Abstract
Xp11 translocation renal cell carcinoma (RCC) harbor various TFE3 gene fusions, and are known to underexpress epithelial immunohistochemical (IHC) markers such as cytokeratin and EMA relative to usual adult type RCC; however, their profile in reference to other IHC markers that are differentially expressed in other subtypes of RCC has not been systematically assessed. Few therapeutic targets have been identified in these aggressive cancers. We created 2 tissue microarrays (TMA) containing five 1.4-mm cores from each of 21 Xp11 translocation RCC (all confirmed by TFE3 IHC, 6 further confirmed by genetics), 7 clear cell RCC (CCRCC), and 6 papillary RCC (PRCC). These TMA were labeled for a panel of IHC markers. In contrast to earlier published data, Xp11 translocation RCC frequently expressed renal transcription factors PAX8 (16/21 cases) and PAX2 (14/21 cases), whereas only 1 of 21 cases focally expressed MiTF and only 5 of 21 overexpressed p21. Although experimental data suggest otherwise, Xp11 translocation RCC did not express WT-1 (0/21 cases). Although 24% of Xp11 translocation RCC expressed HIF-1α (like CCRCC), unlike CCRCC CA IX expression was characteristically only focal (mean 6% cell labeling) in Xp11 translocation RCC. Other markers preferentially expressed in CCRCC or PRCC, such as HIG-2, claudin 7, and EpCAM, yielded inconsistent results in Xp11 translocation RCC. Xp11 translocation RCC infrequently expressed Ksp-cadherin (3/21 cases) and c-kit (0/21 cases), markers frequently expressed in chromophobe RCC. Using an H-score that is the product of intensity and percentage labeling, Xp11 translocation RCC expressed higher levels of phosphorylated S6, a measure of mTOR pathway activation (mean H score = 88), than did CCRCC (mean H score = 54) or PRCC (mean H score = 44). In conclusion, in contrast to prior reports, Xp11 translocation RCC usually express PAX2 and PAX8 but do not usually express MiTF. Although they may express HIF-1α, they only focally express the downstream target CA IX. They inconsistently express markers associated with other RCC subtypes, further highlighting the lack of specificity of the latter markers. TFE3 and Cathepsin K remain the most sensitive and specific markers of these neoplasms. Elevated expression of phosphorylated S6 in Xp11 translocation RCC suggests the mTOR pathway as an attractive potential therapeutic target for these neoplasms.
Keywords: TFE3, renal cell carcinoma, biomarker
Xp11 translocation renal cell carcinomas (RCC) are a recently recognized subset of RCC, characterized by chromosome translocations involving the Xp11.2 breakpoint and resulting in gene fusions involving the TFE3 transcription factor gene that maps to this locus.3 Genes shown earlier to be fused to TFE3 in these neoplasms include ASPL, PRCC, PSF, NONO, and clathrin heavy chain (CLTC).1,2,5,6,12 Xp11 translocation RCC represents the most common type of RCC in children, but is less frequent on a percentage basis in adults.7,20 Morphologically, these neoplasms frequently show papillary architecture and clear cytoplasm, and frequently have associated psammoma bodies. By immunohistochemistry, these neoplasms underexpress epithelial markers such cytokeratin and epithelial membrane antigen (EMA) compared with typical adult type RCC. Recently, Xp11 translocation RCC have been shown to frequently express Cathepsin K, a protease frequently expressed in perivascular epithelioid cell neoplasms (PEComas) but not expressed in adult type RCC.25 The most sensitive and specific immunohistochemical marker for the Xp11 translocation RCC is nuclear labeling for TFE3 protein, which reflects overexpression of the resulting fusion proteins relative to native TFE3.5 The prognosis of the Xp11 translocation RCC is not clear, but cases that present at advanced stage owing to hematogenous metastasis are associated with a poor prognosis.7,28 Few therapeutic targets have been identified thus far in these neoplasms.
In the past few years, several novel markers of RCC have been described. These include tissue-specific transcription factors such PAX2 and PAX8,13,26,29,30 along with cytoplasmic differentiation markers such as hypoxia-inducible protein 2 (HIG2)34 and carbonic anhydrase 9 (CA IX).15,33 Some of these markers have been shown to differentially label the common adult subtypes of renal cell carcinoma, which are clear cell, papillary, and chromophobe RCC. Xp11 translocation RCC have been infrequently been analyzed with these markers, and when they have the results reported have often been different for our experience.
In this study, we formally analyze a series of 21 Xp11 translocation RCC using tissue microarrays (TMA) for a set of immunohistochemical markers recently described for RCC diagnosis or subclassification. We provide an extended immunohistochemical profile of Xp11 translocation RCC, and identify potential therapeutic targets.
MATERIALS AND METHODS
Institutional Review Board Approval
This study was approved by the Institutional Review Board of The Johns Hopkins Hospital.
Tissue Microarrays (TMAs)
We created 2 tissue microarrays (TMA) that harbored 21 different Xp11 translocation RCC, all of which were confirmed by TFE3 immunohistochemistry, with 6 further confirmed by genetics. The latter included 3 PSF-TFE3 RCC, 2 PRCC-TFE3 RCC, and one RCC with a t(X;3)(p11;q23) chromosome translocation.7 As a comparison group and control for labeling, we also included 7 clear cell RCC (CCRCC) and 6 papillary RCC (PRCC) on these TMA. Each TMA contain five 1.4-mm diameter cores from each neoplasm, to account for potential heterogeneity of labeling by the immunohistochemical markers.
Immunohistochemistry Methods
Immunohistochemical labeling was done on the Benchmark XT autostainer (Ventana Medical Systems Inc, Tucson, AZ) using I-View detection kit. The standard antibodies used, vendors, pretreatments, and dilutions were as follows: MiTF (Dako, steam, 1:50), PAX2 (Zymed, catalog#71 to 6000, steam, 1:100), PAX8 (ProteinTech Group, Chicago, IL; steam, 1:100), CA IX (Novacastra NCL-L-CAIX, steam, 1:100), HIG2 (Novacastra, steam, 1:200), Napsin A (Novacastra NCL-L-naps, steam, 1:800), EpCAM (Santa Cruz, sc-25308, steam, 1:200), Ksp cadherin (Invitrogen, San Francisco, CA, steam, 1:100), WT-1 (Santa Cruz, C-19, catalog SC-192, steam, 1:100), Bcl2 (Ventana, catalog#760 to 2693, steam, prediluted), CD117 (Cell Marque CMA768, steam, prediluted) and Estrogen Receptor (Novacastra, 6F11, 1 µg/mL). For HIF-1α, we used the antibody from Novus Biologicals, Littleton, Colorado, (1:1600) using the technique of Tickoo et al.33 For Claudin 7, after a 20-minute pretreatment in citrate buffer we used the antibody from Zymed (318–7378) at 1:100 dilution for 45 minutes, followed by the Dako Polyclonal Envision + secondary for 30 minutes. For Phosphorylated S6, after a 50-minute steam pretreatment in EDTA buffer, we used the antibody from Cell Signaling (#2215) at 1:200 dilution overnight at 4°C, followed by the Dako Polyclonal Envision+ secondary for 30 minutes.
Immunohistochemistry Scoring
For MiTF, PAX2, PAX8, and p21, the percentage of neoplastic cells demonstrating nuclear labeling was recorded, and any labeling was considered a positive result. For Ksp-cadherin, EpCAM, and claudin 7, the percentage of neoplastic cells showing membranous labeling, and whether it was complete or incomplete, was recorded, and any labeling was considered a positive result. For CD117, Napsin A, HIG2, Bcl2, and CA IX, the percentage of neoplastic cells showing cytoplasmic or membranous labeling was recorded, and any labeling was considered a positive result.
For phosphorylated S6, an H score (scale 0 to 300) was calculated for each case based upon the product of the percentage of cells showing cytoplasmic labeling (0 to 100) multiplied by the intensity of labeling (1 = weak; 2 = moderate; 3 = strong). For HIF-1α, the percentage of neoplastic cells showing nuclear labeling was recorded. As described by Tickoo et al,33 cases with 1% to 25% labeling were scored as 1+, 26% to 50% labeling were scored as 2+, and >50% labeling were scored as 3+ (diffusely positive).
RESULTS
Transcription Factors: PAX2/PAX8 and MiTF
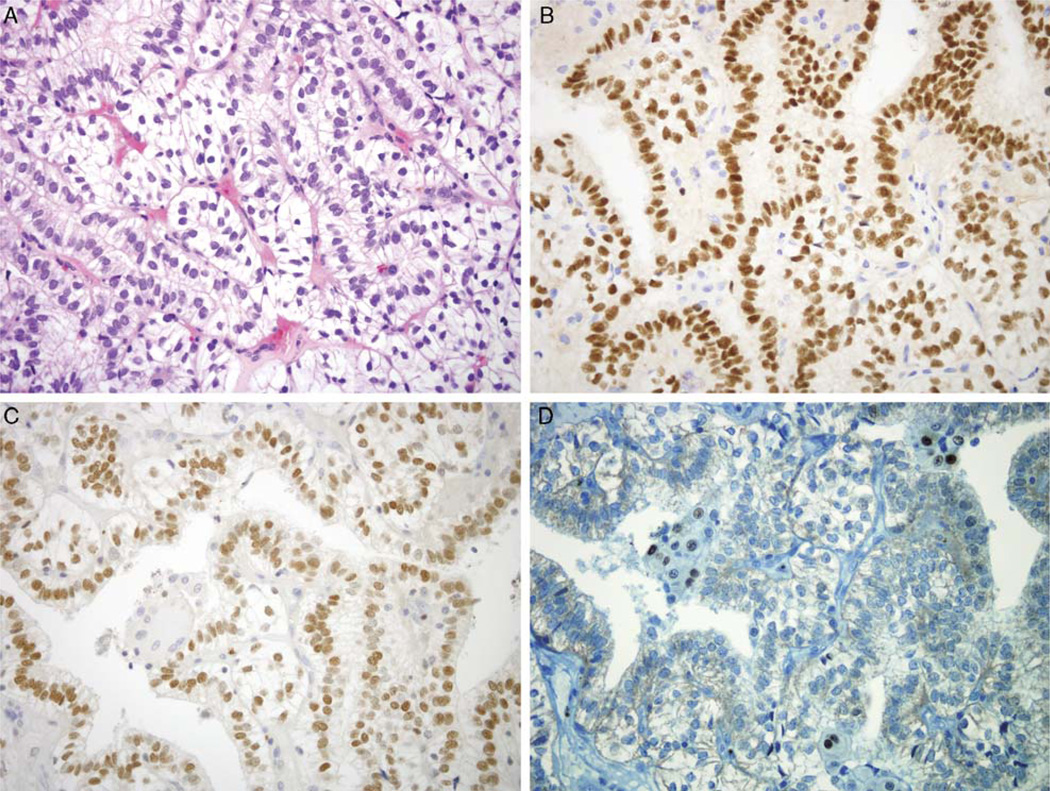
PAX2 and PAX8 are lineage restricted transcription factors that are known to be expressed in the renal and Müllerian systems,13,26,29,30,35 and have been found to be expressed in most clear cell and papillary RCC. We found that 14 of 21 Xp11 translocation RCC showed nuclear labeling for PAX2, whereas 16 of 21 showed nuclear labeling for PAX8. In these cases, labeling was consistently strong, and was seen in a significant proportion of neoplastic cells (mean for PAX2, 45%: mean for PAX8, 58%). As expected, most clear cell and papillary RCC labeled strongly for PAX2 and PAX8 (Table 1). MiTF, a transcription factor that is commonly expressed in melanocytes and their corresponding neoplasms, is in the same subfamily of transcription factors as TFE3 and TFEB and forms heterodimers with them. Only 1 of 21 cases of Xp11 translocation RCC labeled for MiTF; in this case, the labeling was in 5% of neoplastic cells, and of moderate intensity (Fig. 1). As expected, all clear cell and papillary RCC were negative for MiTF.
TABLE 1.
Biomarker Expression in Xp11 Translocation RCC Relative to Clear Cell RCC and Papillary RCC
| Xp11 Translocation RCC (21 Cases) | Clear Cell RCC (7 Cases) | Papillary RCC (6 Cases) | ||||
|---|---|---|---|---|---|---|
| Marker | % Cases Positive | Mean % Labeling | % Cases Positive | Mean % Labeling | % Cases Positive | Mean % Labeling |
| PAX2* | 66 | 45 | 72 | 56 | 100 | 37 |
| PAX8* | 76 | 58 | 72 | 61 | 100 | 67 |
| MiTF* | 5 | 5 | 0 | 0 | 0 | 0 |
| HIF-1α† | 24‡ | 22 | 43‡ | 39 | 0‡ | 0 |
| CA IX† | 57 | 5.7 | 100 | 100 | 66 | 1.8 |
| HIG-2† | 100 | 42 | 100 | 61 | 83 | 11 |
| Napsin A† | 43 | 10 | 43 | 8.5 | 83 | 31 |
| EpCAM* | 43 | 23 | 28 | 5 | 100 | 34 |
| Claudin 7* | 33 | 31 | 14 | 30 | 100 | 70 |
| Ksp-Cadherin* | 14 | 40 | 0 | 0 | 0 | 0 |
| pS6† | 90 | 32.6 | 67 | 23 | 83 | 17 |
| p21* | 24 | 26 | 14 | 90 | 33 | 17 |
| Bcl2* | 24 | 26 | 57 | 37 | 0 | 0 |
| WT-1† | 0 | 0 | 0 | 0 | 0 | 0 |
| CD117† | 0 | 0 | 0 | 0 | 0 | 0 |
| ER† | 0 | 0 | 0 | 0 | 0 | 0 |
Note that the calculated H score is given in the text for pS6.
Mean calculated among positive cases.
Mean calculated among all cases.
Diffusely positive cases.
pS6 indicates phosphorylated S6.
FIGURE 1.
Expression of Transcription Factors in Xp11 Translocation Renal Cell Carcinoma (RCC). A, Hematoxylin and Eosin stain of genetically-confirmed PSF-TFE3 RCC. The neoplasm shows strong nuclear labeling for PAX8 (B) and PAX2 (C), whereas only rare cells label for MiTF (D). All other cases were completely negative for MiTF. All images are 400× magnification.
HIF 1-α/CA IX Pathway
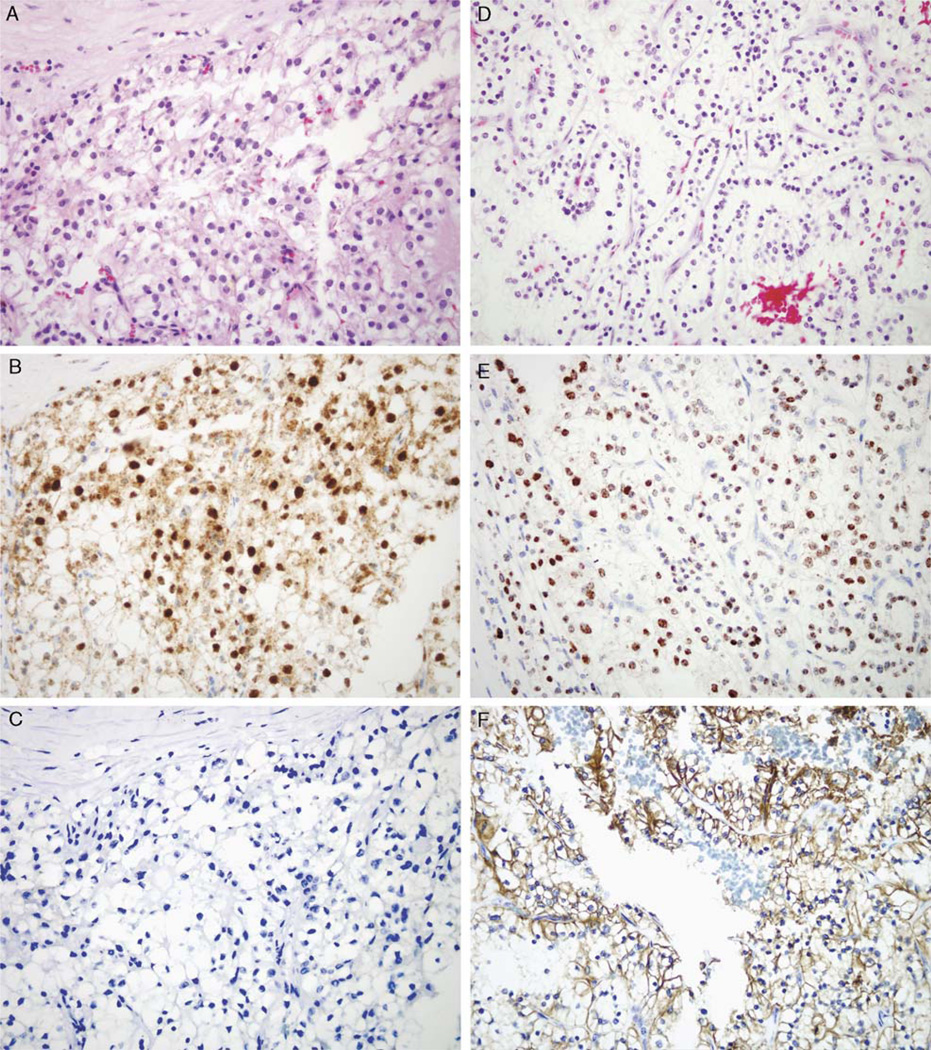
The HIF 1-α/CA IX pathway is aberrantly activated in clear cell RCC. In clear cell RCC, inactivation of the vHL gene allows HIF 1-α to escape degradation, which leads to activation of downstream targets such as vascular endothelial growth factor (VEGF), GLUT-1, and CA IX.14,33 Expression of HIF 1-α and CA IX is typically diffuse in clear cell RCC, and focal in other subtypes of RCC. In the latter, expression is mainly associated with areas of necrosis and thus is likely secondary to focal hypoxia.14,33 We found that only a mean of 5.7% of Xp11 translocation RCC tumor cells labeled for CA IX. By comparison, 100% of clear cell RCC cells labeled for CA IX, while only 1.8% of papillary RCC cells labeled for CA IX, which is consistent with the above published literature.14,33 In an individual case, labeling for CA IX in Xp11 translocation RCC was typically focal and centered around areas of necrosis. Although CA IX labeling was minimal in Xp11 translocation RCC, 5 of 21 (24%) were diffusely (3+) positive for HIF-1α, whereas 9 others showed focal (1+) labeling. By comparison, none of the 6 papillary RCC labeled for HIF-1α, while 3 of 7 (42%) clear cell RCC were diffusely (3+) positive for HIF-1α and 1 other showed focal (1+) labeling (Fig. 2), again consistent with the published literature.14,33
FIGURE 2.
Expression of HIF-1α and CA IX in Xp11 Translocation RCC versus Clear Cell RCC. This genetically-confirmed PRCC-TFE3 RCC (A, Hematoxylin and Eosin stain) shows strong difuse nuclear labeling for HIF-1α (B) but is negative for CA IX (C). In contrast, a conventional (clear cell) adult RCC (D, Hematoxylin and Eosin stain) shows strong diffuse nuclear labeling for HIF-1α (E) and strong diffuse cytoplasmic labeling for the downstream target CA IX (F). All images are 400× magnification.
Clear Cell and Papillary RCC Markers
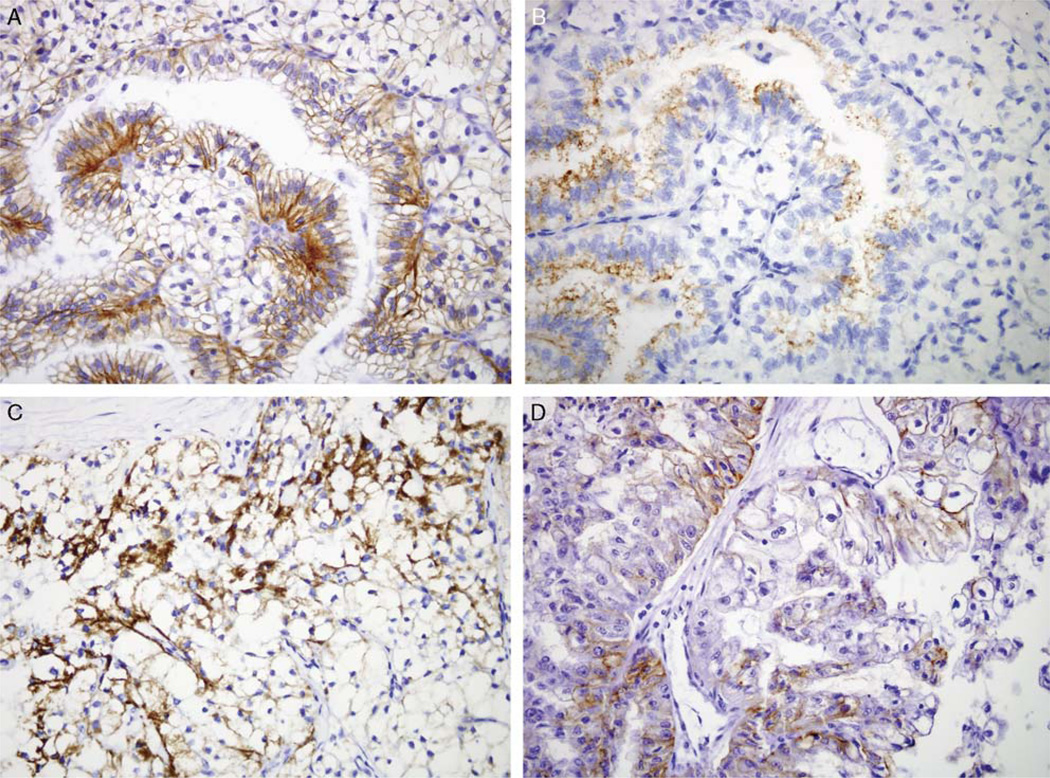
The labeling pattern of Xp11 translocation RCC for markers differentially expressed in clear cell or papillary RCC was not distinctive. For example, Hypoxia-inducible protein 2 (HIG2) is a novel marker of RCC identified by gene expression profiling. HIG2 has been found to be expressed in fetal kidney and RCC, and is secreted into the plasma of patients with RCC; thus, it is a promising diagnostic and screening marker.34 HIG2 labeling was found in a mean of 42% of cells of Xp11 translocation RCC, closer to the 61% mean for clear cell RCC than the 11% mean for papillary RCC. Napsin A is an aspartic proteinase normally expressed in type 2 pneumocytes and alveolar macrophages along with renal convoluted tubules. Napsin A has been found to be expressed more frequently in papillary than clear cell RCC.9 A mean of only 10% of Xp11 translocation RCC cells labeled for Napsin A, similar to the mean of 8.5% of cells in clear cell RCC and much less than the mean of 31% of cells in papillary RCC (Fig. 3). Epithelial Cell Adhesion Molecule (EpCAM) is a cell adhesion molecule expressed in a variety of carcinomas that has drawn great interest as a potential therapeutic target, as humanized anti-EpCAM antibodies are now in clinical trials.36 EpCAM has been shown to be more frequently expressed in papillary RCC than clear cell RCC.38 Nine of 21 cases (43%) of Xp11 translocation RCC labeled for EpCAM, compared with 100% of papillary RCC and 28% of clear cell RCC.
FIGURE 3.
Miscellaneous RCC markers in Xp11 translocation RCC. PSF-TFE3 RCC shows patchy membranous labeling for claudin 7 (A) and cytoplasmic labeling for napsin A (B), accentuated in papillary areas. PRCC-TFE3 RCC shows cytoplasmic labeling for HIG2 (C). Xp11 translocation RCC shows incomplete membranous labeling for Ksp-cadherin (D). All images are 400× magnification.
Chromophobe RCC Markers: Claudin 7, CD117 and Ksp-cadherin
Claudin 7 is a tight junction protein which is preferentially expressed in chromophobe RCC relative to clear cell or papillary RCC.11,17 Seven of 21 cases of Xp11 translocation RCC (33%) labeled focally for claudin 7, more prominently in papillary areas. In comparison, 14% of clear cell RCC labeled for claudin 7, whereas all papillary RCC labeled for claudin 7. However, in all cases, membranous labeling for claudin 7 was incomplete. Ksp-cadherin is normally expressed in distal nephron epithelium, and is typically diffusely expressed in chromophobe RCC and is negative or only focally positive in clear cell or papillary RCC. Kuehn et al reported that 1 of 4 Xp11 translocation RCC labeled diffusely for Ksp-cadherin.21 Xp11 translocation RCC infrequently expressed Ksp-cadherin (3 of 21 cases, 14%), and the cases that did showed incomplete membranous labeling in a mean of 40% of cells (Fig. 3). All clear cell and papillary RCC were negative for Ksp-cadherin. CD117 expression has been shown to distinguish chromophobe RCC from clear cell RCC.37 None of the Xp11 translocation RCC, clear cell RCC and papillary RCC expressed CD117.
Phosphorylated S6
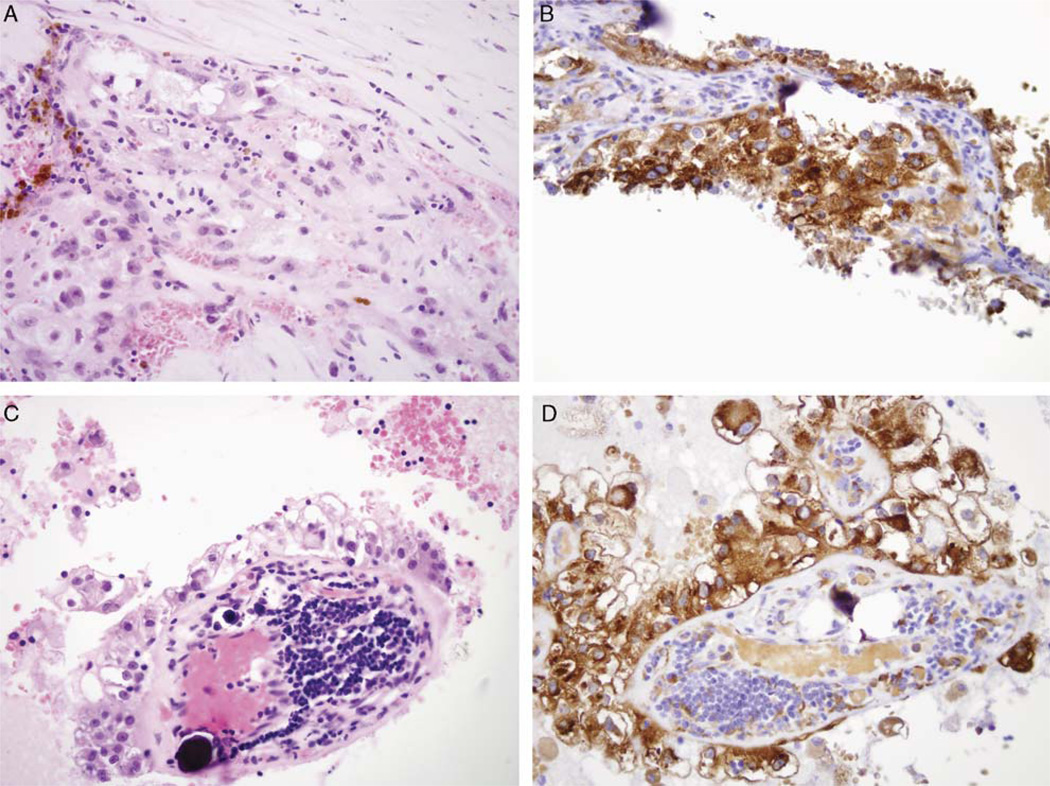
Immunoreactivity for phosphorylated S6 represents a measure of activation of the mammalian target of rapamycin (mTOR) pathway that promotes cell growth, is highly active in many RCC, and can be targeted for therapy.31 The mean H-score for labeling with phosphorylated S6 (pS6) was 88 for the Xp11 translocation RCC, slightly higher than H score of 54 for clear cell RCC and 44 for papillary RCC (Fig. 4). On a percentage basis, the mean percentage of labeling of Xp11 translocation RCC was 32.6%, compared with 23% for clear cell RCC and 17% for papillary RCC.
FIGURE 4.
Expression of phosphorylated S6 (pS6), a marker of mTOR pathway activation, in genetically confirmed Xp11 translocation RCC. Hematoxylin and Eosin stain of t(X;3)(pll;q23) RCC (A) and corresponding pS6 immunohistochemistry (B). Hematoxylin and Eosin stain of PRCC-TFE3 RCC (C) and corresponding pS6 immunohistochemistry (D). All images are 400× magnification.
Other Markers
P21 is a cell-cycle checkpoint protein that has recently been reported to be diffusely expressed in 2 Xp11 translocation RCC.8,16 Sixteen of 21 cases of Xp11 translocation RCC were completely negative for p21, one expressed p21 in 10% of neoplastic cells, whereas 4 expressed p21 in 30% of neoplastic cells. All cases were completely negative for the WT-1 gene product, a target of the TFE3 transcription factor in vitro.18 Only 5 of 21 Xp11 translocation RCC cases were focally immunoreactive for the anti-apoptotic protein Bcl2, which is a target of the related transcription factor MiTF in melanoma and known to be expressed in clear cell RCC.19,24,27 Finally, the involvement of the X chromosome in the chromosomal translocation of the Xp11 translocation RCC, along with a reported bias toward female patients in some studies,7 suggests the possibility of hormonal influence. However, all cases of Xp11 translocation RCC were negative for estrogen receptor.
Summary of Results
The IHC data for this study is summarized in Table 1. For markers that were frequently completely negative (ie, p21), we calculated the mean labeling of the positive cases. For other markers that were infrequently negative (ie, pS6), we calculated the mean labeling of the total number of cases. The individual scores for each Xp11 translocation RCC are presented in Table 2.
TABLE 2.
Xp11 Translocation RCC: Percent Labeling for Biomarkers in Individual Cases
| Case | PAX2 | PAX8 | MiTF | HIF-1α | CA IX | HIG-2 | Napsin A | EpCAM | CL7 | Ksp-Cad | pS6* | p21 | Bcl2 | WT-1 | CD117 | ER |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 70 | 70 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 240 | 30 | 30 | 0 | 0 | 0 |
| 2 | 20 | 20m | 0 | 20 | 5 | 60 | 10 | 5i | 20i | 0 | 20 | 0 | 0 | 0 | 0 | 0 |
| 3 | 90 | 100 | 5m | 5 | 0 | 5 | 20 | 70i | 50i | 0 | 5 | 0 | 50 | 0 | 0 | 0 |
| 4 | 90 | 80 | 0 | 90 | 0 | 70 | 2 | 0 | 0 | 0 | 20 | 0 | 50 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 40 | 0 | 0 | 0 | 0 | 0 |
| 6 | 70 | 70m | 0 | 80 | 5 | 20 | 0 | 5i | 0 | 0 | 270 | 0 | 0 | 0 | 0 | 0 |
| 7 | 50 | 20 | 0 | 10m | 5 | 30 | 0 | 5i | 30i | 0 | 20 | 0 | 50 | 0 | 0 | 0 |
| 8 | 10w | 20 | 0 | 60 | 5 | 50 | 70 | 0 | 0 | 0 | 270 | 30 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 5 | 0 | 50 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
| 10 | 60 | 30 | 0 | 10 | 5 | 20 | 10 | 10i | 0 | 70i | 20 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | 0 | 20 | 60 | 0 | 10i | 0 | 0 | 210 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 60 | 0 | 0 | 5i | 30i | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 10 | 50 | 0 | 0 | 2 | 50 | 80 | 0 | 0 | 20i | 20 | 0 | 0 | 0 | 0 | 0 |
| 14 | 20 | 30 | 0 | 5 | 0 | 60 | 10 | 80c | 80i | 0 | 20 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 20 | 10 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 |
| 16 | 70w | 100m | 0 | 20 | 30 | 90 | 0 | 20c | 0 | 0 | 270 | 10 | 0 | 0 | 0 | 0 |
| 17 | 50 | 50m | 0 | 80 | 0 | 10 | 0 | 0 | 20i | 0 | 20 | 30 | 0 | 0 | 0 | 0 |
| 18 | 0 | 80 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 80m | 0 | 5w | 15 | 100 | 0 | 0 | 0 | 0 | 210 | 0 | 0 | 0 | 0 | 0 |
| 20 | 5w | 80 | 0 | 20 | 5 | 100 | 0 | 0 | 0 | 0 | 40 | 0 | 50 | 0 | 0 | 0 |
| 21 | 20 | 50 | 0 | 50 | 5 | 70 | 5 | 2i | 10i | 0 | 40 | 30 | 0 | 0 | 0 | 0 |
Cases 1 and 2: PRCC-TFE3 RCC; Cases 3–5: PSF-TFE3 RCC; Case 6: t(X: 3)(p11:2q23) RCC.
CL7=claudin 7; Ksp-Cad=Ksp-cadherin.
Membranous labeling: i=incomplete: c=complete.
Intensity (strong unless otherwise specified): w=weak, m=moderate.
H-Score is provided for pS6 labeling.
DISCUSSION
We have extended the known immunohistochemical profile of Xp11 translocation RCC by applying a panel of biomarkers to a series of well-established Xp11 translocation RCC. To our knowledge, this is the first study of this kind on Xp11 translocation RCC. Our results confirm some prior observations regarding these neoplasms, yet refute several others.
The frequent labeling for PAX2 and PAX8 in the Xp11 translocation RCC supports renal tubular differentiation in these frequently cytokeratin-negative neoplasms. It also validates the use of this marker in setting of metastatic disease, which is often the mode of presentation of Xp11 translocation RCC. Although PAX8 labeling has, to our knowledge, not been earlier studied in Xp11 translocation RCC, our results conflict with that of Gupta et al,15 who reported no immunoreactivity for PAX2 in 15 Xp11 translocation RCC. The reason for this difference are not clear, though it could potential reflect differences in case selection. Gupta et al relied entirely on upon their interpretation of TFE3 immunohistochemistry (methodology not provided) to classify their tumors as Xp11 translocation RCC; in contrast, 6 of our immunohistochemically confirmed cases, including those that were positive for PAX2/PAX8, were also molecularly confirmed. Moreover, our cases were all strongly positive for TFE3 using our previously validated methodology.5 Regarding MiTF, we found less frequent labeling than did Ramphal et al,32 who found that 3 of 4 genetically confirmed Xp11 translocation RCC expressed MiTF. On the basis of the results of the Ramphal et al study, one could hypothesize that MiTF is secondarily upregulated in some of these neoplasms by the TFE3 fusion proteins; because it is known that all members of the MiTF/TFE transcription factor subfamily bind to each other, overexpressed TFE3 fusion proteins could in theory compensatorily increase the levels of the MITF to which they bind. However, that upregulation of TFE3, TFEC, or MiTF has not been showed in the related t(6;11) RCC with TFEB gene fusions,4,22 so this mechanism lacks a precedent. Our results of infrequent MiTF expression in Xp11 translocation RCC are more consistent with the latter data.
Regarding the HIF-1α/CA IX pathway, we found only focal labeling in a small percentage of Xp11 RCC for CA IX. The cells which labeled (mean 5.7% in these cases) were typically clustered around areas of necrosis, as it is typical of non clear cell RCC. These results are concordant with those reported by Gupta et al15 using this marker; they found focal labeling for CA IX in 40% of Xp11 translocation RCC. Hence, we believe that difuse labeling for CA IX is helpful in distinguishing clear cell RCC from Xp11 translocation RCC. In contrast, a subset of Xp11 translocation RCC (24%) showed strong nuclear labeling for HIF-1α, similar to clear cell RCC. As CA IX is a downstream target HIF-1á in clear cell RCC, one wonders if this pathway is somehow disrupted in the Xp11 RCC. No prior study has analyzed HIF-1α expression in Xp11 translocation RCC.
Regarding the markers expressed in clear cell and papillary RCC, we did not identify a specific pattern of labeling that distinguishes Xp11 translocation RCC from conventional clear cell or papillary RCC. Xp11 translocation RCC labeled more similarly to clear cell RCC than papillary RCC for markers in this study such as HIG2, EpCAM, Napsin A and Claudin 7. However, prior studies from our group and others10,23 have showed that α-methylacyl-coenzyme A racemase, which is preferentially expressed in papillary RCC compared with clear cell RCC, is consistently expressed in Xp11 translocation RCC. This result highlights the importance of cytogenetics, molecular diagnostics and specific immunohistohemical markers (TFE3, Cathepsin K) relative to standard RCC marker immunohistochemistry in the diagnosis of the Xp11 translocation RCC.
Several of the other miscellaneous markers tested have earlier been implicated in Xp11 translocation RCC. For example, Höcker-Müller et al16 and Barroca et al8 recently reported 2 Xp11 translocation RCC with difuse p21 protein overexpression. In contrast, in our larger series, we found only focal expression of p21 in a minority of Xp11 translocation RCC. Contrary to what in vitro studies might suggest, WT-1 and Bcl2 were infrequently expressed in Xp11 translocation RCC, despite the fact that the related transcription factor MiTF is known to induce BcL2 expression in osteoclasts and melanoma cells27 whereas over expression TFE3 or TFEB in NIH 3T3 cells are known to activate WT-1.18
Finally, our identification of expression of phosphorylated S6 in Xp11 translocation RCC, at levels slightly higher than those seen conventional RCC, suggests that targeting the mTOR pathway may be an effective way to treat the Xp11 translocation RCC. Further studies in this area are clearly warranted.
REFERENCES
- 1.Argani P, Antonescu CR, Couturier J, et al. PRCC-TFE3 renal carcinomas. Morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21) Am J Surg Pathol. 2002;26:1553–1566. doi: 10.1097/00000478-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Argani P, Antonescu CR, Illei PB, et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma. A distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–192. doi: 10.1016/S0002-9440(10)61684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argani P, Ladanyi M. Renal carcinomas associated with Xp11.2 translocations/TFE3 gene fusions. In: Eble JN, Sauter G, Epstein J, et al., editors. Pathology and Genetics of Tumors of the Urinary System & Male Genital Organs. Lyon, France: IARC; 2004. pp. 37–38. [Google Scholar]
- 4.Argani P, Laé M, Hutchinson B, et al. Renal carcinomas with the t(6;11)(p21;q12). Clinicopathologic features and demonstration of the specific Alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA-PCR. Am J Surg Pathol. 2005;29:230–240. doi: 10.1097/01.pas.0000146007.54092.37. [DOI] [PubMed] [Google Scholar]
- 5.Argani P, Lal P, Hutchinson B, et al. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol. 2003;27:750–761. doi: 10.1097/00000478-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Argani P, Lui MY, Couturier J, et al. Cloning of a novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23) Oncogene. 2003;22:5374–5378. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 7.Argani P, Olgac S, Tickoo SK, et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol. 2007;31:1149–1160. doi: 10.1097/PAS.0b013e318031ffff. [DOI] [PubMed] [Google Scholar]
- 8.Barroca H, Castedo S, Vieira J, et al. Altered expression of key cell cycle regulators in renal cell carcinoma associated with Xp11.2 translocation. Path Res Pract. 2009;205:466–472. doi: 10.1016/j.prp.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Bishop JA, Sharma R, Illei PB. Napsin A and thyroid transcription factor-1 expression in carcinomas of the lung, breast, pancreas, colon, kidney, thyroid, and malignant mesothelioma. Hum Pathol. 2010;41:20–25. doi: 10.1016/j.humpath.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Camparo P, Vasiliu V, Molinie V, et al. Renal translocation carcinomas. clinicopathologic, immunohistochemical and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol. 2008;35:656–670. doi: 10.1097/PAS.0b013e3181609914. [DOI] [PubMed] [Google Scholar]
- 11.Choi YD, Kim KS, Ryu S, et al. Claudin-7 is highly expressed in chromophobe renal cell carcinoma and renal oncocytoma. J Korean Med Sci. 2007;22:305–310. doi: 10.3346/jkms.2007.22.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark J, Lu Y-J, Sidhar SK, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 13.Daniel L, Lechevallier E, Giorgi R, et al. Pax-2 expression in adult renal tumors. Hum Pathol. 2001;32:282–287. doi: 10.1053/hupa.2001.22753. [DOI] [PubMed] [Google Scholar]
- 14.Di Cristofano C, Minervini A, Menicagli M, et al. Nuclear expression of hypoxia-inducible factor-1α in clear cell renal cell carcinoma is involved in tumor progression. Am J Surg Pathol. 2007;31:1875–1881. doi: 10.1097/PAS.0b013e318094fed8. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R, Balzwer B, Picken M, et al. Diagnostic implications of transcription factor Pax2 protein and transmembrane enzyme complex carbonic anhydrase IX immunoreactivity in adult renal epithelial neoplasms. Am J Surg Pathol. 2009;33:241–247. doi: 10.1097/PAS.0b013e318181b828. [DOI] [PubMed] [Google Scholar]
- 16.Höcker-Müller J, Babaryka G, Schmid I, et al. Overexpression of cyclin D, D3, and p21 in an infantile renal carcinoma with Xp11.2 TFE3-gene fusion. Path Res Pract. 2008;204:589–597. doi: 10.1016/j.prp.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Hornsby CD, Cohen C, Amin MB, et al. Claudin-7 immunohistochemistry in renal tumors. A candidate marker for chromophobe renal cell carcinoma identified by gene expression profiling. Arch Pathol Lab Med. 2007;131:1541–1546. doi: 10.5858/2007-131-1541-CIIRTA. [DOI] [PubMed] [Google Scholar]
- 18.Huan C, Sashital D, Hailemariam T, et al. Renal carcinoma-associated transcription factors TFE3 and TFEB are leukemia inhibitory factor-responsive transcription activators of E-cadherin. J Biol Chem. 2005;280:30225–30235. doi: 10.1074/jbc.M502380200. [DOI] [PubMed] [Google Scholar]
- 19.Itoi T, Yamana K, Bilim V, et al. Impact of frequent Bcl-2 expression on better prognosis in renal cell carcinoma patients. British Journal of Cancer. 2004;90:200–205. doi: 10.1038/sj.bjc.6601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komai Y, Fujiwara M, Fujii Y, et al. Adult Xp11 translocation renal cell carcinoma diagnosed by cytogenetics and immunohistochemistry. Clin Cancer Res. 2009;15:1170–1176. doi: 10.1158/1078-0432.CCR-08-1183. [DOI] [PubMed] [Google Scholar]
- 21.Kuehn A, Paner GP, Skinnider BF, et al. Expression analysis of kidney-specific cadherin in a wide spectrum of traditional and newly recognized renal epithelial neoplasms: diagnostic and histogenetic implications. Am J Surg Pathol. 2007;31:1528–1533. doi: 10.1097/PAS.0b013e318058818c. [DOI] [PubMed] [Google Scholar]
- 22.Kuiper RP, Schepens M, Thijssen J, et al. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet. 2003;12:1661–1669. doi: 10.1093/hmg/ddg178. [DOI] [PubMed] [Google Scholar]
- 23.Lae M, Argani P, Olshen AB, et al. Global gene expression profiles of renal carcinomas with Xp11 translocations (TFE3 gene fusions) suggest a closer relationship to Alveolar Soft part Sarcoma than to Adult Type Renal Cell Carcinomas. Mod Pathol. 2004;17:163A. (abstract #682). [Google Scholar]
- 24.Lee CT, Genega EM, Hutchinson B, et al. Conventional (clear cell) renal carcinoma metastases have greater Bcl-2 expression than high risk primary tumors. Urol Oncol. 2003;21:179–184. doi: 10.1016/s1078-1439(02)00236-3. [DOI] [PubMed] [Google Scholar]
- 25.Martignoni G, Pea M, Gobbo S, et al. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol. 2009;22:1016–1022. doi: 10.1038/modpathol.2009.58. [DOI] [PubMed] [Google Scholar]
- 26.Mazal PR, Stichenwirth M, Koller A, et al. Expression of aquaporins and PAX-2 compared to CD10 and cytokeratin 7 in renal neoplasms: a tissue microarray study. Mod Pathol. 2005;18:535–540. doi: 10.1038/modpathol.3800320. [DOI] [PubMed] [Google Scholar]
- 27.McGill G, Horstmann M, Widlund H, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 28.Meyer PN, Clark JI, Flanigan RC, et al. Xp11.2 translocation renal cell carcinoma with very aggressive course in five adults. Am J Clin Pathol. 2007;128:70–79. doi: 10.1309/LR5G1VMXPY3G0CUK. [DOI] [PubMed] [Google Scholar]
- 29.Nonaka D, Chiriboga L, Soslow R. Expression of Pax8 as a useful marker in distinguishing ovarian carcinomas from mammary carcinomas. Am J Surg Pathol. 2008;32:1566–1571. doi: 10.1097/PAS.0b013e31816d71ad. [DOI] [PubMed] [Google Scholar]
- 30.Ozcan A, Zhai J, Hanilton C, et al. PAX-2 in the diagnosis of primary renal tumors. Am J Clin Pathol. 2009;131:393–404. doi: 10.1309/AJCPM7DW0XFHDHNY. [DOI] [PubMed] [Google Scholar]
- 31.Pantuck AJ, Seligson DB, Klatte T, et al. Prognostic relevance of the mTOR pathway in renal cell carcinoma. Implications for molecular patient selection for targeted therapy. Cancer. 2007;109:2257–2267. doi: 10.1002/cncr.22677. [DOI] [PubMed] [Google Scholar]
- 32.Ramphal R, Pappo A, Zielenska M, et al. Pediatric renal cell carcinoma. Clinical, pathologic and molecular abnormalities associated with the members of the MiT transcription factor family. Am J Clin Pathol. 2006;126:349–364. doi: 10.1309/98YE9E442AR7LX2X. [DOI] [PubMed] [Google Scholar]
- 33.Tickoo SK, Alden D, Olgac S, et al. Immunohistochemical expression of hypoxia inducible factor-1α and its downstream molecules in sarcomatoid renal cell carcinoma. J Urol. 2007;177:1258–1263. doi: 10.1016/j.juro.2006.11.100. [DOI] [PubMed] [Google Scholar]
- 34.Togashi A, Katagiri T, Ashida S, et al. Hypoxia-Inducible protein 2 (HIG2), a novel diagnostic marker for renal cell carcinoma and potential target for molecular therapy. Cancer Res. 2005;65:4817–4826. doi: 10.1158/0008-5472.CAN-05-0120. [DOI] [PubMed] [Google Scholar]
- 35.Tong G-X, Weeden EM, Hamele-Bena D, et al. Expression of PAX8 in nephrogenic adenoma and clear cell adenocarcinoma of the lower urinary tract. Evidence of related histogenesis? Am J Surg Pathol. 2008;32:1380–1387. doi: 10.1097/PAS.0b013e31816b1020. [DOI] [PubMed] [Google Scholar]
- 36.Trzpis M, McLaughlin PM, de Leij LM, et al. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am J Pathol. 2007;171:386–395. doi: 10.2353/ajpath.2007.070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang HY, Mills SE. KIT and RCC are useful in distinguishing chromophobe renal cell carcinoma from the granular variant of clear cell renal cell carcinoma. Am J Surg Pathol. 2005;29:640–646. doi: 10.1097/01.pas.0000157943.33903.92. [DOI] [PubMed] [Google Scholar]
- 38.Went P, Dirnhofer S, Salvisberg T, et al. Expression of epithelial cell adhesion molecule (EpCam) in renal epithelial tumors. Am J Surg Pathol. 2005;29:83–88. doi: 10.1097/01.pas.0000.146028.70868.7a. [DOI] [PubMed] [Google Scholar]