Abstract
Both the standardized uptake value ratio (SUVR) and the Logan plot result in biased distribution volume ratios (DVR) in ligand-receptor dynamic PET studies. The objective of this study is to use a recently developed relative equilibrium-based graphical plot (RE plot) method to improve and simplify the two commonly used methods for quantification of [11C]PiB PET.
Methods
The overestimation of DVR in SUVR was analyzed theoretically using the Logan and the RE plots. A bias-corrected SUVR (bcSUVR) was derived from the RE plot. Seventy-eight [11C]PiB dynamic PET scans (66 from controls and 12 from mildly cognitively impaired participants (MCI) from the Baltimore Longitudinal Study of Aging (BLSA)) were acquired over 90 minutes. Regions of interest (ROIs) were defined on coregistered MRIs. Both the ROI and pixelwise time activity curves (TACs) were used to evaluate the estimates of DVR. DVRs obtained using the Logan plot applied to ROI TACs were used as a reference for comparison of DVR estimates.
Results
Results from the theoretical analysis were confirmed by human studies. ROI estimates from the RE plot and the bcSUVR were nearly identical to those from the Logan plot with ROI TACs. In contrast, ROI estimates from DVR images in frontal, temporal, parietal, cingulate regions, and the striatum were underestimated by the Logan plot (controls 4 – 12%; MCI 9 – 16%) and overestimated by the SUVR (controls 8 – 16%; MCI 16 – 24%). This bias was higher in the MCI group than in controls (p < 0.01) but was not present when data were analyzed using either the RE plot or the bcSUVR.
Conclusion
The RE plot improves pixel-wise quantification of [11C]PiB dynamic PET compared to the conventional Logan plot. The bcSUVR results in lower bias and higher consistency of DVR estimates compared to SUVR. The RE plot and the bcSUVR are practical quantitative approaches that improve the analysis of [11C]PiB studies.
Keywords: RE plot, [11C]PiB, PET, SUVR, Bias
[11C]PiB is one of the most widely used compounds for in vivo detection of fibrillar β-amyloid deposition in brain (1). The spatial distribution of amyloid deposition in Alzheimer’s disease, Mild Cognitive Impairment (MCI) and cognitively normal adults has been well characterized (2–5). The distribution volume ratio (DVR) or binding potential (BPND) (=DVR-1) is usually used as an index of [11C]PiB specific binding to tissue for quantitative measurement of β-amyloid deposition (6). In ligand-receptor dynamic PET studies, DVR can be estimated by fitting a kinetic model to the tissue kinetics with a metabolite-corrected plasma input function (7, 8), where the tracer kinetic model can be a classical compartment model or a model independent graphical analysis method, and the plasma input function is the tracer radioactivity in plasma, usually measured by serial arterial blood sampling. However, quantitative PET without arterial blood sampling is more practical and various reference tissue models have been evaluated for noninvasive quantification of [11C]PiB dynamic PET (9–14). Importantly, in these studies, the target to reference (cerebellum) tissue tracer concentration ratio was shown to achieve a constant after ~50 min post tracer injection (12, 13) which lends itself to pixel-wise analysis using simplified, non-compartmental approaches (14). Many [11C]PiB studies have used standardized uptake value ratio (SUVR) with a reference tissue because of this method’s simplicity and short scan time, where the SUVR is calculated as target to reference tissue tracer concentration ratio over scan time [T0 T1].
While there are variations in the selection of time windows [T0 T1] for SUVR measurements (15), the main disadvantage of the SUVR is the positive bias resulting in overestimation of DVR (16, 17). In contrast, a commonly used Logan plot demonstrates inherent negative bias which is attributed to noise and the target tissue tracer concentration (18–22). The linearized methods, particularly multilinear reference tissue model (MRTM) (23) provide similar estimates of DVR as a simplified reference tissue model (SRTM) (9) yet with much higher computational efficacy. At the pixel level, PET measurements have high noise and thus underestimation of DVR occurs with both graphical (19) and linearized methods (10, 18, 24). While all of these approaches are associated with bias, bias consistency across clinical groups has not been extensively explored in [11C]PiB studies although bias may be one of the limiting factors for the application of the simplified quantitative methods to this tracer.
A novel, noninvasive and computationally efficient graphical approach, the relative equilibrium-based graphical plot (RE plot), has been recently developed for analysis of tracer kinetics with equilibrium relative to input function (25). For reference tissue input function, a target tissue tracer kinetics attain a relative equilibrium if there is a t* such that the ratio of the target to reference tissue tracer concentration is constant for t ≥ t*. In this study, the RE plot with reference tissue input was used 1) to analyze the overestimation of DVR in the SUVR measurements; 2) to develop a practical method that allows correction of the overestimation bias in the SUVR; and 3) to generate DVR images that avoid the noise-induced underestimation in the estimates from the Logan plot.
MATERIALS AND METHODS
Analysis and Correction of Overestimation of DVR in SUVR Using the RE Plot with Reference Tissue Input
The inconsistent overestimation of DVR in SUVR was first demonstrated by using the Logan plot with plasma input (supplemental data online: Analysis of Overestimation of DVR in SUVR Using the Logan Plot with Plasma Input). In this section, the RE plot with reference tissue input was used to analyze the overestimation of DVR in SUVR. Let C(t) and CREF(t) be the tracer concentrations at time t in target and reference tissues, respectively. In this study, we assume that there is a time t* such that 1) the tissue kinetics attain equilibrium relative to the reference tissue input, i.e., C(t)/CREF(t) is a constant for t ≥ t*; 2) reference tissue input CREF(t) can be approximated by one exponential as CREF(t) = αeβt for t ≥ t*. For tracer kinetics that attain relative equilibrium, the RE plot with reference tissue input (25) described by Eq. 1 has been proposed to simplify and improve the generation of DVR images using the conventional Logan plot with reference tissue input.
| Eq. 1 |
where DVR is the slope of the linear portion of the plots for t ≥ t* and θ is the y-intercept of the linear regression. It has been shown that the RE plot is an unbiased graphical analysis method for estimation of DVR with no noise-induced underestimation of the DVR estimates (25).
By taking derivative of both sides of Eq. 1 in bilinear form and then dividing by CREF(t), we have
| Eq. 2 |
where C′(t) is the derivative of C(t). Note that C(t)/CREF(t) is a constant which is defined as SUVR, and C′(t)/CREF(t) = β for t ≥ t*. Therefore, the relationship between SUVR and DVR is expressed by Eq. 3:
| Eq. 3 |
For the tracer delivered by bolus injection, β is negative. The θ in the RE plot is also negative and its absolute value is positively correlated with the DVR (25). Therefore, the bias term θβ in Eq. 3 is positive, and the overestimation of DVR in SUVR increases as DVR increases.
Eq. 3 shows a linear relationship between θ and SUVR at given β. We can rewrite the linear relationship between θ and SUVR as:
| Eq. 4 |
where λ and μ are constants across all subjects. Based on Eq. 4, the bias-corrected SUVR of estimate of DVR can be calculated usingλ, μ, and β as Eq. 5 below:
| Eq. 5 |
In Eq. 5, β was estimated by fitting one exponential to the reference tissue input CREF(t) for t ≥ t* using linear regression of time t (independent regression variable) versus the natural logarithm of CREF(t) (dependent regression variable) for each dynamic PET scan.
For the ROI based bias correction of SUVR, the ROI-specific λ and μ were estimated by linear regression using Eq. 4 with θ and SUVR from all subjects by assuming that Eq. 4 holds for the whole population for a given ROI. The ROI bcSUVR for each subject was then calculated using Eq. 5 with the ROI-specific λ and μ from the population, and β from the subject’s CREF(t).
For pixel-wise bias correction of SUVR, Eq. 6 below was used to generate bcSUVR images for each subject:
| Eq. 6 |
where and , the ROI specific λi andμi are constants for all subjects obtained as described above for ROI based bias correction, and M is the sample size of the population including both controls and MCI subjects. As in Eq. 5, β was estimated from reference tissue input function CREF(t) (t ≥ t*) for each subject. The simplified pixel-wise bias-correction of SUVR method is based on the following observations: 1) low variation of ROI λs (see Table 1 in the Results section); 2) mean(bcSUVR) = mean(SUVR) − λm mean(βSUVR) − μm mean(β), where the bias correction term μm mean(β) remains constant in control and patient groups because the mean(β) in each group is usually the same, i.e., μm β does not contribute to the correction of inconsistent overestimation of DVR in SUVR. In other words, μm could be biased, but the μm induced bias to bcSUVR contributed to each group is the same.
Table 1.
The statistics of linear regression θ = λSUVR + μ (Eq. 4)
| ROI | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Slope λ | Mean | −38.93 | −37.65 | −38.35 | −41.77 | −42.22 | −38.65 | −38.18 | −38.68 | −42.69 | −39.41 | −40.63 | −39.62 | −45.63 | −37.16 |
| SE | 1.08 | 1.13 | 2.02 | 1.02 | 2.17 | 1.12 | 0.94 | 0.93 | 1.83 | 1.17 | 1.15 | 1.08 | 3.29 | 4.84 | |
| Intercept μ | Mean | 37.26 | 42.38 | 52.73 | 34.84 | 35.52 | 28.44 | 29.00 | 31.18 | 37.27 | 30.22 | 31.69 | 34.49 | 54.64 | 6.28 |
| SE | 1.66 | 1.78 | 2.74 | 1.47 | 2.56 | 1.67 | 1.37 | 1.47 | 2.57 | 1.76 | 2.02 | 1.93 | 6.25 | 9.37 | |
| R-square | 0.94 | 0.94 | 0.83 | 0.96 | 0.83 | 0.94 | 0.96 | 0.96 | 0.88 | 0.94 | 0.94 | 0.95 | 0.72 | 0.44 | |
Regions of interests (ROIs) are numbered as: 1: caudate, 2: putamen, 3: thalamus, 4: lateral temporal, 5: mesial temporal, 6: orbital frontal, 7: prefrontal, 8: superior frontal, 9: occipital, 10: parietal, 11: anterior cingulate, 12: posterior cingulate, 13: pons, 14: white matter. The mean ± SD over all ROIs for λ and μ are −39.97 ± 2.36 and 34.71± 11.49, respectively. The statistical p values of t-test for the difference between the mean of λ (=−39.97) and the λ of each ROI are [0.34 0.04 0.43 0.08 0.30 0.24 0.06 0.17 0.14 0.63 0.57 0.75 0.09 0.56] for the ROIs [1 2 3 4 5 6 7 8 9 10 11 12 13 14].
Human [11C]PiB Dynamic PET
Study Participants
Seventy-eight [11C]PiB dynamic PET studies of nondemented older participants in the neuroimaging substudy of the Baltimore Longitudinal Study on Aging (BLSA) (26) were acquired. In conjunction with each [11C]PiB study, all participants received a detailed physical examination, including medical history updates and laboratory screening, neuropsychological testing, and assessment by the Clinical Dementia Rating (CDR) (27) scale. The CDR scores were typically based on informant (spouse, child, or close friend) interviews conducted by a certified examiner. Cognitive status was determined by consensus diagnosis according to established procedures (28, 29). A diagnosis of mild cognitive impairment (MCI) was made in participants who had cognitive impairment (typically memory) but did not have functional loss in activities of daily living. 66 scans from individuals with CDR = 0 (age range at baseline scan 55.7 – 92.1 years, mean 77.6 ± 6.9) were classified as the normal control group, and 12 scans from individuals with CDR = 0.5 (age range at baseline scan 77.2 – 89.5 years, mean 83.5 ± 4.3) who were classified as the MCI group. In this study, we consider CDR = 0.5 as a very mild cognitively impaired group but only 3 individuals actually met consensus criteria for MCI.
[11C]PiB Dynamic PET
All dynamic PET studies were performed on a GE Advance scanner. The PET scanning was started immediately after intravenous bolus tracer injection of 543.5 ± 29.7 MBq (range 442.9 –605.3 MBq) with high specific activity of 236.6 ± 145.4 MBq/μmol (range 36.1 – 1005.4 GBq/μmol) at the time of injection of [11C]PiB. There were no significant differences in [11C]PiB dose or specific activity among cognitively impaired and cognitively normal older adults (p > 0.05). Dynamic PET data were collected in 3-D acquisition mode according to the following protocol: 4×0.25, 8×0.5, 9×1, 2×3, 14×5 min (90 min total, 37 frames). To minimize head motion during PET scanning, all participants were fitted with thermoplastic face masks for the PET imaging. The reconstruction of dynamic PET images was described in our previous studies (10).
Structural MRI and ROI Definition
Structural magnetic resonance images (MRIs) for anatomic localization were performed on a GE Signa 1.5 Tesla or Phillips Intera 3.0 Tesla MRI scanner using a spoiled gradient recalled acquisition sequence (124 slices with image matrix 256×256, pixel size 0.94×0.94 mm2, slice thickness 1.5 mm). MRIs were typically performed on the same visit, but a major renovation of the MRI research scanners coincided with [11C]PiB imaging studies. While 61 MRI scans were acquired at the time of PET scan, 15 were obtained within 1.8 (SD 0.8; range 1 – 4.1) years of PET scan. Two additional MRI scans were acquired 10.5 and 11.8 years prior to PET scans due to medical issues that prevented concurrent MRI assessments. MRIs were co-registered to the mean of the first 20 min dynamic PET images using SPM2 (Statistical Parametric Mapping software; Wellcome Department of Cognitive Neurology, London, UK) with the mutual information method. In addition to the reference region (cerebellar cortex), 14 ROIs were manually drawn on the coregistered MRIs (10, 12, 13), and copied to the dynamic PET images to obtain ROI TACs for kinetic analysis. The ROIs were also applied directly to parametric images for estimation of DVRs.
Evaluation of the RE Plot and Parameter Estimation
The evaluation of assumptions for the RE Plot was provided in supplemental data online. Single t* value used in the RE and Logan plots, and SUVR (supplemental data online: SUVR Calculation) for all subjects was determined at 52.5 min post tracer injection corresponding to the last 8 time frames from 50 to 90 minutes of dynamic scans. DVRs from the RE plot, the Logan plot with reference tissue input (10, 11, 25) (hereafter the Logan plot) (supplemental data online: The Logan Plot with Reference Tissue Input) and the SUVR were calculated from ROI kinetics. Subsequently, parametric images from pixel-wise kinetics were obtained using the RE and Logan plots, the SUVR, and bcSUVR, and the SRTM. The ROIs from the parametric images were then compared with the DVRs from ROI kinetics. The estimates of DVR from the low noise levels of ROI TACs were used as a reference in this study. Mean parametric images for the control and the MCI groups were then generated using SPM2. The spatial normalization parameters determined by the mean images of the first 20-min of dynamic PET scans were applied to all parametric images (10).
All parameter estimation methods for ROI kinetic analysis and parametric image generation were written in MATLAB (The MathWorks Inc.) and implemented on a Dell PWS690 workstation.
Evaluation of Assumptions for Bias Corrected SUVR
The values of β obtained by fitting one-exponential to the reference tissue input CREF(t) for t ≥ t* =52.5 min were compared between patient and control groups using statistical t test.
To evaluate the assumption that CREF(t) can be approximated by one exponential as CREF(t) = αeβt for t ≥ t* = 52.5 min, CREF(t) was fitted not only by one exponential but also by multi-exponentials as for t ≥ t* = 52.5, where α0 is a constant, n is the number of exponentials, along with {(αi, βi) | 1≤ i ≤ n} were estimated simultaneously by spectral analysis (30–32). To compare the goodness of fit between one-exponential and multi-exponential model fitting, Akaike information criterion (AIC) (16, 33, 34) was calculated. A model associated with the lower AIC is considered the better fitting of the model.
The assumption of the linear relationship between θ and SUVR was evaluated by applying linear regression of θ (y-variable) versus SUVR (x-variable) for each ROI TACs over all subjects, where θ was estimated by the RE plot with t* = 52.5 min.
RESULTS
Validation of Assumptions for Bias Corrected SUVR
We verified the two assumptions used to derive the bias corrected SUVR from the RE plot. The first assumption is that one exponential clearance after t* for reference tissue (cerebellum) kinetics occurs. The cerebellum TACs were well-fitted by an exponential function at t ≥ t* = 52.5 min. The R2 of the linear regression of time t versus the natural logarithm of cerebellum TACs were 0.790 ± 0.196 and 0.867 ± 0.071 for controls and MCI group, respectively. The β values, the one-exponential clearance rates of tracer concentration in cerebellum, were 0.007 ± 0.003 min−1 in controls and 0.008 ± 0.002 in the MCI group. They were not significantly different between these two groups (p = 0.33). The value of Akaike information criterion calculated from one-exponential fitting (−87.47 ± 6.34) was significantly lower than that obtained from multi-exponential fitting (−85.88 ± 6.98; paired t-test, p = 0.009), and the results demonstrate that one-exponential model better predicts or fits reference tissue input CREF(t) for t ≥ t* = 52.5 min.
The second assumption is that there is a linear relationship between SUVR and θ. As shown for a representative ROI, the posterior cingulate cortex, there was a highly linear relationship between SUVR and bias correction factor θ where absolute value of the correction factor θ increases as SUVR increases (supplemental data online, Figure 3). The values for λ and μ, i.e. the slope and the intercept of the linear regression of θ versus SUVR, for all 14 ROIs are listed together with R2 values in Table 1. The percent coefficient of variation (= 100(standard deviation/mean)) of λ was 5.6%. Due to this low variation of λs in different ROIs, there was no significant difference between mean λ and ROI specific λ for all ROIs (p: 0.33 ± 0.23, 0.06 to 0.75, n = 13) except putamen (p = 0.04).
Bias and Bias Correction of SUVR in ROI Kinetics
Inconsistent overestimation of the DVR in SUVR between controls and MCIs is illustrated in Figure 1A. The SUVR overestimation in this figure is calculated as Bias% relative to the DVR estimated from ROI TACs using the Logan plot. The overestimation of DVRs in SUVR ranges from 16 to 32% in the MCI group and is significantly higher (on average 1.73 times higher) than in controls in all but 4 ROIs (mesial temporal, occipital, pons, and white matter).
FIGURE 1.
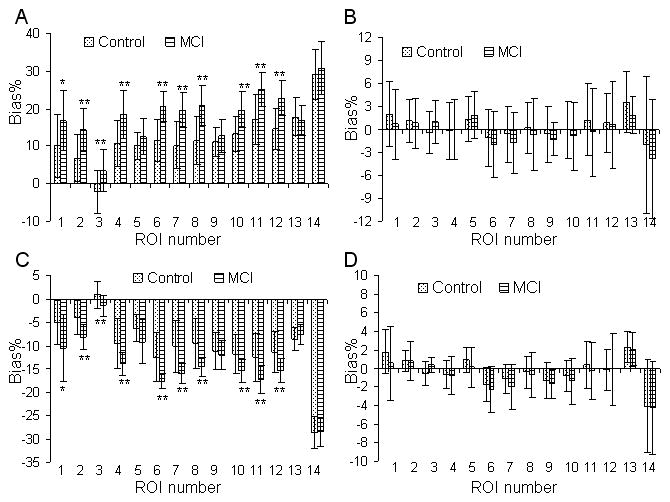
Variability of Bias% in ROI DVR estimates relative to the DVRs from the Logan plot with reference tissue input and ROI TACs between mild cognitive impairment (MCI) and control groups: (A) SUVR, (B) bias-corrected SUVR (bcSUVR), (C) DVR from parametric images generated by the Logan plot, and (D) DVR from the relative equilibrium (RE) plot. The Bias% = 100(DVR/DVR(Logan plot with ROI TACs)-1). Regions of interest (ROIs) are numbered as: 1: caudate, 2: putamen, 3: thalamus, 4: lateral temporal, 5: mesial temporal, 6: orbital frontal, 7: prefrontal, 8: superior frontal, 9: occipital, 10: parietal, 11: anterior cingulate, 12: posterior cingulate, 13: pons, 14: white matter. For the RE plot, the DVRs from ROI TACs are identical to those from DVR images. Note that while group differences in Bias% are detected for SUVR and DVR images generated by Logan plot from high noise pixel TACs, bias-corrected SUVR (bcSUVR) and DVRs from the RE plot do not show any group differences. *: 0.02 ≤ p≤0.05, **: p < 0.01.
The bcSUVR was developed to address the inconsistency bias of the SUVR. Figure 1B shows that this method is associated with minimal bias (Bias% ≤ 2.0% for all ROIs except 3.6% for pons) relative to the DVRs estimated from ROI TACs using the Logan plot, and that there is no difference between the MCI group and control group in Bias% (statistical p value: 0.46 ± 0.28, range 0.08 to 0.94). Figures 2A and 2B show that the linear relationship between the SUVR and DVR from ROI TACs using the Logan plot improved after using the bcSUVR as the slope of the regression line changed from 1.37 to 1.02 and R2 increased from 0.94 to 0.96.
FIGURE 2.
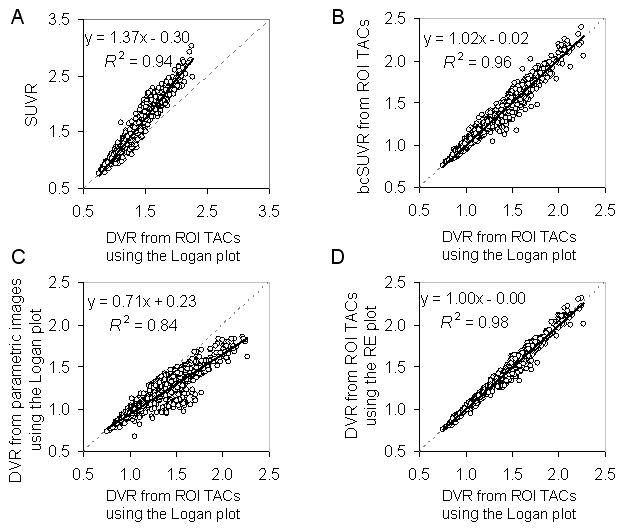
Linear correlations between the DVR estimates from the ROI TACs using the Logan plot and (A) SUVR, (B) bias-corrected SUVR (bcSUVR), (C) DVR from parametric images generated by the Logan plot, (D) DVRs from the parametric images generated by the relative equilibrium (RE) plot.
SUVR, bcSUVR, and DVR Images
The constants for generating the bias corrected SUVR images, λ and μ, are −39.97 and 34.71, respectively (Table 1). The mean images of SUVR, bcSUVR, and DVRs generated by the RE and Logan plots are shown in Figure 3. As expected, there is overestimation in the SUVR images and noise-induced underestimation in the DVR images generated by the Logan plot. Because the ROIs from SUVR images are identical to those from ROI TACs, the overestimation of DVR in SUVR images was the same as shown in Figures 1A and 2A. The bcSUVR images are comparable to images generated by the RE plot. There was a highly linear correlation between the ROI values from bcSUVR images and the bcSUVR calculated from ROI TACs: (bcSUVR image) = 0.96DVR(bcSUVR ROI TACs) + 0.05 with R2 = 0.98. As compared to the DVRs from the RE plot or the Logan plot with ROI TACs, the Bias% of bcSUVR was < ± 6% for ROIs including cortex, striatum and pons, ~−11% in thalamus, and ~8% in white matter. In addition, no significant difference in Bias% of bcSUVR was observed between MCI group and controls. For the Logan plot, the DVR ROI values obtained directly from the parametric DVR images were lower by ~12.2% (excluding white matter) relative to those from the ROI TACs from low noise levels. The underestimation in the DVR images in the MCI group was significantly higher than that of controls in all but 4 ROIs (mesial temporal, occipital, pons, and white matter) (Figure 1C). Note that for the RE plot, the DVR estimates from ROI TACs were identical to those obtained by applying ROIs to DVR images. Therefore, the noise-induced underestimation in the DVR images generated by the Logan plot in both control and MCI groups was removed completely by using the RE plot (Figures 2C, 1D, and 2D).
FIGURE 3.
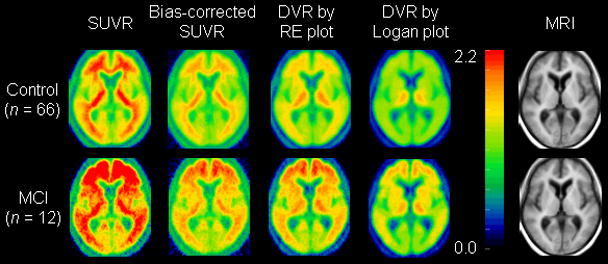
Mean of parametric images generated using SUVR, bias-corrected SUVR (bcSUVR), and DVR images generated by the RE plot and the Logan plot in mild cognitive impairment (MCI) and control groups. Mean MRI is provided for anatomical information.
Comparison of DVR Estimates between Control and MCI Groups
The comparison of ROI estimates of DVR between control and MCI groups is summarized in supplemental data online (Table 2). The DVR estimates from all methods were consistently higher in MCI group than in controls (p < 0.05) for all ROIs except for mesial temporal and occipital cortex, pons, and white matter. In comparison to the DVRs from the Logan plot with ROI TACs, the contrast between DVR in MCI and control groups was increased when quantitated by SUVR, and decreased when parametric images were generated by the Logan plot. Based on the DVRs estimated from the ROI TACs using the Logan plot, the DVRs in ROIs of [frontal (orbital, prefrontal, and superior), cingulate (anterior, and posterior), parietal, lateral temporal, striatum (caudate, putamen), thalamus] were [36.1%, 31.7%, 20.8%, 201%, 26.2%, 10.6%] higher (p<0.05) than those of the control group. In contrast, the difference between MCI and control groups was artificially increased by 8.5% for SUVR, and decreased by 5.6% for the parametric DVR images generated by the Logan plot. As demonstrated in supplemental data online (Table 2) for both control and MCI groups, the DVRs estimated by bcSUVR from ROI TACs, bcSUVR images, and RE plot are similar to those from the Logan plot from ROI TACs. The percent differences in DVRs between MCI and control groups in ROIs of [mesial temporal, pons, and white matter] were consistent across all methods with less than 6% in absolute difference and of no statistical significance (p > 0.30).
DISCUSSION
In this study, we propose that the bcSUVR and the RE plot improve the estimates of DVR from SUVR and the Logan plot, respectively. While the bcSUVR allows simplification of [11C]PiB dynamic PET data acquisition and quantification, the RE plot needs to be applied to the full dynamic PET data. SUVR is associated with positive bias observed with [11C]PiB (13, 15) as well as with neuroreceptor ligands (17, 35, 36). Although this bias has been analyzed by compartmental modeling techniques (16, 17), so far no approaches have been developed for mitigating such bias. The recently developed relative equilibrium-based graphical plot, the RE plot, was used to derive a correction factor that is able to mitigate the known positive bias associated with SUVR (25). Since the method for bcSUVR is based on the parameters of λ and μ estimated from a population, the generalizability of the method was validated by a cross-validation study within our dataset (supplemental data online: Cross-validation for the Bias-corrected SUVR).
Quantification of full dynamic PET data set can also lead to inconsistent bias as demonstrated by the DVR images generated by the Logan plot. Noise-induced underestimation is also observed in the DVR images generated by the SRTM with conventional multi-linear regression method (24). In fact, for ROI [11C]PiB kinetics of low noise levels, it was reported that the SRTM, 2-parameter SRTM, MRTM2, and the Logan plot provide similar DVR estimates (10, 14). In this study, we also found that the DVRs from the SRTM with ROI kinetics were as close to the DVRs from the RE plot as: DVR(SRTM with ROI TACs) = 1.03DV(RE plot) −0.07, R2 = 0.96.
Both the RE plot and the bcSUVR, as well as SUVR, require the presence of relative equilibrium relative to reference tissue kinetics for t ≥ t*. As it may take longer to achieve relative equilibrium for tissues with lower clearance rates or higher DVRs, the t* is likely to be determined by the ROI kinetics from older subjects and MCI patients. Here, we have validated that t* occurs at 52.5 min post tracer injection in populations including MCIs and controls with age ranges from 77.2 – 89.5 years old and the maximum ROI DVR as high as 2.265. The onset of the relative equilibrium t* at 40 to 50 min was demonstrated in other [11C]PiB studies including controls, MCI and AD patients (12, 13, 15). As such, t* = 52.5 min corresponding to dynamic PET scan frames starting from 50 min post tracer injection is an appropriate value for the RE plot and bcSUVR estimation of DVR in [11C]PiB PET studies. Relative equilibrium tracer kinetics were also observed in [11C]raclopride for dopamine D2-like receptor imaging (17, 25), [18F]FDDNP for imaging β-amyloid plaques and neurofibrillary tangles (37), as well as [18F]florbetapir and [18F]florbetaben for imaging β-amyloid (38,39). As such, the RE plot and bcSUVR methods are applicable to these tracer kinetics.
One limitation of this population based approach is that the correction factors λ and μ obtained here may not be applicable to different populations such as the AD patients, to data acquired on different scanners or acquired using different data acquisition or image reconstruction protocols. However, once the bcSUVR method is carefully validated using the RE plot on the full dynamic PET scan data set of a population of interest, it can then be used in simplified PET protocols with short acquisition applied to individuals from that population.
CONCLUSION
We have theoretically analyzed inconsistent overestimation of DVR in SUVR and the noise-induced underestimation of DVR by the Logan plot and then demonstrated it on [11C]PiB studies of nondemented older adults. We propose that bcSUVR derived from the RE plot can simplify both clinical and research [11C]PiB data quantification. The RE plot and bcSUVR are associated with low bias, high consistency, and high computational efficiency in quantification of [11C]PiB retention. Both of these methods improve the quantification of DVR estimates compared to the Logan plot and the SUVR, two frequently used methods for assessment of β-amyloid burden.
Supplementary Material
Acknowledgments
Financial support for the work: This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (NIA), and NIA R&D Contract NO1-AG-0012.
This study was supported in part by the Intramural Research Program, National Institute on Aging, NIH and by N01-AG-3-2124.
Footnotes
DISCLOSURES
We thank the cyclotron, PET, and MRI imaging staff of the Johns Hopkins Medical Institutions; Andrew H. Crabb for data transfer and computer administration.
This work was presented in part at The 56th Society of Nuclear Medicine Conference, June 13–17, 2009, Toronto, Canada, and at The 57th Society of Nuclear Medicine Conference, June 5–9, 2010, Salt Lake City, U.S.A..
References
- 1.Vallabhajosula S. Positron emission tomography radiopharmaceuticals for imaging brain Beta-amyloid. Semin Nucl Med. 2011;41:283–299. doi: 10.1053/j.semnuclmed.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Ziolko SK, Weissfeld LA, Klunk WE, et al. Evaluation of voxel-based methods for the statistical analysis of PIB PET amyloid imaging studies in Alzheimer’s disease. Neuroimage. 2006;33:94–102. doi: 10.1016/j.neuroimage.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 3.Kemppainen NM, Aalto S, Wilson IA, et al. PET amyloid ligand [11C]PIB uptake is increased in mild cognitive impairment. Neurology. 2007;68:1603–1606. doi: 10.1212/01.wnl.0000260969.94695.56. [DOI] [PubMed] [Google Scholar]
- 4.Kemppainen NM, Aalto S, Wilson IA, et al. Voxel-based analysis of PET amyloid ligand [11C]PIB uptake in Alzheimer disease. Neurology. 2006;67:1575–1580. doi: 10.1212/01.wnl.0000240117.55680.0a. [DOI] [PubMed] [Google Scholar]
- 5.Aizenstein HJ, Nebes RD, Saxton JA, et al. Frequent amyloid deposition without significant cognitive impairment among the elderly. Arch Neurol. 2008;65:1509–1517. doi: 10.1001/archneur.65.11.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Innis RB, Cunningham VJ, Delforge J, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- 7.Koeppe RA, Holthoff VA, Frey KA, Kilbourn MR, Kuhl DE. Compartmental analysis of [11C]flumazenil kinetics for the estimation of ligand transport rate and receptor distribution using positron emission tomography. J Cereb Blood Flow Metab. 1991;11:735–744. doi: 10.1038/jcbfm.1991.130. [DOI] [PubMed] [Google Scholar]
- 8.Logan J, Fowler JS, Volkow ND, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(−)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab. 1990;10:740–747. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- 9.Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. Neuroimage. 1996;4:153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Resnick SM, Ye W, et al. Using a reference tissue model with spatial constraint to quantify [11C]Pittsburgh compound B PET for early diagnosis of Alzheimer’s disease. Neuroimage. 2007;36:298–312. doi: 10.1016/j.neuroimage.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Logan J, Fowler JS, Volkow ND, Wang GJ, Ding YS, Alexoff DL. Distribution volume ratios without blood sampling from graphical analysis of PET data. J Cereb Blood Flow Metab. 1996;16:834–840. doi: 10.1097/00004647-199609000-00008. [DOI] [PubMed] [Google Scholar]
- 12.Price JC, Klunk WE, Lopresti BJ, et al. Kinetic modeling of amyloid binding in humans using PET imaging and Pittsburgh Compound-B. J Cereb Blood Flow Metab. 2005;25:1528–1547. doi: 10.1038/sj.jcbfm.9600146. [DOI] [PubMed] [Google Scholar]
- 13.Lopresti BJ, Klunk WE, Mathis CA, et al. Simplified quantification of Pittsburgh Compound B amyloid imaging PET studies: a comparative analysis. J Nucl Med. 2005;46:1959–1972. [PubMed] [Google Scholar]
- 14.Yaqub M, Tolboom N, Boellaard R, et al. Simplified parametric methods for [11C]PIB studies. Neuroimage. 2008;42:76–86. doi: 10.1016/j.neuroimage.2008.04.251. [DOI] [PubMed] [Google Scholar]
- 15.McNamee RL, Yee SH, Price JC, et al. Consideration of optimal time window for Pittsburgh compound B PET summed uptake measurements. J Nucl Med. 2009;50:348–355. doi: 10.2967/jnumed.108.057612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carson RE, Channing MA, Blasberg RG, et al. Comparison of bolus and infusion methods for receptor quantitation: application to [18F]cyclofoxy and positron emission tomography. J Cereb Blood Flow Metab. 1993;13:24–42. doi: 10.1038/jcbfm.1993.6. [DOI] [PubMed] [Google Scholar]
- 17.Logan J, Alexoff D, Kriplani A. Simplifications in analyzing positron emission tomography data: effects on outcome measures. Nucl Med Biol. 2007;34:743–756. doi: 10.1016/j.nucmedbio.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Wallius E, Nyman M, Oikonen V, Hietala J, Ruotsalainen U. Voxel-based NK1 receptor occupancy measurements with [(18)F]SPA-RQ and positron emission tomography: a procedure for assessing errors from image reconstruction and physiological modeling. Mol Imaging Biol. 2007;9:284–294. doi: 10.1007/s11307-007-0096-1. [DOI] [PubMed] [Google Scholar]
- 19.Slifstein M, Laruelle M. Effects of statistical noise on graphic analysis of PET neuroreceptor studies. J Nucl Med. 2000;41:2083–2088. [PubMed] [Google Scholar]
- 20.Logan J, Fowler JS, Volkow ND, Ding YS, Wang GJ, Alexoff DL. A strategy for removing the bias in the graphical analysis method. J Cereb Blood Flow Metab. 2001;21:307–320. doi: 10.1097/00004647-200103000-00014. [DOI] [PubMed] [Google Scholar]
- 21.Fujimura Y, Ikoma Y, Yasuno F, et al. Quantitative analyses of 18F-FEDAA1106 binding to peripheral benzodiazepine receptors in living human brain. J Nucl Med. 2006;47:43–50. [PubMed] [Google Scholar]
- 22.Abi-Dargham A, Martinez D, Mawlawi O, et al. Measurement of striatal and extrastriatal dopamine D1 receptor binding potential with [11C]NNC 112 in humans: validation and reproducibility. J Cereb Blood Flow Metab. 2000;20:225–243. doi: 10.1097/00004647-200002000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Ichise M, Liow JS, Lu JQ, et al. Linearized reference tissue parametric imaging methods: application to [11C]DASB positron emission tomography studies of the serotonin transporter in human brain. J Cereb Blood Flow Metab. 2003;23:1096–1112. doi: 10.1097/01.WCB.0000085441.37552.CA. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Endres CJ, Brasic JR, Huang SC, Wong DF. Linear regression with spatial constraint to generate parametric images of ligand-receptor dynamic PET studies with a simplified reference tissue model. Neuroimage. 2003;18:975–989. doi: 10.1016/s1053-8119(03)00017-x. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Y, Ye W, Brasic JR, Crabb AH, Hilton J, Wong DF. A consistent and efficient graphical analysis method to improve the quantification of reversible tracer binding in radioligand receptor dynamic PET studies. Neuroimage. 2009;44:661–670. doi: 10.1016/j.neuroimage.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resnick SM, Goldszal AF, Davatzikos C, et al. One-year age changes in MRI brain volumes in older adults. Cereb Cortex. 2000;10:464–472. doi: 10.1093/cercor/10.5.464. [DOI] [PubMed] [Google Scholar]
- 27.Morris JC, Storandt M, Miller JP, et al. Mild cognitive impairment represents early-stage Alzheimer disease. Arch Neurol. 2001;58:397–405. doi: 10.1001/archneur.58.3.397. [DOI] [PubMed] [Google Scholar]
- 28.Driscoll I, Resnick SM, Troncoso JC, An Y, O’Brien R, Zonderman AB. Impact of Alzheimer’s pathology on cognitive trajectories in nondemented elderly. Ann Neurol. 2006;60:688–695. doi: 10.1002/ana.21031. [DOI] [PubMed] [Google Scholar]
- 29.Kawas C, Gray S, Brookmeyer R, Fozard J, Zonderman A. Age-specific incidence rates of Alzheimer’s disease: the Baltimore Longitudinal Study of Aging. Neurology. 2000;54:2072–2077. doi: 10.1212/wnl.54.11.2072. [DOI] [PubMed] [Google Scholar]
- 30.Cunningham VJ, Jones T. Spectral analysis of dynamic PET studies. J Cereb Blood Flow Metab. 1993;13:15–23. doi: 10.1038/jcbfm.1993.5. [DOI] [PubMed] [Google Scholar]
- 31.Lawson C, Hanson RJ. Solving Least Squares Problem. Englewood Cliffs, NJ: Prentice Hall; 1974. [Google Scholar]
- 32.Zhou Y, Ye W, Brasic JR, et al. Spectral analysis with a minimal basis function approach for quantification of ligand-receptor dynamic PET study. J Cereb Blood Flow Metab. 2005;25:S634. [Google Scholar]
- 33.Akaike H. An information criteria (AIC) Math Sci. 1976;14:5–9. [Google Scholar]
- 34.Turkheimer FE, Hinz R, Cunningham VJ. On the undecidability among kinetic models: from model selection to model averaging. J Cereb Blood Flow Metab. 2003;23:490–498. doi: 10.1097/01.WCB.0000050065.57184.BB. [DOI] [PubMed] [Google Scholar]
- 35.Carson RE. PET physiological measurements using constant infusion. Nucl Med Biol. 2000;27:657–660. doi: 10.1016/s0969-8051(00)00138-4. [DOI] [PubMed] [Google Scholar]
- 36.Slifstein M. Revisiting an old issue: the discrepancy between tissue ratio-derived binding parameters and kinetic modeling-derived parameters after a bolus of the serotonin transporter radioligand 123I-ADAM. J Nucl Med. 2008;49:176–178. doi: 10.2967/jnumed.107.046631. [DOI] [PubMed] [Google Scholar]
- 37.Wong K, Huang S, Kepe V, Small GW, Barrio JR. Evaluation of Two Graphical Approaches for Regional Analysis and Parametric Mapping of Dynamic [18F]FDDNP PET Studies. IEEE Nuclear Science Symposium Conference Record. 2009:3946–3949. [Google Scholar]
- 38.Rowe CC, Ackerman U, Browne W, et al. Imaging of amyloid beta in Alzheimer’s disease with 18F-BAY94-9172, a novel PET tracer: proof of mechanism. Lancet Neurol. 2008;7:129–135. doi: 10.1016/S1474-4422(08)70001-2. [DOI] [PubMed] [Google Scholar]
- 39.Wong DF, Rosenberg PB, Zhou Y, et al. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F-AV-45 (florbetapir [corrected] F 18) J Nucl Med. 2010;51(6):913–920. doi: 10.2967/jnumed.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


