binding proteins to regulate MDM2 mRNA stability.
The MDM2 gene is expressed as multiple isoforms due to alternative splicing. Importantly, some isoforms are found to be over-expressed in human cancers and associated with prognosis [34, 38, 39]. Of note, some of these isoforms contain the same 3′UTR as the full length MDM2 transcript, which are likely to be regulated by RNPC1. Thus, it would be interesting to look into whether RNPC1 is able to differentially regulate MDM2 variants and whether this regulation plays a role in tumorigenesis.
Most RNPC1 targets identified so far, including p53, p63, p21, HuR, and MDM2 (this study), contain either AU- or U-rich element at their 3′UTR [26–28, 40]. However, we showed recently that RNPC1 stabilizes p73 transcript via a CU-rich element in the 3′UTR of p73 mRNA[41]. In addition, the mechanism and outcome of RNPC1-mediated regulation also varies among its targets. For example, RNPC1 stabilizes p21, p73, and HuR mRNAs [26, 40, 41], but destabilizes p63 and MDM2 mRNAs [27]. Additionally, RNPC1 binds to both 5′ and 3′UTRs of p53 transcript and inhibits p53 mRNA translation [28]. Thus, further studies are needed to determine the common sequence or motif that can be targeted by RNPC1 and to elucidate how RNPC1 differentially regulates its targets. Addressing these questions will shed light on the biological function of RNPC1.
We have previously reported that RNPC1 is a target of p53 family proteins, which in turn inhibits p53 mRNA translation via binding to p53 5′ and 3′UTRs [26, 28]. In the current study, we found that RNPC1 can shorten the half-life of MDM2 transcript independent of p53. These data indicate that RNPC1 is a key player of the p53-MDM2 circuitry as a target of p53 and a negative regulator of p53 and MDM2. Thus, we postulate that under nonstressed conditions, RNPC1 and MDM2 restrain p53 expression by targeting p53 mRNA translation and p53 protein degradation, respectively. However, in response to various types of stress, p53 is activated and then induces RNPC1 and MDM2 expression. On one hand, induction of RNPC1 destabilizes MDM2 transcript and results in a decreased expression of MDM2, releasing p53 from MDM2-mediated proteasomal degradation. On the other hand, RNPC1 exerts as a negative regulator of p53 (Fig. 6F). Although the biological function of these regulations under a physiological condition remains to be determined, these data uncover a novel mechanism by which MDM2 is regulated by RNPC1 via mRNA stability, which may be important for fine-tuning the p53-MDM2 circuitry.
Material Methods
Reagents
Anti-MDM2 antibodies, SMP14 and 2A10, were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and EMD (Rockland, MA), respectively. Anti-HA was purchased from Covance (San Diego, CA). Anti-p53 (1C12) was purchased from Cell signaling (Danvers, MA). Anti-actin, proteinase inhibitor cocktail, RNase A, protein A/G beads were purchased from Sigma (St. Louis, MO). Rabbit RNPC1 antibody was made as previously described [26]. Scrambled small interfering RNA (siRNA) (GGC CGA UUG UCA AAU AAU U) and siRNA against RNPC1 (GCC AUU GGG GTG CAG CAG UU) were purchased from Dharmacon RNA Technologies (Chicago, IL).
Generation of expression and reporter vectors
pcDNA3-HA-RNPC1, pcDNA3-RNPC1, and pBabe-H1-siRNPC1 vectors were generated as described previously [26]. pGL3 control vector and pGL3 vector that contains a full length of p21 3′UTR [42] was a gift from Drs. S. Lee and P. Leedman (Harvard Medical School, Boston, MA, and Western Australian Institute for Medical Research, Perth, Western Australia, Australia).
To generate a luciferase reporter carrying MDM2 3′UTR-A from nt 1782 to 3433, a DNA fragment was amplified using cDNA samples from MCF7 cells as a template with a forward primer, 5′ TTG ACC TGT CTA TAA GAG AAT TAT ATA TTT C 3′, and a reverse primer, 5′ GTC TTA CGG GTA AAT GGT GGC T 3′. The DNA fragment was initially cloned into pGEMT vector and then inserted into pGL3 vector through SpeI and ApaI sites to generate pGL3-MDM2-3UTR-A. To generate pGL3-MDM2-3UTR-B that contains MDM2 3′UTR from nt 3412 to 4880, the same strategy was used except that the primers used were forward primer, 5′ AGC CAC CAT TTA CCC GTA AGA C 3′, and reverse primer 5′CAG GCA AAC CTT ATT CGG CTC 3′. To generate pGL3-MDM2-3UTR-C that contains the MDM2 3′UTR from nt 4860 to 5921, the same strategy were used except that the primers were forward primer, 5′GAG CCG AAT AAG GTT TGC CTG 3′, and reverse primer, 5′ CAG ATT CTG CTT GGT TCT AGC TTC 3′.
To generate pcDNA4 vector that carries a 2xFLAG-tagged MDM2, a cDNA that contains 2xFLAG MDM2 coding region [43] was inserted into pcDNA4 vector through KpnI and EcoRI to generate pcDNA4-2xFLAG-MDM2. To generate pcDNA4 vectors expressing 2xFLAG-tagged MDM2 along with a portion of MDM2 3′UTR (MDM2 3′UTR-A, or MDM2 3′UTR-B, or MDM2 3′UTR-C), a DNA fragment from MDM2 3′UTR was cloned downstream of MDM2 open reading fram (ORF) through NotI site to generate pcDNA4-2xFLAG-MDM2-3UTR-A, pcDNA4-2xFLAG-MDM2-3UTR-B, and pcDNA4-2xFLAG-MDM2-3UTR-C, respectively. To generate pcDNA4-2xFLAG-MDM2-3UTR-D vector, the MDM2 3′UTR-D fragment was amplified by PCR using cDNA sample from MCF7 cells with a forward primer 5′GAA GCT AGA ACC AAG CAG AAT CTG 3′ and a reverse primer, 5′GAA CAC CTT CCA AGT GCT GGT G 3′. This fragment was initially cloned into pGEMT vector, and then inserted into pcDNA4-2xFLAG-MDM2 through NotI site.
Cell culture
RKO, HCT116, p53−/−HCT116, H1299, MCF7, PC3, and SW480 cells were cultured in DMEM supplemented with 10% fetal bovine serum as previously described [28]. Primary MEFs were isolated as previously described [28] and cultured in DMEM supplemented with 10% fetal bovine serum, 1x nonessential amino acids, and 55 μM beta-mercaptoethanol. RKO, HCT116, p53−/−HCT116, H1299, MCF7, and SW480 cell lines, which inducibly express RNPC1a, HA-tagged RNPC1a, HA-tagged RNPC1b, or shRNA against RNPC1, were generated by using a Tet-on inducible system as described previously [28, 44]. Briefly, to generate stable cell lines that inducibly express RNPC1a or HA-tagged RNPC1a, pcDNA4-RNPC1a or pcDNA4-HA-RNPC1a was transfected into parental cells expressing a tetracycline repressor (pcDNA6). The RNPC1a-expressing cells were selected with zeocin and confirmed by Western blot analysis. To generate stable cell lines that inducibly express HA-tagged RNPC1b, the same strategy was applied except that pcDNA4-HA-RNPC1b was used. To generate stable RKO cell lines that inducibly express HA-tagged ΔRNP1 and ΔRNP2, the same strategy was applied except that pcDNA4-HA-ΔRNP1 or pcDNA4-HA-ΔRNP2 was used. To generate SW480 cell line that can inducibly expresses an shRNA against RNPC1, pBabe-H1-siRNPC1, which was generated previously [26], was transfected into SW480 cells expressing tetracycline repressor. The cells were selected with puromycin and confirmed by Western blot analysis. For induction, tetracycline (250–500 ng/mL) was added to culture medium.
Western blot analysis
Western blot analysis was performed as previously described [45]. Briefly, Cells were washed and collected from plates in PBS solution, resuspended with 2x SDS sample buffer, and boiled for 5 min. Proteins were then resolved in an 8–12% SDS-PAGE gel and transferred to a nitrocellulose membrane, followed by ECL detection. The level of protein was quantified using Labworks software (UVP, Upland, CA).
REMSA probe generation and labeling
RNA probes were generated and 32P-labeled by in vitro transcription using PCR products containing T7 promoter and various regions from the MDM2 3′ UTR as a template. The primers for generating probe A were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GTT GAC CTG TCT ATA AGA GAA TTA TAT ATT TC 3′, and reverse primer, 5′ GTC TTA CGG GTA AAT GGT GGC T 3′. The primers for generating probe B were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GAG CCA CCA TTT ACC CGT AAG AC 3′, and reverse primer, 5′ CAG GCA AAC CTT ATT CGG CTC 3′. The primers for generating probe C were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GGA GCC GAA TAA GGT TTG CCT G 3′, and reverse primer, 5′ CAG ATT CTG CTT GGT TCT AGC TTC 3′. The primers for generating probe D were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GGA AGC TAG AAC CAA GCA GAA TCT G 3′, and reverse primer, 5′ GAA CAC CTT CCA AGT GCT GGT G 3′. The primers for generating probe A1 were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GTT GAC CTG TCT ATA AGA GAA TTA TAT ATT TC 3′, and reverse primer, 5′ CAC GGT GAA ACC CTG TCT CTA C 3′. The primers for generating probe A2 were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GTT AGC CAG GAT GGT CTC GAT C 3′, and reverse primer, 5′ CCA GCC TGG CCA ACA TGG TG 3′. The primers for generating probe A3 were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GTC ACG AAC TCC TGA CCT CAA G 3′, and reverse primer, 5′ TTT GGG AGG CTG AGG TGA GTA G 3′. The primers for generating probe A4 were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GAT GCT GGG ATT ACA GAT GTG 3′, and reverse primer, 5′ GTC TTA CGG GTA AAT GGT GGC T 3′. The primers for generating probe C1 were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GGA GCC GAA TAA GGT TTG CCT G 3′, and reverse primer, 5′ AGG TCC TCA AAG CAT TAT TTG GAG 3′. The primers for generating probe C2 were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GCC AAA GGT AAA AGT ACT AAT CCC 3′, and reverse primer, 5′ GAG AAA AAC ACT AAA TCA AGA TGG 3′. The primers for generating probe C3 were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GCC ATA TGT GAA TTG TAT ATA CTT AGG 3′, and reverse primer, 5′ CAG ATT CTG CTT GGT TCT AGC TTC 3′. The primers for generating p21 probe were forward primer, 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GGC GAG GGC CTC AAA G 3′, and reverse primer, 5′ GCA GGG GGC GGC CAG GGT AT 3′.
Recombinant protein purification and REMSA
Recombinant proteins were expressed in bacteria BL21 and purified by glutathione sepharose beads. REMSA was performed as described previously [27]. Briefly, 32P-labeled probes were incubated with recombinant protein in a binding buffer (10 mM HEPES-KOH at pH 7.5, 90 mM potassium acetate, 1.5 mM magnesium acetate, 2.5 mM DTT, 40 U of RNase inhibitor [Ambion]) at 30°C for 30 min. RNA-protein complexes were resolved on a 5% acrylamide gel and radioactive signals were detected by autoradiography.
RNA isolation, RT–PCR, and quantitative PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to user’s manual. cDNA was synthesized using MMLV reverse transcriptase from Promega according to the manufacturer’s instructions. The PCR program used for amplification was (i) 94°C for 5 min, (ii) 94°C for 45 s, (iii) 58°C for 45 s, (iv) 72°C for 1 min, and (v) 72°C for 10 min. From steps 2 to 4, the cycle was repeated 25 times for human GAPDH and mouse actin or 30 times for RNPC1 and MDM2. The primers used to amplify human GAPDH were forward primer, 5′ CGG GAA ACT GTG GCG TGA TG 3′, and reverse primer, 5′ GGA GTG GGT GTC GCT GTT GAA GT 3′. The primers to amplify human RNPC1 were forward primer 5′ ACT ACC GAC GCC TCG CTC AG 3′, and reverse primer, 5′ CCC AGA TAT GCC AGG TTC AC 3′. The primers to amplify mouse RNPC1 were forward primer 5′ GAC GCA TCG CTC AGA AAG T 3′, and reverse primer, 5′ GAG GAG TCA GCC CGT AGG T 3′. The primers to amplify human MDM2 were forward primer, 5′ GAA CTT GGT AGT AGT CAA TCA GC 3′, and reverse primer, 5′ GCC TGA TAC ACA GTA ACT TGA TA 3′. The primers to amplify mouse MDM2 were forward primer, 5′ ATG AGG TCT ATC GGG TCA CAG T 3′, and reverse primer, 5′ CAC ATC CAA GCC TTC TTC TGC 3′. The primers to amplify mouse actin were forward primer, 5′ CCC ATC TAC GAG GGC TAT 3′, and reverse primer, 5′ AGA AGG AAG GCT GGA AAA 3′.
Quantitative PCR (qPCR) was performed in 20-μl reactions using 2X qPCR SYBR Green Mix (ABgene, Epsom, UK) with 5 μM primers. Reactions were run on a realplex (Eppendorf, Germany) using a two-step cycling program: 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, 68°C for 30 s. A melt curve (57–95°C) was generated at the end of each run to verify the specificity. The primers for human GAPDH were forward primer, 5′-CCC AGC CTC AAG ATC ATC AGC AAT G 3′, and reverse primer, 5′ATG GAC TGT GGT CAT GAG TCC TT 3′. The primers for human MDM2 were forward primer, 5′ TGC CAA GCT TCT CTG TGA A 3′, and reverse primer, 5′ CGA TGA TTC CTG CTG ATT GA 3′. The primers for the mouse MDM2 and actin were the same as shown above.
RNA Immunoprecipitation Followed by RT-PCR (RNA-CHIP)
RNA immunoprecipitation and RT-PCR (RIP) was performed as previously described [46]. Briefly, cells (2 × 107) were uninduced or induced to express RNPC1 for 24h. Cell extracts were prepared with immunoprecipitation buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes, 1 mM DTT, and 0.5% NP-40), and then incubated with 2 μg of anti-HA or anti-RNPC1 or isotype control IgG at 4 °C overnight. The RNA-protein immunocomplexes were brought down by protein A/G beads, followed by RT-PCR analysis.
Luciferase assay
Dual-luciferase reporter assay was performed according to the manufacturer’s instructions (Promega, Madison, WI, USA). Briefly, 5ng of Renilla luciferase vector (pRL-CMV, Promega), an internal control, and 200 ng of a pGL3 reporter that contains various region of MDM2 3′UTR or p21 3′UTR were cotransfected into H1299 cells (5 × 104) along with pcDNA3 or a pcDNA3 vector expressing HA-tagged RNPC1. Thirty-six hours post-transfection, luciferase activity was measured with the dual luciferase kit and Turner Designs luminometer. The fold change in relative luciferase activity is a ratio of the luciferase activity induced by RNPC1 divided by that induced by empty pcDNA3 vector.
Supplementary Material
Acknowledgments
This work is supported in part by NIH grants R01 CA076069 and R01 CA102188
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Cahilly-Snyder L, et al. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet. 1987;13(3):235–44. doi: 10.1007/BF01535205. [DOI] [PubMed] [Google Scholar]
- 2.Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10(6):1565–9. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Momand J, et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 4.Oliner JD, et al. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362(6423):857–60. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 5.Haupt Y, et al. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 6.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420(1):25–7. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 7.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 8.Jones SN, et al. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378(6553):206–8. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 9.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 10.Bouska A, Eischen CM. Murine double minute 2: p53-independent roads lead to genome instability or death. Trends Biochem Sci. 2009;34(6):279–86. doi: 10.1016/j.tibs.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Ganguli G, Wasylyk B. p53-independent functions of MDM2. Mol Cancer Res. 2003;1(14):1027–35. [PubMed] [Google Scholar]
- 12.Barak Y, et al. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12(2):461–8. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zauberman A, et al. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23(14):2584–92. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trotta R, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3(2):145–60. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 15.Moumen A, et al. Met acts on Mdm2 via mTOR to signal cell survival during development. Development. 2007;134(7):1443–51. doi: 10.1242/dev.02820. [DOI] [PubMed] [Google Scholar]
- 16.Xiong L, et al. A novel role for IGF-1R in p53-mediated apoptosis through translational modulation of the p53-Mdm2 feedback loop. J Cell Biol. 2007;178(6):995–1007. doi: 10.1083/jcb.200703044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang S, et al. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275(12):8945–51. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 18.Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19(11):1473–6. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 19.Lohrum MA, et al. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3(6):577–87. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23(23):8902–12. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279(43):44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 22.Dai MS, et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24(17):7654–68. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin A, et al. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24(17):7669–80. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35(3):316–26. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong X, et al. Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene. 2011;30(15):1798–811. doi: 10.1038/onc.2010.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20(21):2961–72. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Jun Cho S, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc Natl Acad Sci U S A. 2010;107(21):9614–9. doi: 10.1073/pnas.0912594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, et al. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 2011;25(14):1528–43. doi: 10.1101/gad.2069311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 2010;38(7):2256–67. doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto S, et al. RNA-binding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes Cells. 2009;14(11):1241–52. doi: 10.1111/j.1365-2443.2009.01347.x. [DOI] [PubMed] [Google Scholar]
- 31.Leveille N, et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23(15):5113–21. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai H, et al. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278(46):45946–53. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- 34.Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell. 2002;2(1):9–15. doi: 10.1016/s1535-6108(02)00091-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, et al. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2012 doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 36.Xiao J, et al. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 2011;30(24):5021. doi: 10.1038/emboj.2011.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh M, et al. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest. 2009;119(12):3530–43. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukas J, et al. Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res. 2001;61(7):3212–9. [PubMed] [Google Scholar]
- 39.Sigalas I, et al. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med. 1996;2(8):912–7. doi: 10.1038/nm0896-912. [DOI] [PubMed] [Google Scholar]
- 40.Cho SJ, et al. The RNA-binding Protein RNPC1 Stabilizes the mRNA Encoding the RNA-binding Protein HuR and Cooperates with HuR to Suppress Cell Proliferation. J Biol Chem. 2012;287(18):14535–44. doi: 10.1074/jbc.M111.326827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan W, et al. P73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability. Mol Cell Biol. 2012 doi: 10.1128/MCB.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giles KM, et al. The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J Biol Chem. 2003;278(5):2937–46. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- 43.Jung YS, Qian Y, Chen X. DNA polymerase eta is targeted by Mdm2 for polyubiquitination and proteasomal degradation in response to ultraviolet irradiation. DNA Repair (Amst) 2012;11(2):177–84. doi: 10.1016/j.dnarep.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harms KL, Chen X. Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer Res. 2007;67(7):3145–52. doi: 10.1158/0008-5472.CAN-06-4397. [DOI] [PubMed] [Google Scholar]
- 45.Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20(25):3193–205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- 46.Peritz T, et al. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1(2):577–80. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.