Abstract
Background
To examine the relationship between autism spectrum disorders (ASD) and specific language impairment (SLI), family studies typically take a comparative approach where families with one disease are examined for traits of the other disease. In contrast, the present report is the first study with both disorders required to be present in each family to provide a more direct test of the hypothesis of shared genetic etiology.
Methods
We behaviorally assessed fifty-one families including at least one person with ASD and at least one person with SLI (without ASD). Pedigree members were tested using 22 standardized measures of language and intelligence. Since these extended families include a non-shared environmental contrast, we calculated heritability, not just familiality, for each measure twice: 1) baseline heritability analysis compared to 2) heritability estimates after statistically removing ASD subjects from pedigrees.
Results
Significant increases in heritability on four supra-linguistic measures (including Pragmatic Judgment) and a composite language score, but not on any other measures, were observed when removing ASD subjects from the analysis indicating differential genetic effects that are unique to ASD. Non-genetic explanations such as effects of ASD severity or measurement error or low score variability in ASD subjects were systematically ruled out, leaving the hypothesis of non-additive genetics effects as the potential source of the heritability change caused by ASD.
Conclusions
While the data suggest genetic risk factors common to both SLI and ASD, there are effects that appear unique to ASD, possibly caused by non-additive gene-gene interactions of shared risk loci.
Keywords: autism, specific language impairment, heritability, epistasis, gene-gene, interaction, shared etiology
Introduction
Autism Spectrum Disorders (ASD) are a group of complex developmental disabilities that may include problems with 1) social interaction, 2) communication and 3) restricted interests and/or stereotypies. When there is profound impairment in all three areas, the individual is classified with autistic disorder while deficits in one area paired with lesser impairments in two and/or three are considered as part of the autism spectrum. Specific Language Impairment (SLI) is a neurodevelopmental disorder that is characterized by significant limitations in language abilities occurring in the absence of any other frank neurological disorder or environmental cause. Both disorders share variable limitations of communication, making etiological overlap between the disorders possible. Initial studies indicated similar patterns of impairment in SLI and ASD, including in structural language (semantics and syntax) and phonological short-term memory (PSTM) in a subgroup of children with autism (1, 2). Since that intial work, numerous studies have examined what may be grossly binned as three competing hypotheses explored below: 1) incidental etiological overlap, where overlap is induced by the definitions of the two disorders, 2) familiality of the two disorders but not necessarily due to shared genetics and, 3) shared genetics.
Incidental Etiological Overlap
A recent review proposed that the SLI and ASD may have incidental overlap without meaningful shared etiology (3). This conclusion assumes that structural language, such vocabulary and grammar affected in SLI, and social use of language, such as pragmatics and supralinguistic tasks know to be impaired in ASD, form two quantitative dimensions for language competence. Therefore, in this system, the hallmark deficits of SLI and ASD are on different axes. Persons with SLI (low structural language) will display the full range of social language ability and persons with ASD (low social language ability) will display the full range structural language. Some overlap of SLI and ASD is inevitable in this bivariate system but not productively considered shared etiology, as the two dimensions may be independent. If SLI and ASD were unrelated, the prevalence of one estimated from a sample ascertained for the other would match the population estimate. Two such studies have been reported where families ascertained for SLI were examined for cases of ASD. The prevalence of autism was estimated to be 2.5 and 4.3 times higher than the prevalence in the general population (4). These studies, though limited in sample size, indicate that SLI and ASD are not independent.
Familiality
Most family studies recruit nuclear families ascertained through autism probands and then compare relatives of the autism proband to relatives in control families. Assuming that autism liability may not manifest in relatives as a distinct clinical entity, researchers assess differences in quantitative assessments of autism symptomatology. Quantitative differences relative to control families are defined as the broader autism phenotype (BAP). Several autism domains have well replicated BAP features including social and communication behaviors (5-8) and restricted interests and rigidity (9). Pragmatics, or how language context contributes to meaning, is the most replicated language domain within the BAP where parents of children with autism have lower mean scores than controls (8, 10, 11). Since SLI is defined by structural language deficits, not pragmatic deficits, it is not unexpected that while pragmatics is consistently shown to be part of the BAP, results from structural language deficit studies have not generally been supportive of common etiology. Phonological short-term memory (PSTM) deficits, a common marker for SLI, has not been consistently found in ASD families (10, 12), regardless of the structural language impairment status of the autism proband (impaired versus unimpaired). Comparisons of SLI relatives with relatives of typically developing probands, or with mental retardation probands, yield similar negative findings (5, 13). To date, there is no compelling evidence that ASD and SLI are jointly familial.
Shared Genetic Markers
In 2008, a series of three papers provided converging evidence from independent study designs/methods that implicate CNTNAP2 in autism susceptibility (14-16). This striking result has since been followed up in SLI proband families with significant results for association of SLI with CNTNAP2 (17). It is not known if the risk mutations are identical in the two disorders. CNTNAP2 is also associated with communication ability at age 2 in a general population sample (18). Further studies are needed to fully assess if alleles within CNTNAP2 affect pragmatic aspects of language. It is unclear how much of the shared genetic liability between ASD and SLI is accounted for by CNTNAP2, these data provide evidence that shared genetic etiology is possible and remains to be fully explained. CNTNAP2 has also been implicated in other cognitive and neurological disorders from intellectual disability to stuttering (19-26), making the ASD- SLI connection harder to disentangle.
Hypothesis
The present study presents a complementary study design that extends the ASD-SLI overlap literature in a novel direction. Our project was designed to address the question of shared genetic etiology in ASD and SLI by directly testing genetic overlap in pedigrees containing both disorders. We have ascertained nuclear families with both ASD and SLI in mutually exclusive persons, then collected direct measurement of language in all family members, both affected and unaffected, including as many extended family members as possible. The extended family design is useful for quantitative genetic heritability studies since extended families have relatives that share genetics but not environment (such as cousins), thus providing the key contrast that allows for dissociation of shared environment from additive genetics. We assessed the effect of ASD on heritability estimates by comparing an analysis using all subjects to one where ASD subjects are removed. We hypothesized that measures known to be associated with the language profile in autism, such as supralinguistic skills including pragmatics, would show differential heritability indicating genetic effects unique to autism. We were also specifically interested in non-word repetition and overall structural language ability, hypothesizing that both would be associated with differential heritability potentially indicating genetic effects unique to SLI.
Methods
Families
Fifty-one families were ascertained through a proband with autism and with the additional requirements that each family have at least one additional family member meeting the study criteria for SLI and no less than five participants (affected and unaffected) per family (x̄=6.9, SD=2.8, range=5-20). Families were recruited from the greater New Jersey area for a total of 234 subjects with at least some quantitative language phenotypic data including 27 persons with ASD, 55 with SLI and 152 unaffected. Subjects gave informed consent conforming to the guidelines for treatment of human subjects governed by the Institutional Review Board at Rutgers University.
Autism proband criteria
To be identified as an autism proband, the following criteria were met: A diagnosis of autistic disorder on at least two of the following three measures: 1) Autism Diagnostic Interview - Revised (ADI-R) (27, 28) score of “autism,” 2) Autism Diagnostic Observation Schedule (27, 29) score of “autism,” 3) DSM-IV, autistic disorder.
Specific language impairment proband criteria
To be identified as an SLI proband, a the following inclusionary/exclusionary criteria were met:
A core standard score of <= 85 on the age appropriate version of the Clinical Evaluation of Language Fundamentals (CELF-4) (30, 31) and <= PIQ.
Performance IQ (PIQ) >= 80 on the Wechsler Abbreviated Scale for Intelligence (WASI) (32).
Hearing within normal limits [positive identification of 500 Hz at 30 dB (SPL), and 10 00, 2000, and 4000 Hz at 20 dB (SPL)].
No motor impairments or oral structural deviations affecting speech or non-speech movement of the articulators.
No history of autism spectrum disorders or frank neurological disorders such as intellectual disability or brain injury, as determined by parental interview.
Native English speaker with English as the primary language spoken at home.
Measures
All SLI probands, non-ASD non-SLI family members, and higher functioning family members with ASD received age appropriate measures of language (Means and SD by diagnostic status in Table 1). The standardized language battery included: a. CELF-4 and CELF Preschool (30, 31)- Core standard scores were derived from 3-5 subtests scaled scores (age depending) that addressed areas of language comprehension, expression, and structure including Word Structure, Recalling Sentences, Formulating Sentences, Word Classes (Expressive and Receptive) and Word Definitions. b. The Comprehensive Assessment of Spoken Language (CASL) (33)- The supralinguistic core of subtests addressed metalinguistic language skills including abstraction, inference, and a subtest on the pragmatic aspects of language. These areas are of great relevance to older children, adults, and higher functioning individuals with autism who may be challenged by meaning that cannot be accessed directly through lexical and grammatical information. Subtests include Meaning from Context, Nonliteral Language, Ambiguous Sentences and Pragmatic Judgment (which has been shown to correlate (34) with the Vineland Adaptive Behavior Scales (35)). c. The Comprehensive Test of Phonological Processing (CTOPP) (36), the Elision subtest was used to measure deletion and phonological manipulation of sounds in words while the Non-word Repetition task measured phonological short-term memory; both have a strong documented relationship with oral language abilities.
Table 1.
Summary statistics for affected and unaffected family members
| ASD Subjects Mean (SD) |
SLI Subjects Mean (SD) |
Other Subjects Mean (SD) |
|
|---|---|---|---|
| N | 27+ | 55 | 152 |
| Age (years) | 13.9 (9.8) | 24.4 (19.9) | 32.8 (19.1) |
| Sex (% Male) | 72% | 71% | 47% |
| CELF | |||
| Core Score | 71.5 (26.5) | 69.2 (18.2) | 107 (12) |
| Formulating Sentences | 5.7 (3.8) | 5.2 (3.1) | 11.3 (2.5) |
| Repeating Sentences | 4.7 (3.4) | 3.8 (2.3) | 9.9 (2.6) |
| Word Classes Receptive | 7.8 (2.8) | 6.9 (2.9) | 12 (1.6) |
| Word Classes Expressive | 5.7 (3.1) | 5.5 (2.5) | 11.2 (2.5) |
| Word Classes Total | 6.5 (2.6) | 6 (2.4) | 11.6 (2) |
| Word Definitions | 8.4 (2.4) | 8.1 (2.5) | 12.9 (2.4) |
| CASL | |||
| Ambiguous Sentences | 76.6 (12.3) | 74.8 (8.4) | 98.3 (14.2) |
| Meaning from Context | 71.1 (16.3) | 74.3 (11.4) | 103.5 (11.1) |
| Non-literal Language | 70.5 (21) | 70.2 (15.1) | 100 (14.7) |
| Pragmatic Judgment | 68.9 (21.2) | 71.3 (16.9) | 99.2 (10.9) |
| CTOPP | |||
| Elision | 8 (3.9) * | 5.5 (3.4) * | 9.6 (2.8) |
| Nonword Repetition | 8.6 (2.8) | 7.4 (3.1) | 9 (2.5) |
| WASI | |||
| PIQ | 93 (16.5) | 90.9 (13.9) | 108.9 (12.1) |
There are 64 subjects with ASD in the sample, but only 27 had quantitative language data
ASD mean is different from SLI mean, p < .05 after Bonferonni correction
Descriptive data for subjects with ASD capable of taking the language battery, SLI subjects, and all other family members are included in Table 1. Tested ASD and SLI subjects only differed on Elision (p < .05) where the average of SLI subjects is 2.5 points lower than ASD. On Elision, ASD subjects did not differ from all other family members. Families had an average 1.1 persons with language impairment and 1.3 with ASD (0.43 for subjects with ASD that had at least some language data). The total sample was 58% male as is consistent with an increased male risk of ASD and SLI. Average age of all subjects was 30 (range 5-80) with subgroups listed in Table 2. While all ASD subjects were in the child generation, when interpreting the average age of SLI subjects it should be noted that 8 were parents with the remainder in the child generation (bimodal distribution). While scores on our standard measures use normative data that does not exceed 18 years old, and in some cases 21 years old, there was no evidence of ceiling effects in this sample when administered to ages beyond the normative data.
Table 2.
Relationships used in heritability analysis by SOLAR
| Relative pairs | Number |
|---|---|
| Parent-offspring | 261 |
| Sibling | 104 |
| Grandparent-grandchild | 24 |
| Avuncular | 61 |
| Half sibling | 4 |
| Great grandparent-grandchild | 2 |
| First cousin | 39 |
| First cousin, once removed | 3 |
| Total | 498 |
Statistical Analysis
Additive genetic heritability was estimated using procedures described in detail previously (37). Heritability was calculated by the SOLAR package v4.3.1 (38) through maximum likelihood procedures using information from the entire pedigree jointly (see supplemental for more methodologic details). The 234 participants that had behavioral data comprised 498 not-mutually exclusive relative pairs with 133 having shared genetics but no shared environment (Table 2), indexing the pedigree complexity relative power of the sample. This sample size is roughly comparable to our previous study of heritability in SLI pedigrees (39).
Tests of heritability changes based on the presence of ASD in the pedigree were conducted by comparing the likelihood of two models with the likelihood ratio test (LRT). For the baseline heritability model we tested age, sex and PIQ as covariates (kept in model if p < .05). Baseline analysis used all available pedigree data, which we denote as ASD+. The likelihood of the baseline model was compared to the likelihood of a model that included ASD status as a covariate, which we denote ASD−. ASD status was coded as a binary variable where persons with ASD were coded as 0 and unaffected persons were coded as 1. All pedigree members were coded in this way. This procedure statistically controls for variation caused by ASD status, which has the net effect of removing ASD from the pedigree in a way that is less wasteful of valid information than artificially setting scores from ASD persons to missing values. Changes in heritability between the baseline model and comparison model are informative about the statistical fit of an additive genetics model for ASD. A parallel set of analyses controlling for SLI status was also conducted. We applied a Bonferroni correction for 14 likelihood ratio tests requiring a critical p-value < .0035 to reach overall (p < .05) significance.
Interpretation of the model
The statistical model (see supplemental for details) assumes only additive effects. Therefore data that is fully inconsistent with an additive model would therefore be expected to show no heritability. Data that is consistent with additive genetics would simply yield the heritability of the trait. In the present study, we tested for the presence of an in-between case. If heritability increases when excluding ASD subjects, this is an indication that subjects with ASD were reducing the additive heritability (i.e., an additive model is not the best fit for the data). To reject the additive genetic model in favor of a non-additive model, complicating features of the data must be ruled out (outliers, non-normal data, possible mediator variables). Non-additive models include dominance (where carriers of ASD do not have intermediate phenotypes) and gene-gene interactions, also called epistasis.
Results
Baseline
Results for language measures are presented in Table 3. Estimates using the entire pedigree are called ASD+ and are used as a baseline reference for further comparisons. All measures showed significant evidence for an additive genetic component (h2 = 0.24 to 0.86).
Table 3.
Heritability (SE) of Language and Reading Measures with/without ASD
| ASD+ | ASD− | Δ h 2 | Covariates | |
|---|---|---|---|---|
| CELF | ||||
| Core Score | 0.43 (0.19) | 0.53 (0.18) | 0.10 * | sex, PIQ |
| Formulating Sentences | 0.24 (0.18) | 0.24 (0.19) | 0.00 | sex, PIQ |
| Repeating Sentences | 0.62 (0.17) | 0.65 (0.16) | 0.03 | PIQ |
| Word Classes Expressive | 0.34 (0.17) | 0.37 (0.17) | 0.03 | sex, PIQ |
| Word Classes Receptive | 0.47 (0.20) | 0.48 (0.19) | 0.01 | sex, PIQ |
| Word Classes Total | 0.36 (0.20) | 0.40 (0.19) | 0.04 | sex, PIQ |
| Word Definitions | 0.64 (0.16) | 0.65 (0.15) | 0.01 | age, sex, PIQ |
| CASL | ||||
| Ambiguous Sentences | 0.38 (0.21) | 0.46 (0.19) | 0.08 * | PIQ |
| Meaning from Context | 0.59 (0.17) | 0.70 (0.14) | 0.11 * | PIQ |
| Non-literal Language | 0.39 (0.16) | 0.45 (0.16) | 0.06 * | PIQ |
| Pragmatic Judgment | 0.38 (0.18) | 0.44 (0.20) | 0.06 * | PIQ |
| CTOPP | ||||
| Elision | 0.46 (0.23) | 0.48 (0.25) | 0.02 | age, PIQ |
| Nonword Repetition | 0.86 (0.13) | 0.86 (0.13) | 0.00 | age, sex |
| WASI | ||||
| PIQ | 0.43 (0.20) | 0.43 (0.20) | 0.00 | sex |
ASD+ h2 different from ASD−, p<.05 after Bonferroni correction
Removing ASD Subjects
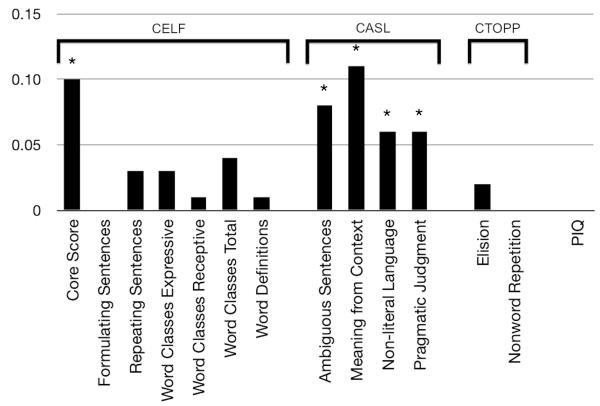
The effect of ASD status was statistically controlled to (operationally) remove all subjects with ASD (ASD− in Table 3). Examining of the distribution of all heritability estimates, ASD+ v. ASD-, a paired t-test was significant (t_13 = −3.94, p < .05), indicating differences in heritability estimates between conditions; since the changes in heritability cannot be thought of as truly independent observations, this result should be considered suggestive as the t-test may be anti-conservative. Post hoc likelihood ratio tests comparing the model likelihoods for ASD+ v. ASD− indicated five heritabilities were significantly different (p < .05), including all four supralinguistic tasks and the overall language score from the CELF (Figure 1). In each case, heritability was higher when controlling for ASD (i.e., ASD− > ASD+). The largest change occurred in Meaning from Context, which increased 11.5 percentage points. The three other supralinguistic measures also showed significant increases in heritability of 5, 7, and 9 percentage points. There was also a significant increase in heritability for overall language ability as measured by the CELF (10 percentage points) but not on any individual subscales, though semantics (Word Classes Total, 4%) and syntax (Recalling Sentences, 3%) both trended toward significance.
Figure 1.
Barchart of changes in heritability caused by including ASD status as a covariate. Significant changes are. While the correlation between heritability estimates from the baseline models versus the comparison models was high overall (ρ=0.97,P<.05), several measures clearly deviate from the overall trend that are statistically significant (denoted with asterisks). The barchart demonstrates the signal-to-noise ratio of the results.
Examination of Moderator Effects Potentially Causing Heritability Changes
If scores from persons with ASD were associated with greater measurement error, then controlling for ASD should reduce the standard error of the heritability estimates (noise reduction). The average of the SE’s for models with significant heritability differences were identical in ASD+ and ASD− to 3 decimal places (0.181). This was also true of models that did not show heritability changes. Significant ASD effects on heritability were not accompanied by significant changes in standard errors for the models. The observed slight fluctuations in SE’s between ASD+ v. ASD− models appeared random (sign test, p = 0.14).
While mean scores in the ASD subjects were lower than the corresponding population means, the distribution of scores did not appear to play a role in heritability changes (Figure 2). Inclusion of outliers could affect heritability estimation. While the data from ASD subjects did not contain outliers, as defined by the absolute value of a score > 3 SD from the population mean, we did examine if moderately extreme scores > 2 SD had an effect. A total of 8 datapoints across all 5 measures were moderately extreme, according to this definition, and removal of these scores by treating them as missing data did not change the 5 heritabilities for the ASD− models. Further, all heritability changes remained significant (LRT, p < .05).
Figure 2.
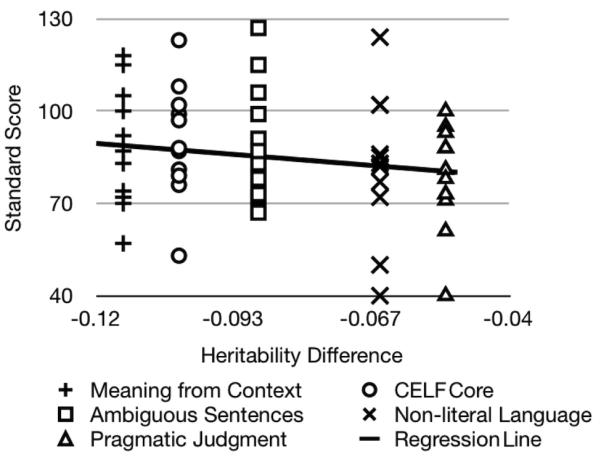
Scatter plot of scores from ASD subjects (y-axis) on changes in heritability (x- axis). While the average for each measure is below population levels, it is clear that changes in heritability are not driven strictly by universally low performance of ASD subjects. Regression of ASD proband scores on changes in heritability (line in figure) yielded non-significant effects of both trait and quantitative value of the heritability difference.
We also examined ASD severity as a possible moderator of the heritability changes whereby increasing severity results in less reliable quantitative language scores. For this analysis we applied ADOS calibrated severity scores for Modules 1-3 as covariates (40). Since Module 4 does not have calibrated severity scores, we also incorporated information about that module as an additional binary covariate (not administered = 0, administered = 1) to account for the lack of quantitative severity scores. Estimates of heritability were highly similar when ADOS severity was included as a covariate in the model, failing to support severity of impairment as a moderator of heritability changes.
Removing Language Impaired Subjects
To examine if the genetic etiology of ASD was similar to SLI, we compared (baseline) SLI+ analysis to SLI− analysis for all traits except the CELF core score, which by definition would be censored, in the analytical sense, since all SLI subjects scored below a fixed threshold, thus removing one tail of the distribution; Note that calculation of heritability on censored data in general pedigrees has not been developed in the literature. Considering the remaining 13 traits, only three of the supralinguistic measures showed significant differential heritability (Ambiguous Sentences, Meaning from Context, Non-literal Language) while Pragmatic Judgment did not (Figure 3).
Figure 3.
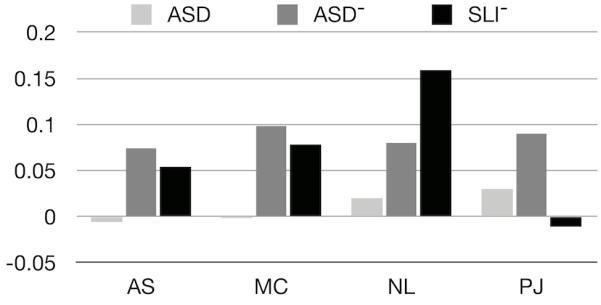
Changes in supralinguistic measure heritability relative to a control condition, where unaffected individuals were randomly removed form the analysis using the same statistical procedure as for ASD and SLI subjects. Our baseline analysis, ASD (all subjects), is equivalent to randomly removing individuals from the analysis as shown by the slight deviations from the control baseline (lightest gray). Both ASD− and SLI− show large deviations on the supralinguistic measures relative to the control condition. ASD− and SLI− induce similar changes in heritability for both AS (ambiguous sentences) and MC (meaning from context) while NL (non-literal language) also shows changes in heritability but SLI− is greater than ASD−. A notable difference is in PJ (pragmatic judgment) where SLI− is consistent with the control condition (i.e., no effect of SLI on PJ heritability).
Discussion
When controlling for ASD in families ascertained for both ASD and SLI subjects, heritability increases are seen for five out of fourteen measures, by as much as 11 percentage points. Changes occurred in the global language measure and all four supralinguistic measures. These results cannot be accounted for by potentially misleading properties of the data such outliers, reduced variability in ASD subjects, measurement error or severity of the ASD phenotype. We conclude that the heritability increases caused by exclusion of ASD family members are not consistent with a simple additive-genetics model of heritability. The data, therefore, suggest that non-additive genetic effects contribute to ASD. SLI heritabilities also show similar non-additive effects for three of the supralinguistic measures excluding pragmatics. Taken together, the results provide empirical support for a hypothesis initially articulated and validated through an extensive computer simulation study (41), which stated that gene-gene interaction can account for molecular genetic findings shared between ASD and SLI while still allowing for phenotypic difference that give rise to the different diagnoses. The present dataset is not large enough to support direct estimation of the additional gene-gene interaction variance component parameters (additive-additive, additive-dominant, dominant-dominant); though this could be accomplished with a larger sample size in future studies.
The lack of non-additive effects for pragmatics in SLI while present in ASD indicates that some genetic effects are unique to ASD, though only in this one domain of language. Pragmatic language has previously been shown to be heritable in twins ascertained for SLI using the Children’s Communications Checklist self/teacher report (42), but this is the first study to estimate the genetic variance on a wider range of supralinguistic skills through direct quantitative assessment. Such higher order linguistic tasks are associated with the limited ability of individuals with ASD to understand language abstraction and also the well-appreciated pragmatic deficits in ASD. All four supralinguistic scales from the CASL showed significant heritability, though inclusion of ASD individuals in the analysis significantly diminishes that heritability, implying that some differing etiologies are influencing the performance on these tasks in ASD versus non-ASD family members. While diminished, the heritabilities for these traits are still substantial when considering all family members (ASD and non-ASD), implying that there are also genetic loci in common across all family members that contribute to these abilities. It is interesting to note that in our sample, mean scores from persons with ASD are not significantly different than those of SLI subjects and both groups are lower than population means. Reduced supralinguistic skills are not a defining characteristic of SLI so this observation is unexpected and novel relative to the SLI and ASD literatures. This is the first study to ascertain families with both disorders and it is possible that, since supralinguistic skills are heritable, selecting a family into the study where at least one person has ASD (and so is expected to have supralinguistic deficits), essentially selects for risk loci that may produce such deficits in family members without ASD. Under those selection demands, the subjects with SLI may be at greatest risk for reduced performance.
The Core standard score of the CELF is less heritable when including ASD subjects in the analysis while only a few individual subscales displayed that trend, and none were significant. It is possible that the nature of the composite CELF Core standard score, which weights data from multiple subscales, has greater variability and/or reliability than the individual subscales. If true, then the observed heritability results would be expected as the subscales have less reliability and/or variability for the analysis. Yet without strong results from the subscales, the standard score on the CELF is indeed quite broadly representative of language skills making interpretation challenging. Further studies with greater sample sizes will be needed to identify which aspects of structural language are key to the observed decreased heritability.
We hypothesized that heritability differences would be observed in PSTM since this is a cognitive domain where ASD and SLI have notably divergent presentations in the types of errors that tend to manifest with low performance (43, 44). Further, PSTM deficits are quite common in SLI, but occur only a subgroup of subjects with ASD. Our data indicate that the same genetics that influences PSTM in subjects with SLI (and the rest of the pedigree) also influences PSTM in ASD, leaving the heritability estimates essentially the same. PSTM performance was previously shown to differentiate ASD and SLI; not in overall performance as scored in the CTOPP, but only when errors by syllable length were considered (43, 44). It is possible that the quantitative score studied here is simply not suitable for detection of such qualitative differences in performance, making genetics that are unique to ASD possible.
Overall the presented heritability estimates are consistent with the literature with one qualified exception. Our non-word repetition heritability estimate from the CTOPP is higher than commonly reported for that measure (37, 45). However, the Children’s Test of Nonword Repetition (46), a similar measure of PSTM, was estimated to have heritability > 1.0 in twins with SLI (47). This estimate is higher than genetically possible due to the nature of their chosen statistical formulation but does indicate genetic effects much larger than 0 in SLI subjects. It is therefore possible that our CTOPP non-word repetition heritability is reasonable for pedigrees selected for SLI and autism.
Several aspects of statistical modeling should be considered when interpreting the data. The nature of pedigrees precludes holding age constant across all subjects. And while we applied age as a covariate to capture age effects, it is still possible that age effects, such as changes in additive genetics throughout the lifespan, remain. Recently, we applied these pedigree methods to a collection of SLI pedigrees without ASD and found the results to be quite comparable to the literature on twin children, presumably due to generally modest sizes of age-related changes in heritability (37). Here too, the heritability estimates are very similar to previous literature on twin children.
Heritability of the ASD diagnosis as a categorical trait shows wide variability, with high heritability (48-50), generally greater than 80 in previous studies, while one recent study indicates a more modest effect of genetics (51). Additionally, ASD symptoms show considerable variation, even when considering the differences in study designs and sample sizes (50, 52-54). In contrast, our quantitative phenotype heritability results are consistent with the language genetics literature, which generally show much less variability (55) than has been shown in ASD (50, 52-54). The magnitude of the heritability changes observed in our study should therefore be interpreted relative to the stability of the language genetics literature rather than the variability associated with a categorical diagnosis of autism. Due to the limited number of persons with ASD in our pedigrees (many families have only 1) as a consequence of our ascertainment protocol, analysis of ASD as a categorical trait is not powerful enough to inform the ongoing debate about the heritability of the categorical ASD diagnosis. Lastly, while epistasis appears to be a likely explanation for how ASD differs from SLI despite shared genetics, it was not possible to formally estimate the necessary additional parameters with the current sample size. Additional studies with much larger samples are necessary to test an epistasis model for ASD and SLI.
Supplementary Material
Acknowledgements
We would like to thank the participating families whose time and cooperation made this work possible and Drs. Barbie Zimmerman-Bier and Kapila Seshadri for help in recruiting and assessing families for this study. Family collection was supported by the National Institute of Mental Health R01MH070366 and RC1MH088288.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
Dr. Brzustowicz serves as a consultant for the Janssen Pharmaceutical Companies of Johnson & Johnson, and serves on the Scientific Advisory Board of Motif BioSciences. Dr. Brzustowicz is named on the following patents: Millonig JH, Brzustowicz LM, Gharani N, “Compositions and Methods for Diagnosing Autism”, US Patent #7,629,123, December 8, 2009; Brzustowicz LM, Firestein BL, “Methods and Compositions for the Diagnosis and Treatment of Schizophrenia”, US Patent #8,067,158, November 29, 2011. Dr. Buyske is named on the following patent application: Buyske S, Stenroos ES, Johnson WG “Association of GSTM1 with autism and assays and methods based thereon”, US Patent Application 12/069,988, filed February 11, 2009.
All other authors reported no biomedical financial interests or potential conflicts of interest.
References
- 1.Joseph RM, Tager-Flusberg H, Lord C. Cognitive profiles and social-communicative functioning in children with autism spectrum disorder. J Child Psychol Psychiatry. 2002;43:807–821. doi: 10.1111/1469-7610.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kjelgaard MM, Tager-Flusberg H. An Investigation of Language Impairment in Autism: Implications for Genetic Subgroups. Lang Cogn Process. 2001;16:287–308. doi: 10.1080/01690960042000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomblin B. Co-morbidity of autism and SLI: kinds, kin and complexity. Int J Lang Commun Disord. 2011;46:127–137. doi: 10.1111/j.1460-6984.2011.00017.x. [DOI] [PubMed] [Google Scholar]
- 4.Conti-Ramsden G, Simkin Z, Botting N. The prevalence of autistic spectrum disorders in adolescents with a history of specific language impairment (SLI) J Child Psychol Psychiatry. 2006;47:621–628. doi: 10.1111/j.1469-7610.2005.01584.x. [DOI] [PubMed] [Google Scholar]
- 5.Bishop DV, Maybery M, Wong D, Maley A, Hill W, Hallmayer J. Are phonological processing deficits part of the broad autism phenotype? Am J Med Genet B Neuropsychiatr Genet. 2004;128B:54–60. doi: 10.1002/ajmg.b.30039. [DOI] [PubMed] [Google Scholar]
- 6.Losh M, Adolphs R, Poe MD, Couture S, Penn D, Baranek GT, et al. Neuropsychological profile of autism and the broad autism phenotype. Arch Gen Psychiatry. 2009;66:518–526. doi: 10.1001/archgenpsychiatry.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy M, Bolton PF, Pickles A, Fombonne E, Piven J, Rutter M. Personality traits of the relatives of autistic probands. Psychol Med. 2000;30:1411–1424. doi: 10.1017/s0033291799002949. [DOI] [PubMed] [Google Scholar]
- 8.Ruser TF, Arin D, Dowd M, Putnam S, Winklosky B, Rosen-Sheidley B, et al. Communicative competence in parents of children with autism and parents of children with specific language impairment. J Autism Dev Disord. 2007;37:1323–1336. doi: 10.1007/s10803-006-0274-z. [DOI] [PubMed] [Google Scholar]
- 9.Losh M, Childress D, Lam K, Piven J. Defining key features of the broad autism phenotype: a comparison across parents of multiple- and single-incidence autism families. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:424–433. doi: 10.1002/ajmg.b.30612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitehouse AJ, Barry JG, Bishop DV. The broader language phenotype of autism: a comparison with specific language impairment. J Child Psychol Psychiatry. 2007;48:822–830. doi: 10.1111/j.1469-7610.2007.01765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehouse AJ, Coon H, Miller J, Salisbury B, Bishop DV. Narrowing the broader autism phenotype: a study using the Communication Checklist-Adult Version (CC-A) Autism. 2010;14:559–574. doi: 10.1177/1362361310382107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindgren KA, Folstein SE, Tomblin JB, Tager-Flusberg H. Language and reading abilities of children with autism spectrum disorders and specific language impairment and their first-degree relatives. Autism Res. 2009;2:22–38. doi: 10.1002/aur.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilowsky T, Yirmiya N, Shalev RS, Gross-Tsur V. Language abilities of siblings of children with autism. J Child Psychol Psychiatry. 2003;44:914–925. doi: 10.1111/1469-7610.00175. [DOI] [PubMed] [Google Scholar]
- 14.Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakkaloglu B, O’Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitehouse AJ, Bishop DV, Ang QW, Pennell CE, Fisher SE. CNTNAP2 variants affect early language development in the general population. Genes Brain Behav. 2011 doi: 10.1111/j.1601-183X.2011.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregor A, Albrecht B, Bader I, Bijlsma EK, Ekici AB, Engels H, et al. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med Gen et. 2011;12:106. doi: 10.1186/1471-2350-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrin AL, Giacheti CM, Maximino LP, Abramides DV, Zanchetta S, Rossi NF, et al. Identification of a microdeletion at the 7q33-q35 disrupting the CNTNAP2 gene in a Brazilian stuttering case. Am J Med Genet A. 2010;152A:3164–3172. doi: 10.1002/ajmg.a.33749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, Collins AL, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Gen et. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Molecular psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Molecular psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 25.Peter B, Raskind WH, Matsushita M, Lisowski M, Vu T, Berninger VW, et al. Replication of CNTNAP2 association with nonword repetition and support for FOXP2 association with timed reading and motor activities in a dyslexia family sample. J Neurodev Disord. 2011;3:39–49. doi: 10.1007/s11689-010-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newbury DF, Paracchini S, Scerri TS, Winchester L, Addis L, Richardson AJ, et al. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav Genet. 2011;41:90–104. doi: 10.1007/s10519-010-9424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, et al. Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. Autism Dev Disord. 1989;19:185–212. doi: 10.1007/BF02211841. [DOI] [PubMed] [Google Scholar]
- 28.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 29.Gotham K, Risi S, Pickles A, Lord C. The Autism Diagnostic Observation Schedule: revised algorithms for improved diagnostic validity. J Autism Dev Disord. 2007;37:613–627. doi: 10.1007/s10803-006-0280-1. [DOI] [PubMed] [Google Scholar]
- 30.Semel E, Wiig EH, Secord WA. Clinical evaluation of language fundamentals, fourth edition (CELF-4) The Psychological Corporation/A Harcourt Assessment Company; Toronto: 2003. [Google Scholar]
- 31.Wiig EH, Secord WA, Semel E. Clinical evaluation of language fundamentals—Preschool, second edition (CELF Preschool-2) The Psychological Corporation/A Harcourt Assessment Company; Toronto: 2004. [Google Scholar]
- 32.Wechsler D. Wechsler Abbreviated Scale of Intelligence (WASI) San Antonio, TX: 1999. [Google Scholar]
- 33.Carrow-Woolfolk E. Circle Pines. AGS; MN: 1999. Comprehensive Assessment of Spoken Language. [Google Scholar]
- 34.Reichow B, Salamack S, Paul R, Volkmar FR, Klin A. Pragmatic Assessment in Autism Spectrum Disorders: A Comparison of a Standard Measure With Parent Report. Commun Disord Q. 2008;29:169–176. doi: 10.1177/1525740108318697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sparrow SS, Balla DA, Cicchetti DV. Vineland Adaptive Behavior Scales. American Guidance Service; Circle Pines, MN: 1984. expanded edition. [Google Scholar]
- 36.Wagner RK, Torgesen JK, Rashotte CA. Comprehensive test of phonological processing. PRO-ED; Austin, TX: 1999. [Google Scholar]
- 37.Logan J, Petrill SA, Flax J, Justice LM, Hou L, Bassett AS, et al. Genetic Covariation Underlying Reading, Language and Related Measures in a Sample Selected for Specific Language Impairment. Behav Genet. doi: 10.1007/s10519-010-9435-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Logan J, Petrill SA, Flax J, Justice LM, Hou L, Bassett AS, et al. Genetic covariation underlying reading, language and related measures in a sample selected for specific language impairment. Behav Genet. 2011;41:651–659. doi: 10.1007/s10519-010-9435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotham K, Pickles A, Lord C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J Autism Dev Disord. 2009;39:693–705. doi: 10.1007/s10803-008-0674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishop DV. Overlaps between autism and language impairment: phenomimicry or shared etiology? Behav Genet. 2010;40:618–629. doi: 10.1007/s10519-010-9381-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop DV, Maybery M, Wong D, Maley A, Hallmayer J. Characteristics of the broader phenotype in autism: a study of siblings using the children’s communication checklist-2. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:117–122. doi: 10.1002/ajmg.b.30267. [DOI] [PubMed] [Google Scholar]
- 43.Riches NG, Loucas T, Baird G, Charman T, Simonoff E. Non-word repetition in adolescents with Specific Language Impairment and Autism plus Language Impairments: a qualitative analysis. J Commun Disord. 2011;44:23–36. doi: 10.1016/j.jcomdis.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Whitehouse AJ, Barry JG, Bishop DV. Further defining the language impairment of autism: is there a specific language impairment subtype? J Commun Disord. 2008;41:319–336. doi: 10.1016/j.jcomdis.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Raskind WH, Hsu L, Berninger VW, Thomson JB, Wijsman EM. Familial aggregation of dyslexia phenotypes. Behav Genet. 2000;30:385–396. doi: 10.1023/a:1002700605187. [DOI] [PubMed] [Google Scholar]
- 46.Gathercole SE, Willis CS, Baddeley AD, Emslie H. The Children’s Test of Nonword Repetition: a test of phonological working memory. Memory. 1994;2:103–127. doi: 10.1080/09658219408258940. [DOI] [PubMed] [Google Scholar]
- 47.Bishop DV, North T, Donlan C. Nonword repetition as a behavioural marker for inherited language impairment: evidence from a twin study. J Child Psychol Psychiatry. 1996;37:391–403. doi: 10.1111/j.1469-7610.1996.tb01420.x. [DOI] [PubMed] [Google Scholar]
- 48.Taniai H, Nishiyama T, Miyachi T, Imaeda M, Sumi S. Genetic influences on the broad spectrum of autism: study of proband-ascertained twins. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:844–849. doi: 10.1002/ajmg.b.30740. [DOI] [PubMed] [Google Scholar]
- 49.Rosenberg RE, Daniels AM, Law JK, Law PA, Kaufmann WE. Trends in autism spectrum disorder diagnoses: 1994-2007. Journal of autism and developmental disorders. 2009;39:1099–1111. doi: 10.1007/s10803-009-0723-6. [DOI] [PubMed] [Google Scholar]
- 50.Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167:1357–1363. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- 51.Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, et al. Genetic Heritability and Shared Environmental Factors Among Twin Pairs With Autism. Archives of general psychiatry. 2011 doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hallmayer J. Chromosomes 1, 2, and 7 workshop. American journal of medical genetics. 1999;88:219–223. [PubMed] [Google Scholar]
- 53.Robinson EB, Koenen KC, McCormick MC, Munir K, Hallett V, Happe F, et al. Evidence that autistic traits show the same etiology in the general population and at the quantitative extremes (5%, 2.5%, and 1%) Archives of general psychiatry. 2011;68:1113–1121. doi: 10.1001/archgenpsychiatry.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson EB, Koenen KC, McCormick MC, Munir K, Hallett V, Happe F, et al. A multivariate twin study of autistic traits in 12-year-olds: testing the fractionable autism triad hypothesis. Behav Genet. 2012;42:245–255. doi: 10.1007/s10519-011-9500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stromswold K. The heritability of language: a review and metaanalysis of twin adoption and linkage studies. Language. 2001;77:647–723. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.