Abstract
Copy number variations (CNVs) have been reported to be causal suspects in a variety of psychopathologic traits. We investigate whether de novo and/or inherited CNVs contribute to the risk for Attention Problems (APs) in children. Based on longitudinal phenotyping, 50 concordant and discordant monozygotic (MZ) twin pairs were selected from a sample of ∼3200 MZ pairs. Two types of de novo CNVs were investigated: (1) CNVs shared by both MZ twins, but not inherited (pre-twinning de novo CNVs), which were detected by comparing copy number (CN) calls between parents and twins and (2) CNVs not shared by co-twins (post-twinning de novo CNVs), which were investigated by comparing the CN calls within MZ pairs. The association between the overall CNV burden and AP was also investigated for CNVs genome-wide, CNVs within genes and CNVs outside of genes. Two de novo CNVs were identified and validated using quantitative PCR: a pre-twinning de novo duplication in a concordant-unaffected twin pair and a post-twinning deletion in the higher scoring twin from a concordant-affected pair. For the overall CNV burden analyses, affected individuals had significantly larger CNVs that overlapped with genes than unaffected individuals (P=0.008). This study suggests that the presence of larger CNVs may increase the risk for AP, because they are more likely to affect genes, and confirms that MZ twins are not always genetically identical.
Keywords: copy number variation, twin, Attention Problem, ADHD
Introduction
Copy number variations (CNVs) are polymorphisms in the number of copies of chromosomal segments (duplications and deletions) ranging from 1 kb to several Mb and have been recognized as a major contributor to human genetic variability. CNVs collectively encompass a larger part of the genome than single-nucleotide polymorphisms (SNPs).1, 2, 3 Mutation rates for CNVs are two to four times higher than those of point mutations and affect larger segments of the genome.4, 5 CNVs have been shown to correlate with changes in gene expression levels.6, 7, 8, 9 Changes in copy number (CN) can also lead to the generation of new combinations of exons between different genes, causing protein changes in structure and modified protein activities.10, 11 Therefore, CNVs are likely to be involved in phenotypic variation, including disease susceptibility, especially when they are large and affect multiple genes. CNVs can be either inherited or de novo, with the assumption that de novo CNVs are more likely to have deleterious effects.12 CNVs have been linked to several neuropsychiatric disorders including schizophrenia, autism and attention-deficit hyperactivity disorder (ADHD).13, 14, 15, 16
We investigated whether there is an association between CNVs (de novo and inherited) and Attention Problems (AP) in a selected sample of concordant and discordant monozygotic (MZ) twin pairs. The AP scale has been shown to be predictive for ADHD. Children who score low on the AP scale of the Child Behavior Checklist (CBCL) have a non-ADHD diagnosis in 96% of the cases, and children with a high AP score have a positive diagnosis for ADHD in 36% (girls) and 59% (boys) of cases.17 In addition, the sensitivity and specificity of the measure is increased if longitudinal scores on AP are considered. Heritability estimates for AP and ADHD in children are about 70% and 75%, respectively,18, 19 and ∼75% of the covariance between the AP scale and ADHD has been estimated to be explained by genetic influences.20 Previous work that included part of the current MZ sample showed structural21 and functional22 brain differences in addition to significant behavior differences among the discordant twin pairs.23
In this study, MZ twins discordant and concordant for AP are examined for the presence of two types of de novo CNVs (1) pre-twinning de novo CNVs: CNVs that emerged during parental meiosis, and are therefore shared by the MZ twins, but not by the parents (parental genotypes were available for more than half of the subjects) and (2) post-twinning de novo CNVs: CNVs that undergo a CN change in mitosis during the development of one of the twins, causing a discordance between the MZ twins. Post-twinning de novo mutations could result in a genetic discordance in all tissues (due to a premorula mutation, most likely at the two-cell stage) or somatic mosaicism (due to mutation at the four-cell stage or later).24 De novo CNVs have been demonstrated in MZ twins25 and are one mechanism by which phenotypic discordance in MZ twins may be explained. Validation of de novo CNVs identified through a genome-wide scan is important because of the tendency to discover false positive mutations when using SNP microarray technology.26 In this study, we employ the use of quantitative PCR (qPCR) to confirm the de novo CNVs indentified from the genome-wide scan for CNVs. In addition, the association between the genome-wide CNV burden and AP is investigated for CNVs genome-wide, CNVs overlapping with genes and CNVs outside of genes (for the de novo and inherited CNVs pooled together).
Materials and methods
Subjects
A total of 50 MZ twin pairs were selected from the Netherlands Twin Register (NTR).27 Selection was based on longitudinal maternal reports from the AP scale of the CBCL.28 The AP scale has been used to identify children at risk for clinical ADHD and consists of 11 items (eg, ‘cannot sit still, restless or hyperactive', ‘cannot concentrate, pay attention for long', ‘impulsive or acts without thinking', and so on). Normative scores are provided for the AP scale, which allows for determining whether a child is at risk for ADHD based on gender and age-specific T-scores.23 The AP scale was collected at ages 7, 10 and 12 years and eligible twin pairs were selected from a total sample of 3228 MZ twin pairs. A total of 1966 MZ twin pairs (birth cohorts 1986–1994) had measures from at least two time points and an additional 1256 pairs had longitudinal ratings from all three time points. Children were identified as affected if they had a T-score >60 at all available time points and a T-score of at least 65 at one or more time points. Children were classified as unaffected if they had a T-score of <55 at all time points. A T-score of 65 represents the clinical cut-off for ADHD.17 The criterion of longitudinal discordance in MZ twins represents a severe selection measure, as only 18 of the 1966 pairs with longitudinal data available meet this criterion. There were 52 concordant high-scoring twin pairs (both twins affected), 962 concordant low-scoring twin pairs (both twins unaffected) and 18 discordant twin pairs (one affected and one unaffected). The twins were only selected on AP and not on the presence or absence of any other disorders. AP was not measured for the parents. DNA samples were available for 50 MZ pairs: 17 concordant-high (6 male and 11 female pairs), 22 concordant-low (8 male and 14 female pairs) and 11 discordant (4 male and 7 female pairs) twin pairs and 36 parent pairs. The study was approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Center, Amsterdam, and an Institutional Review Board certified by the US Office of Human Research Protections (IRB number IRB-2991 under Federal-wide Assurance-3703; IRB/institute codes, NTR 03-180).
Genotyping
Twins and their parents provided buccal swabs for DNA extraction. Methods for buccal swab collection, genomic DNA extraction and zygosity testing have been described previously.29 Genotyping was performed on the Affymetrix Human Genome-Wide SNP 6.0 Array according to the manufacturer's protocol (Affymetrix, Santa Clara, CA, USA). This array contains 906 600 SNPs and 940 000 CN probes. Of the CN probes, 800 000 are evenly spaced across the genome and the rest across 3700 known CNV regions. A total of 172 individuals were genotyped (50 MZ twin pairs and 36 parent pairs). Twins were randomly distributed across plates with respect to AP scores and twins from the same twin pair were genotyped on separate plates. Parents were genotyped together, but not on the same plate as their offspring. Quality control (QC) was done according to the protocol and resulted in a total sample size of 153 individuals comprising 45 complete twin pairs (21 concordant low, 10 discordant and 14 concordant high). Of these 45 complete twin pairs, 25 sets had DNA from both parents who passed QC, 4 complete twin pairs had DNA from one parent who passed QC, the unpaired twins had DNA from one parent who passed QC and 1 unpaired twin had DNA from both parents who passed QC. CNVs were called with the Birdsuite30 and PennCNV31 algorithms. CN segments were only included in further analyses if the following conditions were met: (1) the CN calls agreed between both algorithms, (2) the overlapping part of the segments from both algorithms was >100 kb and (3) the segment was not in a centromere. Because calling algorithms can produce artificially split CNV calls, adjacent CNV calls were merged after visual inspection of LogR ratio (LRR) and B-allele frequency (BAF) plots, if the gap in between was ≤50% of the entire length of the newly merged CNV (see Supplementary Figure 1 for LRR and BAF plots of all these CNVs). The CNV calling and QC procedures are described in more detail in the Supplementary Information.
Pre-twinning de novo CNV detection
CN calls from the 25 MZ twin pairs who had both parents who passed QC were examined to detect possible pre-twinning de novo CNV events. These segments were identified with a script written in Perl (scripts are available in the Supplementary Material), where segments with the same start and end positions between both twins and both parents, as well as overlapping segments, were compared. If the overlapping segments showed the same CN between twins and a discrepancy with the parental CN calls and the overlap was >100 kb, the overlapping part was included as a de novo CNV segment. In order to judge whether a CNV is inherited or de novo, allele-specific CN information is needed from the parents. Because allele-specific CN calls were not available, the allele-specific CNs were assumed to be as follows: if CN=2, each allele is assumed to have a CN of 1 (1–1), if CN=3, 1–2 is assumed, if CN=4, 2–2 is assumed, if CN=1, 1–0 is assumed, if CN=0, 0–0 is assumed. If possible de novo CNVs were detected, these were tested for confirmation using qPCR (see Supplementary Information for more details on the qPCR replication).
Post-twinning de novo CNV detection
The CN calls, passing the above per sample and per CNV QC thresholds, of the 45 complete MZ twin pairs were analyzed to detect possible post-twinning de novo CNV events. These segments were identified with a program written in Perl (scripts are available in the Supplementary Material), where segments with the same start and end positions between twins, as well as overlapping segments, were compared. If two overlapping segments showed a different CN between twins and a size >100 kb, the overlapping part was identified as a de novo CNV segment. Putative de novo CNVs were tested for confirmation using qPCR (see Supplementary Information for more details on the qPCR replication).
Statistical analysis for genome-wide CNV burden and AP
Genome-wide CNV burden linked to AP was analyzed with permutation tests in Plink32 in the 45 complete twin pairs and 4 unpaired twins. Phenotypes were not permuted between males, females or related individuals, thereby correcting for sex and twin relations. The amount of CNV events, as well as the average size, was tested for association with AP status. This was done for three groups of CNV events with any deviation from the expected CN (CN=0, 1, 3 or 4): CNVs genome-wide, CNVs that overlap with genes and CNVs that do not. Significant results were followed by post-hoc tests, by testing gains (CN=3 or 4), losses (CN=1 or 0), losses of one copy (CN=1), losses of two copies (CN=0), gains of one copy (CN=3) and gains of two copies (CN=4). Inherited as well as de novo CNVs were included in the analysis (de novo CNVs that were not validated by qPCR were removed from the analysis). For the male participants, the CNs of the X and Y chromosomes were transformed by adding one copy to the observed CN, in order to include the sex chromosomes with the autosomes in the permutation analysis (ie, the expected CN of 1 was turned into a CN of 2, like in the autosomes). This transformation was not applied to the pseudoautosomal regions (PARs), because these already have an expected CN of 2.
Results
De novo CNVs
A total of 26 de novo CNV events were identified from the microarray data: 8 pre-twinning and 18 post-twinning CNVs. CNV qPCR targets for 18 regions in the human genome were identified, which would validate all 26 de novo CNVs. The primer- and probe-binding sites for qPCR were selectively chosen in regions within the CNV for which (1) there is no polymorphic SNP, (2) there is no homology to other regions in the genome and (3) there are no common repetitive elements. Based on these criteria, primers and probes could only be selected for 11 of the 18 CNV targets, allowing for testing the validity of 17 of the 26 de novo CNVs using the qPCR method (3 pre-twinning and 14 post-twinning CNVs).
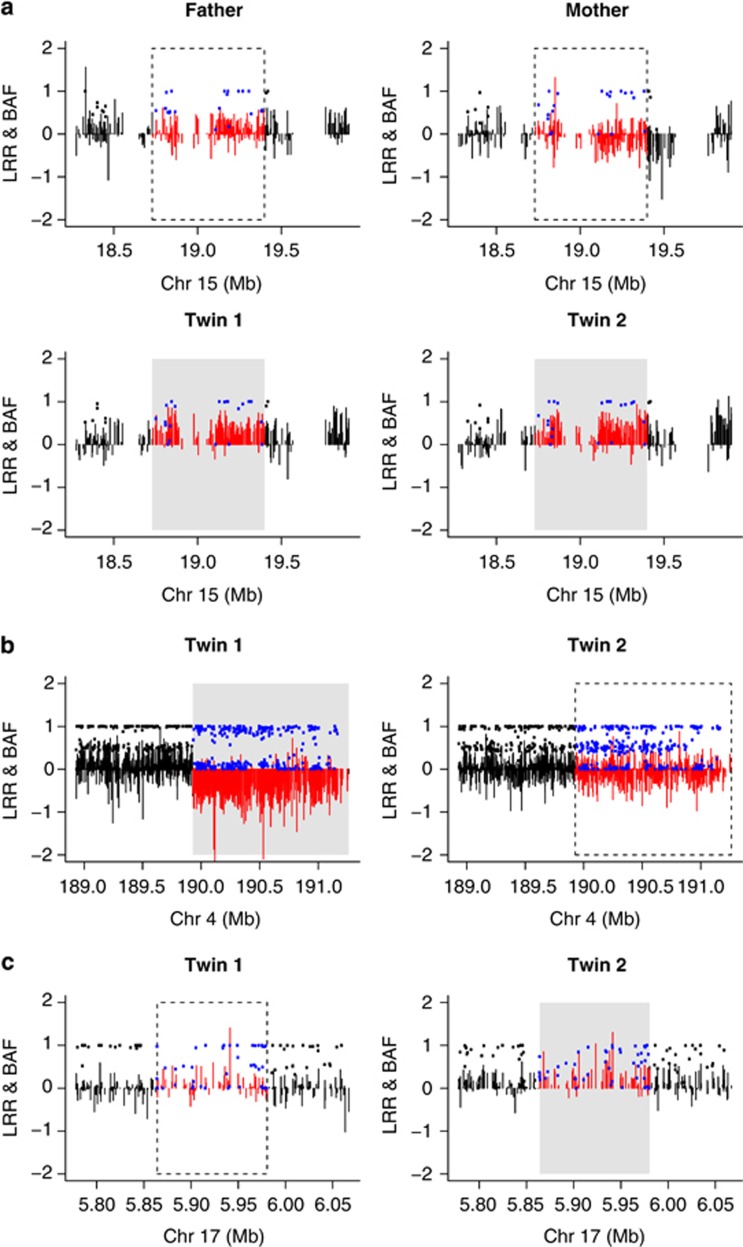
Of the three possible pre-twinning de novo CNVs that could be included in the qPCR replication study, one was validated on chromosome 15q11.2 in a male concordant-unaffected twin pair (see Supplementary Table 1, Figure 1 and Supplementary Figure 2a). In this pedigree, both the microarray and qPCR data show that both parents have a CN of 2 in this region and that both twins have a CN of 3. Of the 16 putative post-twinning de novo CNVs that were included in the qPCR replication study, qPCR experiments validated 1 de novo CNV event, a 1.3-Mb deletion in a male concordant-high twin pair, in the higher scoring co-twin (see Supplementary Table 2, Figure 1b and Supplementary Figure 2b). In addition, a 116-kb duplication was not validated nor rejected by the qPCR experiments in the affected twin of a male discordant pair (see Supplementary Table 2, Figure 1c and Supplementary Figure 2c).
Figure 1.
The pre- and post-twinning de novo CNVs. Each plot shows LRR (vertical bars) and BAF (solid points). The LRR and BAF are shown in color in the region of the CNV (red and blue, respectively) and in black in the flanking regions. The actual deletion/duplication is highlighted by a gray rectangle, whereas a CN call of 2 is highlighted by a dashed rectangle. (a) Depicts the region of the pre-twinning de novo duplication in family 34 for both parents and both twins (both unaffected for AP). The duplication is mainly characterized by an increase in LRR in the twins compared with the parents. The clustering of BAF does not show striking differences between the twins and the parents, most likely because there are relatively few SNP probes in this region (CN probes do not have BAF values). (b) Shows the region of the post-twinning deletion in family 5 for both twins (both affected with AP). The deletion is characterized by a decrease in LRR and an altered clustering of BAF, only seen in twin 1 (the oldest twin). (c) shows the region of the possible post-twinning duplication in family 33 for both twins (discordant), where twin 2 is affected with AP. Although both calling algorithms called a de novo duplication, the LRR and BAF values do not show striking differences when inspected visually, which is why extra qPCR experiments were conducted for this region.
The 1.3-Mb deletion was initially called as two separate CNVs of 848 and 334 kb by Birdsuite and PennCNV. The qPCR targets were designed for both these regions and the gap in between. All the three qPCR experiments resulted in a deletion for the oldest twin, and a CN of 2 for the youngest, confirming that this is indeed one large deletion that was artificially split by the calling algorithms.
Interestingly, qPCR was not able to reject or validate the microarray-supported hypothesis of a 116-kb de novo CNV duplication in the affected twin of a discordant pair on 17p13.2. Despite both the calling algorithms supporting a duplication in this region, the LRR and BAF plots (Figure 1c) were visually ambiguous, so it was decided to add a second qPCR target to this region 30 kb downstream. The qPCR experiments did not unequivocally validate or refute the presence of a duplication in this region (Supplementary Figure 2c). The experiment was repeated three different times for each target assay, with four sample replicates in each experiment. In each instance, the calculated CN for the affected twin was greater than that of unaffected twin (2.34 vs 1.91 and 2.40 vs 1.97 for the chr17:5921845 and chr17:5951803 targets, respectively).
Genes located within each of the de novo CNV regions are summarized in Table 1. Figure 1 shows the LRR and BAF plots and Supplementary Figure 2 displays the qPCR replication data of the de novo CNV regions. In addition, Supplementary Figure 3 places each of these de novo CNVs in a more global context by showing all of the cataloged structural variations from the Database of Genomic Variations (DGV).
Table 1. Genes within each confirmed de novo CNV region. Genes that are in the RefSeq database http://www.ncbi.nlm.nih.gov/gene as well as in the Ensembl database http://www.ensembl.org are reported.
| Gene ID | Description | Tissue expressed (transcripts per million) a | Gene ontology (functioning of gene products) b |
|---|---|---|---|
| chr15: 18 728 578–19 399 146 | |||
| HERC2P3 | Hect domain and RLD 2 pseudogene 3, non-coding RNA | Brain (69); Thyroid (42); Uterus (38); Thymus (36); Testis (24); Pharynx (24); Mammary Gland (13); Stomach (10); Placenta (10); Eye (9); Blood (8); Connective Tissue (6); Prostate (5); Embryonic Tissue (4); Intestine (4); Kidney (4); Liver (4); Skin (4) | Molecular functions: metal ion binding; ubiquitin-protein ligase activity. |
| chr4: 189 928 060–191 261 904 | |||
| HSP90AA4P | Heat shock protein 90kDa alpha (cytosolic), class A member 4, pseudogene, non-coding RNA | Ascites (24); Skin (4) | Cellular components: cytoplasm.Molecular functions: ATP binding; nucleotide binding; unfolded protein binding.Biological processes: protein folding; response to stress. |
| FRG1 | FSHD region gene 1, mRNA | Bone Marrow (143); Pharynx (72); Pituitary Gland (60); Blood (56); Lymph Node (54); Salivary Gland (49); Ovary (48); Parathyroid (48); Eye (47); Muscle (46); Liver (33); Mammary Gland (32); Stomach (31); Uterus (30); Adrenal Gland (30); Embryonic Tissue (27); Lung (26); Prostate (21); Testis (21); Cervix (20); Connective Tissue (20); Pancreas (18); Kidney (14); Skin (14); Bone (13); Heart (11); Intestine (8); Brain (4); Placenta (3) | Cellular components: cajal body; catalytic step 2 spliceosome; nuclear speck; nucleolus; nucleus.Biological processes: nuclear mRNA splicing, via spliceosome; RNA splicing; rRNA processing. |
| TUBB4Q | Tubulin, beta polypeptide 4, member Q, pseudogene, mRNA | Skin (4) | Cellular components: cytoplasm; cytoskeleton; microtubule. Molecular functions: GTP binding; GTPase activity; nucleotide-binding; structural molecule activity Biological processes: ‘de novo' posttranslational protein folding; cellular protein metabolic process; microtubule-based movement; protein folding; protein polymerization. |
| FRG2 | FSHD region gene 2, mRNA | NA | Cellular components: nucleus. |
| DUX4 family | double homeobox 4, mRNA | NA | Cellular components: nucleus. |
| Molecular functions: sequence-specific DNA binding; sequence-specific DNA binding transcription factor activity. | |||
| chr17: 5 864 185–5 980 521 | |||
| WSCD1 | WSC domain containing 1, mRNA | Muscle (120); Umbilical Cord (73); Pituitary Gland (60); Eye (56); Testis (48); Brain (45); Kidney (37); Ovary (29); Thyroid (21); Bone Marrow (20); Vascular (19); Embryonic Tissue (18); Spleen (18); Placenta (17); Lung (14); Mouth (14); Bone (13); Connective Tissue (13); Heart (11); Pancreas (9); Blood (8) | Cellular components: integral to membrane (ie, penetrating at least one phospholipid bilayer of a membrane); membrane (ie, double layer of lipid molecules that encloses all cells, and many organelles; may be a single or double lipid bilayer; also includes associated proteins)Molecular functions: sulphotransferase activity. |
Abbreviations: FSHD, facioscapulohumeral muscular dystrophy; NA, tissue-specific gene expression data not available.
Data from NCBI UniGene http://www.ncbi.nlm.nih.gov/unigene.
Data from the Gene Ontology Project http://www.geneontology.org.
Genome-wide CNV burden and AP
There was a nominally significant association with AP and the average size of CNVs within genes, where the affected individuals had larger CNV events than the unaffected group (>120 kb more on average, P=0.00830, cf. a level of 0.00833 (=0.05/6) maintains a family-wise type-I error of 0.05, Table 2). The post-hoc tests showed that each type of CNV showed the same trend (a larger average CNV size in the affected group, Table 3), except for the CNVs with deletions of two copies (CN=0), which was the least common type, occurring only seven times (five events in affected individuals and two events in unaffected individuals). None of these types showed a significant signal, suggesting that the significant effect of burden is due to the combined effect of both losses and gains. The average size of the CNVs did not differ significantly between affected and unaffected individuals for the regions outside of genes. The number of CNVs also did not show significant differences, both within and outside of genes.
Table 2. Results for permutation tests for the number of CNVs genome-wide and their size vs AP.
| CNV events | Mean number of CNVs – unaffected | Mean number of CNVs – affected | Empirical P-values (number of CNVs vs AP) | Average size of CNVs (kb) – Unaffected | Average size of CNVs (kb) – Affected | Empirical P-values (size of CNVs vs AP) |
|---|---|---|---|---|---|---|
| CNVs genome-wide (CN=0, 1, 3 and 4) | 4.528 | 3.805 | 0.961 | 242.2 | 280.0 | 0.058 |
| CNVs overlapping genes (CN=0, 1, 3 and 4) | 2.566 | 1.854 | 0.989 | 266.6 | 388.7 | 0.008 |
| CNVs outside of genes (CN=0, 1, 3 and 4) | 2.094 | 2.000 | 0.638 | 210.7 | 197.2 | 0.738 |
Abbreviations: AP, Attention Problems; CN, copy number; CNV, copy number variation.
Table 3. Results for post-hoc permutation tests for the size of different types of CNVs genome-wide vs AP.
| Average size of CNVs (kb) - unaffected | Average size of CNVs (kb) – affected | Empirical P-values | |
|---|---|---|---|
| Losses (CN=0 and 1) | 269.0 | 330.0 | 0.198 |
| Deletion: 1 copy (CN=1) | 264.1 | 361.8 | 0.099 |
| Deletion: 2 copies (CN=0) | 356.1 | 266.8 | 0.687 |
| Gains (CN=3 and 4) | 330.0 | 434.5 | 0.104 |
| Duplication: 1 copy (CN=3) | 335.5 | 420.6 | 0.149 |
| Duplication: 2 copies (CN=4) | 193.6 | 441.1 | 0.226 |
Abbreviations: AP, Attention Problems; CN, copy number; CNV, copy number variation.
Discussion
This study investigated the importance of the number and size of CNVs for AP in ‘identical' twins. The presence of de novo CNV mutations and effects of genome-wide CNV burden were examined.
The pre-twinning de novo CNVs were examined for a subset of the sample (25 twin pairs) that had genomic DNA from both parents available and who passed QC. One pre-twinning de novo CNV mutation was detected that resulted in both MZ twins having a duplication (CN=3) on chromosome 15q11.2. This region contains the gene HERC2P3, which is expressed in the human brain (Table 1). However, both individuals in this twin set scored in the normal range for AP. We assume this to be a de novo pre-twinning CNV event, but we recognize the possibility of a rare condition that one of the parents carries two copies for one allele and zero copies on the other allele, in which case this would not be a de novo CNV event.
A post-twinning de novo deletion of ∼1.3 Mb on 4q35.2 was confirmed with three qPCR experiments in a concordant-affected twin pair. The twin with the deletion had a higher AP score, 20% lower birth weight than the co-twin, scored in the clinical range for the DSM-oriented CBCL scale for conduct problems and performed worse at school according to longitudinal parental and teacher reports. The 4q35.2 subtelomeric deletions found in this twin have been suggested to contribute to co-morbid psychiatric illness and mental retardation.33 The deletion contains the FRG1 gene, which is expressed in the human brain. In addition, chromosome 4q35 contains a polymorphic D4Z4 macrosatellite repeat, consisting of 10–100 tandem 3.3-kb D4Z4 repeats. An identical copy of the DUX4 gene (double homeobox) is located in each of the 3.3-kb repeat elements. Contractions in this polymorphic region have been implicated in facioscapulohumeral muscular dystrophy (FSHD).34 The DUX4 protein has been shown to function as a transcriptional activator of the paired-like homeodomain transcription factor 1 (PITX1),35 which is expressed in the pituitary gland and brain. DUX4 is a nuclear protein also capable of acting as a pro-apoptotic protein, inducing cell death through caspase 3/7 activity when overexpressed.36 Although FRG1 and DUX4 have been highly implicated in the pathophysiology of FSHD, our findings and the molecular mechanisms of these proteins make them possible targets for follow-up study on how they may have an impact on the developing brain.
The microarray supported hypothesis of a 116-kb duplication on 17p13.2 in the affected twin of a discordant pair could not be validated or rejected using qPCR (Supplementary Figure 2c). The algorithm for predicting CN is based on the delta CT of the reference target (in this case RNaseP) to the CNV target of interest. Although experimental variation can affect the calculated CN of the genomic DNA in a qPCR experiment (eg, technical reproducibility, genomic DNA quality, and so on),37, 38 in all instances (12 replicates for two assay targets) the affected twin had a larger calculated CN for this region of the genome. Considering that the genomic DNA was normalized and the fact that these samples are MZ twins makes interpretation of the data difficult. We hypothesize that the duplication in 17p13.2 is a somatic mutation resulting in mosaicism of the affected twin. Somatic mosaicism is generally defined as the presence of genetically distinct populations of cells for a given tissue in the same organism. It has been suggested that somatic mosaicism in pathogenic genes may be relatively common.25 We cannot conclusively determine this hypothesis, but it was only possible to detect/suspect this by examining MZ twin pairs. Regions in 17p13.2 have been associated with autism spectrum disorder.39, 40, 41 The WSCD1 gene from the duplication in 17p13.2 in the affected twin of the discordant pair is expressed in the brain and is involved in the phospholipid bilayer of the membrane (Table 1), which has been suggested to have a major role in the high degree of comorbidity between ADHD, dyspraxia and autism spectrum disorders,42 which have all been reported by the parents and teachers of the carrier of the putative de novo duplication. The unaffected co-twin had an above-average IQ and had no health or other problems reported.
Each of the de novo CNVs identified in this study has been compared with the catalog of structural variants from the DGV (Supplementary Figure 3). There have been several duplications and deletions reported for the pre-twinning de novo CNV on 15q11.2 and the post-twinning deletion on 4q35.2. Interestingly, a slightly larger deletion of 4q35.2 was identified from the Vrije University Hospital clinical database in a child with autism, ADHD and developmental delay without dysmorphism (Petra Zwijnenburg, personal communication). There have not been any duplications reported in the Database for Genomic Variation for the putative de novo CNV in the affected twin of a discordant pair on 17p13.2.
The CNVs that were not identified as de novo were assumed to be inherited and were included with the de novo CNVs in the genome-wide CNV burden association analysis. The association analysis of genome-wide CNV burden and AP showed that CNVs that overlap with genes were larger in size in affected than in unaffected subjects (P=0.008). Deletions and duplications showed the same trend, but no significant signals, indicating that both contributed to the main effect. The CNVs that were larger in subjects with high AP scores were scattered across the genome. This suggests that AP might be influenced by many CNVs with small effects, which has been recently revealed to be the case for SNP effects on complex traits as well.43 Because the majority of human genes are expressed in the cortex,44 randomly located CNVs affecting genes are likely to have an effect on highly heritable cognitive traits, such as AP. An alternative hypothesis is that neuropsychiatric disorders are caused by rare and highly penetrant CNVs, which often disrupt the balance of dosage-sensitive genes.13, 45, 46 Studying the genes affected by this disruption may provide important insights into the susceptibility of disease.
Rare events, such as de novo CNVs, are hard to detect when the tools used to measure them are relatively noisy, as is the case with CNV signals from microarray chips that are currently available. In this study, this could be especially problematic when trying to detect post-twinning de novo CNVs by comparing twin pairs that were genotyped on separate plates. Stringent QC procedures might not be enough to distinguish real signal from noise, which made replication with qPCR a necessary step to validate the presence of these apparent mutations. In order to accurately detect de novo CNVs, it is important to confirm the mutation using a molecular assay more sensitive to CN alterations than the microarrays used to initially screen for them. qPCR has been shown to be highly effective in the validation of CNVs from microarray data.26, 47, 48 The outcome of this study shows that even when only considering large CNVs (>100 kb), there can still be a substantial amount of false positives among the few CN differences between the MZ twins, reflecting the difficulty in measuring CNVs accurately. We excluded the source DNA (buccal-derived) as a major factor. In a different sample of twin families in which blood- and buccal-derived DNAs were collected, we have shown that the CNV calls between blood and buccal sources did not show a greater discordance than those from the same source (eg, both samples from blood), indicating that buccal-derived DNA is suitable for the microarray chip used in the present study (Paul Scheet and Erik A Ehli et al, unpublished data). The validated de novo CNV, however, confirms that MZ twins are not always 100% genetically identical and that these differences are detectable. An important question remains: how common are these post-twinning de novo mutations? To answer this question in more detail, high-throughput CNV-calling methods are needed with higher resolutions and accuracy than the microarray chips currently available. Most heritability studies rely on the assumption that MZ twins are 100% identical.49, 50 Our study largely supports this assumption, but also suggests that the rare post-twinning de novo events may lead to phenotypic discrepancies. As a result, the classical twin design may slightly underestimate the genetic effects of a trait. If CNV discordance between MZ twins contributes to phenotypic discordance, the CNV effect on the phenotype would be inadvertently attributed to unique environmental effects in a classical twin study design.
In conclusion, this study found that CNVs that overlap with genes tend to be larger in individuals that consistently score high on AP and who may also have associated elevations in other behavioral problems. Also, two de novo CNVs were detected: a pre-twinning duplication and a post-twinning deletion that resulted in a discordance in CN between the MZ twins. Replication studies with larger sample sizes are needed to validate the effect of the size of CNVs on AP and to investigate the effects of the regions where the de novo CNVs were found.
Acknowledgments
Funding was obtained from the Netherlands Organization for Scientific Research (MagW grants 480-04-004; 463-06-001; ZonMW 91210020); Spinozapremie (56-464-14192), Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL), the VU University's Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); the European Science Council (ERC Advanced, 230374), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), and the National Institutes of Health (NIMH, RO1 MH58799-03). Genotyping and analyses were funded by the NIMH Grand Opportunity grant 1RC2 MH089995-01. AA was supported by CSMB/NCA. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003), the Dutch Brain Foundation and the department of psychology and education of the VU University Amsterdam. We wish to thank all participating twin families. We would also like thank Petra Zwijnenburg from the Department of Pediatrics at the Vrije University Hospital for her time discussing CNVs in the Hospital clinical databases that were relevant to this study.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Kidd JM, Cooper GM, Donahue WF, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- van Ommen GJB. Frequency of new copy number variation in humans. Nat Genet. 2005;37:333–334. doi: 10.1038/ng0405-333. [DOI] [PubMed] [Google Scholar]
- Aldred PMR, Hollox EJ, Armour JAL. Copy number polymorphism and expression level variation of the human -defensin genes DEFA1 and DEFA3. Hum Mol Genet. 2005;14:2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Armour JAL, Barber JCK. Extensive normal copy number variation of a [beta]-defensin antimicrobial-gene cluster. Am J Hum Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzmeier RM, Ganz T. Human defensin gene copy number polymorphisms: Comprehensive analysis of independent variation in [alpha]-and [beta]-defensin regions at 8p22-p23. Genomics. 2005;86:423–430. doi: 10.1016/j.ygeno.2005.06.003. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2005;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Rotger M, Saumoy M, Zhang K, et al. Partial deletion of CYP2B6 owing to unequal crossover with CYP2B7. Pharmacogenet Genom. 2007;17:885–890. doi: 10.1097/FPC.0b013e3282ef5cd1. [DOI] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Cook EH, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- Merikangas AK, Corvin AP, Gallagher L. Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends Genet. 2009;25:536–544. doi: 10.1016/j.tig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Cubells JF. Copy number variants: a new molecular frontier in clinical psychiatry. Curr Psychiatry Rep. 2011;13:129–137. doi: 10.1007/s11920-011-0183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Zaharieva I, Martin A, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks EM, Hudziak JJ, Dolan CV, Ferdinand RF, Boomsma DI. The relations between DISC-IV DSM diagnoses of ADHD and multi-informant CBCL-AP syndrome scores. Compr Psychiatry. 2006;47:116–122. doi: 10.1016/j.comppsych.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Perlis RH, Doyle AE, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Derks EM, Hudziak JJ, Boomsma DI. Genetics of adhd, hyperactivity, and attention problems. In Handbook of behavior genetics. New York: Springer Verlag; 2009. [Google Scholar]
- Derks EM, Hudziak JJ, Dolan CV, van Beijsterveldt TCEM, Verhulst FC, Boomsma DI. Genetic and environmental influences on the relation between attention problems and attention deficit hyperactivity disorder. Behav Genet. 2008;38:11–23. doi: 10.1007/s10519-007-9178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Ent D, Lehn H, Derks EM, et al. A structural MRI study in monozygotic twins concordant or discordant for attention/hyperactivity problems: evidence for genetic and environmental heterogeneity in the developing brain. Neuroimage. 2007;35:1004–1020. doi: 10.1016/j.neuroimage.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Ent D, van Beijsterveldt CE, Derks EM, et al. Neuroimaging of response interference in twins concordant or discordant for inattention and hyperactivity symptoms. Neuroscience. 2009;164:16–29. doi: 10.1016/j.neuroscience.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehn H, Derks EM, Hudziak JJ, Heutink P, van Beijsterveldt T, Boomsma DI. Attention problems and attention-deficit/hyperactivity disorder in discordant and concordant monozygotic twins: evidence of environmental mediators. J Am Acad Child Adolesc Psychiatry. 2007;46:83–91. doi: 10.1097/01.chi.0000242244.00174.d9. [DOI] [PubMed] [Google Scholar]
- Vadlamudi L, Dibbens LM, Lawrence KM, et al. Timing of de novo mutagenesis--a twin study of sodium-channel mutations. N Engl J Med. 2010;363:1335–1340. doi: 10.1056/NEJMoa0910752. [DOI] [PubMed] [Google Scholar]
- Bruder CEG, Piotrowski A, Gijsbers AACJ, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Imamura A, Tasaki S, et al. Failure to Confirm CNVs as of Aetiological Significance in Twin Pairs Discordant for Schizophrenia. Twin Res Hum Genet. 2010;13:455–460. doi: 10.1375/twin.13.5.455. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, de Geus EJC, Vink JM, et al. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- Achenbach T. Manual for the CBCL/4-18 and 1991 Profile. Burlington: University of Vermont, Department of Psychiatry; 1991. [Google Scholar]
- Willemsen G, de Geus EJC, Bartels M, et al. The Netherlands Twin Register Biobank: a resource for genetic epidemiological studies. Twin Res Hum Genet. 2010;13:231–245. doi: 10.1375/twin.13.3.231. [DOI] [PubMed] [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard B, Hollox E, Malloy MP, et al. A 4q35. 2 subtelomeric deletion identified in a screen of patients with co-morbid psychiatric illness and mental retardation. BMC Med Genet. 2004;5:21. doi: 10.1186/1471-2350-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutekom JCTV, Wljmenga C. Tlenhoven EAEV et al: FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2:2037. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- Dixit M, Ansseau E, Tassin A, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA. 2007;104:18157. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaljow V, Marcowycz A, Ansseau E, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Cukier HN, Pericak-Vance MA, Gilbert JR, Hedges DJ. Sample degradation leads to false-positive copy number variation calls in multiplex real-time polymerase chain reaction assays. Anal Biochem. 2009;386:288–290. doi: 10.1016/j.ab.2008.11.040. [DOI] [PubMed] [Google Scholar]
- Fernandez-Jimenez N, Castellanos-Rubio A, Plaza-Izurieta L, et al. Accuracy in copy number calling by qpcr and prt: a matter of dna. PloS One. 2011;6:e28910. doi: 10.1371/journal.pone.0028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer A, Giacobini MB, Eriksson M, et al. Copy number variation characteristics in subpopulations of patients with autism spectrum disorders. Am J Med Genet. 2010;156:115–124. doi: 10.1002/ajmg.b.31142. [DOI] [PubMed] [Google Scholar]
- Cannon DS, Miller JS, Robison RJ, et al. Genome-wide linkage analyses of two repetitive behavior phenotypes in Utah pedigrees with autism spectrum disorders. Mol Aut. 2010;1:1–13. doi: 10.1186/2040-2392-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joober R, El-Husseini A. Synaptic abnormalities and candidate genes in autism. In molecular mechanisms of synaptogenesis. New York: Springer Verlag; 2006. [Google Scholar]
- Richardson AJ, Ross M. Fatty acid metabolism in neurodevelopmental disorder: a new perspective on associations between attention-deficit/hyperactivity disorder, dyslexia, dyspraxia and the autistic spectrum. Prostaglandins Leukot Essent Fatty Acids. 2000;63:1–9. doi: 10.1054/plef.2000.0184. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Inoue K, Lupski JR. Genetics and genomics of behavioral and psychiatric disorders. Curr Opin Genet Dev. 2003;13:303–309. doi: 10.1016/s0959-437x(03)00057-1. [DOI] [PubMed] [Google Scholar]
- Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Weaver S, Dube S, Mir A, et al. Taking qPCR to a higher level: Analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods. 2010;50:271–276. doi: 10.1016/j.ymeth.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Zhang D, Qian Y, Akula N, et al. Accuracy of CNV Detection from GWAS Data. PloS One. 2011;6:e14511. doi: 10.1371/journal.pone.0014511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3:872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- Plomin R, DeFries J, McClearn G, McGuffin P. Behavioral genetics. Vol. 3. New York: Worth; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.