Abstract
Structure–affinity relationships for ligand binding at the human A2A adenosine receptor have been probed using site-directed mutagenesis in the transmembrane helical domains (TMs). The mutant receptors were expressed in COS-7 cells and characterized by binding of the radioligands [3H]CGS21680, [3H]NECA, and [3H]XAC. Three residues, at positions essential for ligand binding in other G protein-coupled receptors, were individually mutated. The residue V(3.32) in the A2A receptor that is homologous to the essential aspartate residue of TM3 in the biogenic amine receptors, i.e., V84(3.32), may be substituted with L (present in the A3 receptor) but not with D (in biogenic amine receptors) or A. H250(6.52), homologous to the critical N507 of rat m3 muscarinic acetylcholine receptors, may be substituted with other aromatic residues or with N but not with A (Kim et al. J. Biol. Chem. 1995, 270, 13987–13997). H278(7.43), homologous to the covalent ligand anchor site in rhodopsin, may not be substituted with either A, K, or N. Both V84L(3.32) and H250N(6.52) mutant receptors were highly variable in their effect on ligand competition depending on the structural class of the ligand. Adenosine-5′-uronamide derivatives were more potent at the H250N(6.52) mutant receptor than at wild type receptors. Xanthines tended to be close in potency (H250N(6.52)) or less potent (V84L-(3.32)) than at wild type receptors. The affinity of CGS21680 increased as the pH was lowered to 5.5 in both the wild type and H250N(6.52) mutant receptors. Thus, protonation of H250-(6.52) is not involved in this pH dependence. These data are consistent with a molecular model predicting the proximity of bound agonist ligands to TM3, TM5, TM6, and TM7.
Introduction
The principal mechanism by which caffeine and other alkylxanthines act as physiological stimulants is by acting as competitive antagonists of the ubiquitous neuromodulator adenosine.1,2 Adenosine has a depressant action on the central and peripheral nervous systems and the cardiovascular, renal, and immune systems, as well as other systems. Four pharmacologically distinct subtypes of adenosine receptors, A1, A2A, A2B, and A3, have been cloned.3,4 These receptors are members of the superfamily of rhodopsin-like G protein-coupled receptors (GPCRs).
Site-directed mutagenesis has been used in an attempt to identify amino acid residues in the transmembrane helical domains (TMs) of the human A2A adenosine receptor that are essential for ligand recognition.5–7 A rhodopsin-based model5 of the human A2A adenosine receptor has been proven to be highly consistent with mutagenesis results regarding orientation of individual amino acid residues within the central ligand-binding cavity, thus implicating residues in TM5 (fifth helical transmembrane domain), TM6, and TM7 of the A2A adenosine receptor in ligand recognition. The points of ligand recognition in A1 adenosine receptors have also been probed either through single-amino acid replacements8–10 or through the construction of chimerical receptors.11,12 In addition to mutagenesis we have designed unique ligand probes for characterizing adenosine receptors, including the affinity label PAPA-APEC, which provided the first evidence that A1 and A2A adenosine receptors were distinct molecular species,13 and the fluorescent label FITC-APEC.14,15 PAPA-APEC has been shown to covalently cross-link to TM5 of the A2A adenosine receptor.16 In mutagenesis studies of adenosine receptors, amino acid residues both within the transmembrane helical domains and in the second extracellular loop,5,11 which joins TM4 and TM5, have been shown to be essential for ligand binding.
A recent review has noted striking similarity in structure–function relationships among the diverse GPCRs that bind small, non-peptide ligands,17 suggesting conservation of an overall molecular architecture. A dendrogram based on the sequence alignment of many GPCRs18 clearly shows that the adenosine receptors are more closely related to the biogenic amine receptors than to any other class. This study illustrates that there is homology not only in sequence but also in the conservation of function in ligand binding with respect to specific positions on the transmembrane helices. Thus, some homologous locations on the helices appear to be important for ligand binding in both adenosine and biogenic amine receptors, even though the amino acid residues at the site may be entirely different and the ligands are structurally very different (Figure 1).
Figure 1.
Generalized features of GPCRs,17 showing residue locations in TM3, TM5, and TM7 that are important for ligand binding in adenosine receptors and in biogenic amine receptors. Glycosylation occurs on the N-terminal segment (NT) in the biogenic amine receptors and on the second extracellular loop (E2) in adenosine A2A receptors. A palmitoylation site occurs in biogenic receptors at the C-terminal segment (CT) and in the adenosine A1 and A3 receptors. A sodium-binding site occurring at a D residue (2.50) modulates agonist binding. The third intracellular loop (I3), which directly contacts G proteins, contains an α-helical region. Adenosine is shown in the central cavity of the transmembrane region, which has been opened for illustration purposes. Mutagenesis of human A2A receptors indicates that the adenine moiety is largely associated with aromatic residues in TM5, TM6, and TM7.5 TM3 and TM7 contain residues, mutation of which selectively affects agonists, that are proposed to coordinate the ribose moiety.6
| residue | adenosine A2A | β2 adrenergic | m3 muscarinic | D2 dopamine |
|---|---|---|---|---|
| 3.32 | V | D | D | D |
| 6.52 | H | F | N | F |
| 7.43 | H | Y | Y | Y |
In the present study we have introduced single-amino acid replacements at three positions of the human A2A adenosine receptor that are also known as important ligand recognition elements in biogenic amine receptors: positions 3.32, 6.52, and 7.43, using an extended notation that allows immediate comparison to structural studies of other receptors.17,19 (Residue identifiers17,19 specify the helix (X.) and the position relative to a key conserved residue in that helix, which is designated (X.50).) In the human A2A adenosine receptors these positions correspond (in order) to V84 in TM3, to H250 in TM6, and to H278 in TM7. Position 3.32 corresponds to the essential D present in the biogenic amine receptors which forms the counterion to the charged amino groups of (endogenous) ligands and has not previously been mutated for any adenosine receptor subtype. Residues at positions 6.52 and 7.43 were found to influence ligand affinity in m3 muscarinic receptors. In A1 and A2 adenosine receptor subtypes both are conserved H residues. In A1 and A2A adenosine receptors these H residues have been shown to be involved in ligand recognition.5,8
Results
Ligand Binding Properties of Mutant Human A2A Adenosine Receptors
Mutant human A2A adenosine receptors containing single-amino acid replacements at three positions within the transmembrane helical regions were prepared using PCR techniques and expressed in COS-7 cells. Amino acids mutated are indicated using the residue identifiers as described in van Rhee and Jacobson,17 which specify the helix (X.) and the position relative to a key conserved residue in that helix, which is designated (X.50). For example, for the helices mutated in the present study, the key conserved residues are R102(3.50), P248(6.50), and P285(7.50), V84(3.32), H250(6.52), and H278(7.43), respectively, all of which are conserved between A1 and A2 receptors and were individually substituted. The following mutant receptors were prepared: V84A(3.32), V84D(3.32), V84L(3.32), H250N(6.52), H278K(7.43), and H278N(7.43). Other H250(6.52) and H278(7.43) receptor mutants had been prepared in a previous study.5 Initially, radioligand binding experiments5,6,20,21 using fixed concentrations of the agonist [3H]CGS21680 (80 nM), the agonist [3H]NECA (100 nM), or the antagonist [3H]XAC (30 nM) were carried out on the wild type and mutant receptors. The mutant receptors V84A(3.32), V84D(3.32), H278K(7.43), and H278N(7.43) displayed negligible specific binding (less than 10% of that of wild type receptors) of all three radioligands. The mutant receptors V84L(3.32) and H250N(6.52) displayed levels of specific binding similar to that of wild type for all three radioligands at the indicated concentrations. Precise saturation and competition studies were carried out on these mutants using [3H]CGS21680 (Table 1).
Table 1.
Binding Characteristics of Wild Type and Mutant Human A2a Adenosine Receptors Using the Agonist Radioligand [3H]CGS21680a
| constructb
|
||||
|---|---|---|---|---|
| compound | WT | V84L | H250N | |
| Bmax (pmol/mg) | [3H]CGS21680 | 5.69 ± 0.51 | 3.16 ± 0.15** | 1.67 ± 0.11** |
| Kd (nM) | [3H]CGS21680 | 36.4 ± 2.7 | 27.3 ± 3.2** | 7.60 ± 1.49** |
| Ki (nM) | agonists | |||
| DPMA | 38.1 ± 0.8 | 18.9 ± 8.1** | 35.9 ± 4.4 | |
| NECA | 21.6 ± 4.5 | 21.5 ± 9.5 | 5.24 ± 4.69** | |
| IB-MECA | 370 ± 91 | 143 ± 28* | 32.0 ± 13.7** | |
| antagonists | ||||
| CGS15943 | 0.142 ± 0.047 | 0.842 ± 0.433 | 0.457 ± 0.225 | |
| XAC | 7.89 ± 1.17 | 45.6 ± 12.6** | 15.6 ± 6.4** | |
| galangin | 16700 ± 900 | 18100 ± 5000 | 33000 ± 5800 | |
| nifedipine | 24400 ± 6300 | 22200 ± 1700 | 22200 ± 1900 | |
| CPX | 226 ± 28 | 788 ± 93** | 291 ± 104 | |
| BTH4 | 106000c | 72600 ± 44000 | 54300 ± 4400 | |
| amiloride | 12000 ± 4100 | 11600 ± 2400 | 3280 ± 1330* | |
Agonist and antagonist binding affinities (Ki values, structures in Figure 2 and Jacobson et al., 1992) were determined in [3H]CGS21680 (15 nM) competition binding studies at pH 6.8 using membrane homogenates prepared from transiently transfected COS-7 cells, as described in the Experimental Section. Data are presented as means ± SD of three independent experiments, unless indicated, each performed in duplicate. Each sample contained 7–11 μg of membrane protein/tube. Ki values were calculated from IC50 values using the KaleidaGraph program. All constructs contain an HA tag sequence at the N-terminus (Kim et al., 1995).
P ≤ 0.01,
P ≤ 0.05 vs wild type receptors.
Constructs that showed <3% of specific binding of [3H]CGS21680 (15 nM) found for HA-tagged wild type receptors were V84A, V84D, H278N, and H278K mutant receptors. The expression levels at these four mutants showed levels comparable to HA-tagged wild type receptors. Kd was determined in saturation experiments using [3H]CGS21680 at the V84L and H250N mutant receptors transfected in COS-7 cells.
n = 2 (105 and 107 μM).
The V84D(3.32) mutant receptor was intended to resemble the biogenic amine receptors. Since we could not detect specific radioligand binding with adenosine radioligands, the construct was probed for specific binding using an extremely high-affinity muscarinic acetylcholine receptor antagonist, [3H]QNB.22 Not unexpectedly, no specific binding was observed at a radio-ligand concentration of 10 nM.
To eliminate improper delivery of the receptors to the plasma membrane as a probable cause of the lack of high-affinity binding in membranes, a hemagglutinin (HA) epitope tag was incorporated at the N-terminus of all of the constructs, allowing immunological detection.5 In this manner using an ELISA method the V84A(3.32), V84D(3.32), H278K(7.43), and H278N(7.43) mutant receptors were found to be properly expressed on the surface of the COS-7 cells (Table 2). To estimate approximate levels of receptor protein present in the plasma membrane (percent with respect to the HA-tagged wild type receptor), a standard curve was constructed from different batches of transfected COS-7 cells expressing varying levels of wild type HA-tagged A2A adenosine receptors (see the Experimental Section).5 The ELISA procedure does not interfere with the intactness of the plasma membrane barrier.5 Thus the assay is specific for receptors having the extracellular N-terminus properly oriented in the membrane.
Table 2.
ELISA of A2A Receptor Mutants on the Surface of COS-7 Cells
| mutant | expression levela (%) |
|---|---|
| V84A | 98.2 ± 4.0 |
| V84D | 99.4 ± 6.7 |
| H278N | 95.7 ± 5.6 |
| H278K | 97.5 ± 4.4 |
Table shows expression level as a percentage of HA-tagged A2A wild type (100%), n = 8. Expression level was determined using a standard curve (Kim et al., 1995). None of the values listed were significantly different from control.
Saturation experiments were carried out with the wild type receptor and the V84L(3.32) and H250N(6.52) mutant receptors (Table 1) using the agonist [3H]-CGS21680 to label a high-affinity state. In Scatchard analysis for each of these receptor constructs, only a single high-affinity affinity state was detected. The Kd value for [3H]CGS21680 binding at the V84L(3.32) mutant receptor was only slightly lower than for the wild type receptor, while at the H250N(6.52) mutant receptor affinity of the radioligand was increased nearly 5-fold. The Bmax values of these two mutant receptors were somewhat diminished.
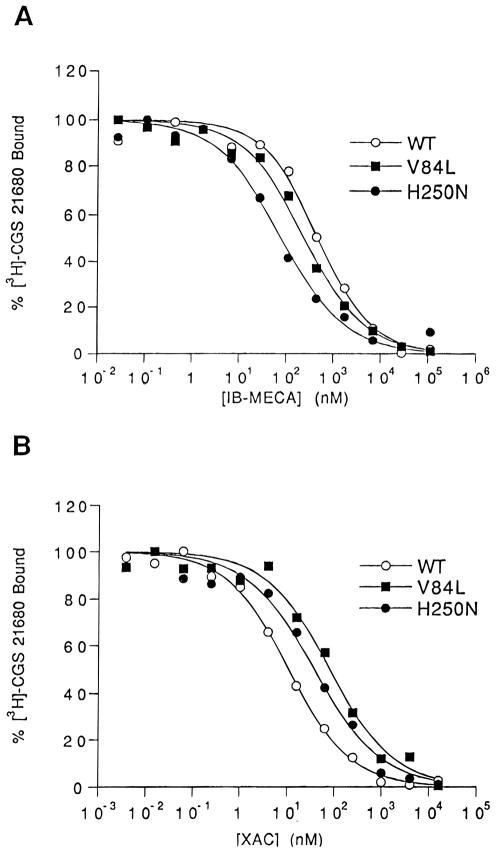
The agonists selected for competition (Figure 2 and Table 1) included adenosine derivatives2 modified at the N6-position (DPMA), the 5′-position (NECA), the 5′- and 2-positions (CGS21680), and the 5′- and N6-positions (IB-MECA). Agonist competitors at V84L(3.32) mutant receptors retained (NECA) or increased (DPMA, 2-fold; IB-MECA, 2.6-fold) their affinity. At H250N(6.52) mutant receptors, IB-MECA (Figure 3A) and NECA were 12- and 4-fold more potent, respectively, than at wild type receptors.
Figure 2.
Structure of the agonists and antagonists used in this study. Structure numbering is included for adenosine and xanthine derivatives.
Figure 3.
Representative competition curves for displacement of binding of [3H]CGS21680 at wild type and mutant human A2A receptors by IB-MECA (A) and XAC (B).
The diverse set of adenosine antagonists2 shown in Figure 2 was studied in competition for [3H]CGS21680 binding. Included in the set of adenosine antagonists were two xanthines, the xanthine amine congener (XAC) and 8-cyclopentyl-1,3-dipropylxanthine (CPX); a potent and nonselective non-xanthine (CGS15943); amiloride; nifedipine;23 and two recently reported antagonists, i.e., the flavonol galangin24 and the tetrahydrobenzothiophenone BTH4,25 both of which do not contain nitrogen atoms. Among antagonist competitors of binding at V84L(3.32) mutant receptors there was variation in the affinity depending on the chemical class. At V84L(3.32) mutant receptors CPX and XAC (Figure 3B) had reduced affinity (4–6-fold) compared to wild type receptors. However nifedipine, galangin, amiloride, and BTH4 displayed wild type affinity for V84L(3.32) mutant receptors. At H250N(6.52) mutant receptors most antagonists displayed approximately wild type affinity, except XAC, which displayed a 2-fold decrease in affinity, and amiloride, which displayed a 4-fold gain in affinity. Thus, the loss of affinity of the xanthine antagonists XAC and CPX was more prevalent in the V84L(3.32) than in the H250N(6.52) mutant receptor, while a gain of affinity was present in only one case in the H250N(6.52) mutant receptor.
pH Dependence of Ligand Binding
The affinity of CGS21680 increases as the pH is lowered to 5.5 in both the wild type and H250N(6.52) mutant receptors (Table 3). Thus, it is highly unlikely that protonation of H250(6.52) is involved in the pH dependence of the wild type receptor.26,27
Table 3.
Saturation Binding Characteristics of Wild Type and Mutant Human A2A Adenosine Receptors as a Function of pH, Using the Agonist Radioligand [3H]CGS 21680a
| construct
|
||
|---|---|---|
| WT | H250N | |
| pH 5.5 | ||
| Bmax (pmol/mg) | 5.60 ± 0.29 | 1.47 ± 0.07 |
| Kd (nM) | 22.0 ± 5.4* | 4.30 ± 1.80** |
| pH 6.8 | ||
| Bmax (pmol/mg) | 5.69 ± 0.51 | 1.67 ± 0.11 |
| Kd (nM) | 36.4 ± 2.7 | 7.60 ± 1.50** |
| pH 7.5 | ||
| Bmax (pmol/mg) | 4.57 ± 0.39 | 1.00 ± 0.04 |
| Kd (nM) | 74.0 ± 12.8** | 11.6 ± 2.0** |
| pH 8.4 | ||
| Bmax (pmol/mg) | 6.94 ± 7.30 | 0.827 ± 0.220 |
| Kd (nM) | 632 ± 716 | 99 ± 38** |
Data are presented as means ± SD of three independent experiments, each performed in duplicate. Each sample contained 7–11 μg of membrane protein/tube.
P < 0.01,
P < 0.05 vs pH 6.8 (WT column) or corresponding pH for wild type receptors (H250N column).
Discussion
Three amino acid residues of the human A2A adenosine receptors, V84(3.32), H250(6.52), and H278(7.43), are among those sites that appear to be involved in ligand recognition based on site-directed mutagenesis and radioligand binding with both agonists and an antagonist (Figure 1). The combination of ELISA and radioligand binding results indicates that the V84(3.32) and H278(7.43) residues are particularly critical, either through direct contact or indirect influence, for high-affinity binding of ligands. Position 3.32 corresponds to the essential D in TM3 of the biogenic amine receptors,17 whereas position 6.52 corresponds to the conserved N in TM6 of muscarinic acetylcholine receptors that was shown to be important for antagonist binding.22 H278(7.43) of the human A2A adenosine receptor is homologous to K296(7.43) in bovine rhodopsin, the retinal anchor site, and Y533(7.43) in the human m3 muscarinic acetylcholine receptor, which is generally conserved as Y in biogenic amine receptors, and substitution of this residue with either K or N proved to be highly detrimental to ligand binding. Mutations of H278(7.43) have been made in both A1 receptors (L)8–10 and A2A receptors (A, Q, F, or Y in ref. 5; N or K in this study), and none are tolerated for high-affinity ligand recognition. The requirement for H278-(7.43) therefore appears to be absolute.
In a previous study5 H250(6.52) was substituted with other aromatic residues (as occur in α- and β-adrenergic, serotonin, and dopamine receptors), which resulted in preservation of high-affinity radioligand binding, and with A (as in rhodopsin), which resulted in loss of high-affinity binding. In the present study this residue was replaced with N (as in m1–m5 muscarinic receptors). A gain of affinity was observed and most pronounced for the agonists bearing a 5′-uronamide moiety (CGS21680, NECA, and IB-MECA) at the H250N(6.52) mutant receptor. Amiloride,28 which has a single carbonyl group proximal to the aromatic ring, was also more potent than at wild type receptors. Xanthines and CGS15943 tended to be close in potency at H250N(6.62) mutant receptors and wild type receptors. The patterns of changes in ligand affinity suggest that structural classes may be discerned.
A pH dependence of binding of CGS21680 has been noted previously.26,27 It was speculated that this effect was related to the protonation of one of the two histidine residues in the binding pocket.29 Since one of the receptor mutants in the present study, H250N(6.52), was functional yet lacking a second histidine residue, we also probed the participation of histidine residues in the observed phenomena of the pH dependence of radioligand affinity. In this study we have shown that even in the absence of a histidine residue at position 6.52, a pH dependence is still observed. Therefore, we speculate that the remaining H278(7.43) is the most likely candidate to be involved in this effect. We were unable to directly test the hypothesis that the H278-(7.43) residue is the site of protonation responsible for the pH dependence of radioligand binding, because mutations at this position were not tolerated.
Xanthines were less potent at V84L(3.32) mutant receptors than at wild type receptors, but the affinity of the other antagonists seemed to be unaffected by the mutation. The pattern of changes in ligand affinity with respect to the V84L(3.32) mutant parallels that of the H250N(6.52) mutant. Thus, it is possible to discern structural classes of ligands based on whether a particular mutation leads to enhanced or diminished affinity. There are other examples of the importance of residues occupying position 3.32 in binding of nonbiogenic amine ligands. This was shown, for example, in NK1,30 rhodopsin,31 and TRH receptors.32 Mutation of several hydrophilic residues, T88(3.36) and Q89(3.37), of the human A2A adenosine receptor has also been shown to affect ligand binding,6 although these residues are located more than one helical turn from V84(3.32) in the cytosolic direction.
The structure–function parallels of the data presented above are strongest with mutagenesis studies performed with receptors for biogenic amines. For instance, in the rat 5HT2 serotonin receptor the D155N-(3.32) mutation led to a decreased affinity for both agonists and antagonists,33 and similar effects were observed for the D107N(3.32) mutant of human H1 histamine receptors.34 Moreover, in the hamster β2 adrenergic receptor the D113E(3.32) mutation not only decreased the affinity for agonists and antagonists but also revealed certain antagonists to be partial agonists.35 The more disparate mutation D113S(3.32) in the hamster β2 adrenergic receptor displayed a general decrease in affinity for antagonists but selectively changed the affinity order of agonists.36 In human D2 dopamine receptors a D114C(3.32) mutant was shown to display decreased affinity toward antagonists and in addition strongly reacted with a thiol reagent37 indicating that the position is exposed to the solvent in the cavity formed by the transmembrane bundle. This, in turn, is highly consistent with a role for V84(3.32) in the binding of ligands to the human A2A adenosine receptor.
The parallels between structure–function profiles for A2A and biogenic amine receptors are not limited to TM3 but extend into TM6 and TM7. The change of a polar to an acidic residue in the rat m3 muscarinic mutant receptor, N507D(6.52), led to an overall decrease in antagonist affinity and changed the rank order of affinity for agonists.22 The change from an aromatic residue to an alanine in the human D2 dopamine receptor, F390A(6.52), resulted in an across the board decrease of affinity,38 as was demonstrated for the H250A(6.52) mutant of the human A2A adenosine receptor.5 In the case of the Y316A(7.43) mutant of the hamster β2 adrenergic receptor, ligand binding was decreased to such an extent that impaired processing was inferred to have caused this effect.39 In this study, however, we have shown that although ligand binding is absent from H278K(7.43) and H278N(7.43) human A2A adenosine receptors, the receptors are still expressed at the cell surface with the N-termini properly oriented (Table 2). Wess et al.40 demonstrated that the Y533F(7.43) mutation in the rat m3 muscarinic receptor considerably decreased ligand affinity but that the receptors were properly expressed at the cell surface. This is consistent with our observations and suggests that the contribution of an amino acid located at position 7.43 in the receptor (possibly specific hydrogen bonding with ligands for either biogenic amine or adenosine receptors) is both essential and a general principle.
In our earlier study of H278(7.43) mutants of the human A2A adenosine receptor (Figure 1), we formed a molecular model that contained a conjugated hydrogen bond between S281(7.46), H278(7.43), and the 5′-position of agonists.5 Our current mutagenesis work on H278K(7.43) and H278N(7.43) mutant receptors remains consistent with this hypothesis. Also, the demonstration of pH dependency with the H250N(6.52) mutant similar to the wild type indirectly supports the importance of the only other transmembrane histidine, H278. Replacement of H278 by the positively charged K did not result in high-affinity ligand binding, neither did the replacement of this amino acid by N, a residue with both hydrogen bond donor and acceptor functionality, result in high-affinity ligand binding. Only in the wild type H278(7.43) receptor is there a possibility that a protonated residue can act as a proton relay system,7,41 thereby sustaining the integrity of both the agonist and the antagonist ligand-binding domain.
Furthermore, molecular modeling is consistent with the proximity of bound agonist ligands to TM3, TM5, TM6, and TM7, as has been proposed for muscarinic and other biogenic amine receptors.19,42,43
The coordinates of the human A2A receptor model5 are available from the Protein Database (Brookhaven National Laboratory) using the URL http://www.pdb.bnl.gov/cgi-bin/browse under the ID code 1mmh. As noted previously,5 the critical residue F182 (5.43) corresponds to a serine essential for agonist binding in catecholamine receptors.
Molecular modeling has been used to gain insight into the relative orientation of adenosine derivatives17 and xanthines2,41,44 in the adenosine receptor binding site. Incorporation of XAC into an A2A adenosine receptor model based on a bacteriorhodopsin template41 has been carried out according to the hypothesis of ‘N6/C8′ overlay investigated by van der Wenden et al.44 The amine-functionalized chain of XAC is oriented towards the exofacial side of the receptor. Similar results were obtained when XAC was docked according to this hypothesis into our NECA-occupied A2A receptor model based on the structure of rhodopsin (5 and unpublished), and moreover the model predicts that both the 8-substituent and the N3 propyl group of potent 1,3-dialkyl-xanthine antagonists are oriented toward TM3, whereas the N1 propyl group is in proximity to TM7. In addition to the present and previous mutagenesis results5,6,12 and the molecular modeling, the proximity of TM3 to the ligand-binding site was further emphasized in photo-affinity labeling experiments.45 An azido group located on the N3 substituent of a xanthine was found to cross-link with TM3.
Further modeling studies (in progress) will be required to analyze the microscopic basis for the differences between different antagonists binding to mutant receptors. According to the computer model of adenosine antagonists by van Rhee et al.,25 XAC and CGS15943 have a similar mode of orientation in the ligand-binding domain of the receptor, in which the furyl substituent of CGS15943 occupies approximately the same space as the phenyl ring of XAC. Both of the xanthine antagonists and CGS15943 lost affinity in the V84L mutant, whereas the affinity of ligands with a very different structure and lower wild type affinity (galangin, nifedipine, and BTH4) is unchanged.
The desired, long term objective of receptor mutagenesis is to be able to design and synthesize more selective ligands based on knowledge of ligand recognition in receptor binding. This goal will require refinements in molecular modeling methodology as well as in biophysical methods for obtaining corroborative evidence for specific interactions. The present study has supported the view that there is a common architecture in general underlying GPCR structure–activity. Specifically, adenosine receptors bear a resemblance to biogenic amine receptors, to which they are closely related by sequence homology.17,18 Moreover, distinct differences in the properties of diverse ligands binding to human A2A adenosine receptors, mutated at critical positions, have been identified.
Experimental Section
Materials
Human A2A adenosine receptor cDNA (pSVLA2A) was provided by Dr. Marlene A. Jacobson (Merck Research Labs, West Point, PA). Taq polymerase for the polymerase chain reaction (PCR) was purchased from Perkin Elmer Cetus (Emeryville, CA) and adenosine deaminase from Boehringer Mannheim Biochemicals (Indianapolis, IN). All other enzymes used in this study were obtained from New England Biolabs (Beverly, MA). DPMA, NECA, CGS15943, XAC, nifedipine, amiloride, and 2-chloroadenosine were purchased from Research Biochemicals International (Natick, MA). Galangin was purchased form Apin Chemicals Ltd. (Oxfordshire, U.K.), and BTH4 (ethyl 3-(benzylthio)-4-oxo-4,5,6,7-tetrahydrobenzo-[c]thiophene-1-carboxylate) was purchased from Maybridge Chemicals (Trevillet, U.K.). The Sequenase Kit was from United States Biochemical (Cleveland, OH). All oligonucleotides used were synthesized by Bioserve Biotechnologies (Laurel, MD). A monoclonal antibody (12CA5) against a hemagglutinin epitope (HA) was purchased from Boehringer Mannheim Biochemicals (Indianapolis, IN), and goat anti-mouse IgG (γ-chain specific) antibody conjugated with horse-radish peroxidase, FBS, and o-phenylenediamine dihydrochloride were purchased from Sigma Chemical Co. (St. Louis, MO). DEAE-dextran was obtained from Pharmacia-LKB (Piscataway, NJ). [3H]CGS 21680 and [3H]NECA were purchased from DuPont NEN (Boston, MA), and [3H]XAC was custom synthesized by the same supplier.
Plasmid Construction and Site-Directed Mutagenesis
The coding region of pSVLA2A was subcloned into the pCD cDNA expression vector,46 yielding pCDA2A. All mutations were introduced into pCDA2A using standard PCR mutagenesis techniques.47 The accuracy of all PCR-derived sequences was confirmed by dideoxy sequencing of the mutant plasmids.48
Epitope Tagging
A 9-amino acid sequence derived from the influenza virus hemagglutinin (HA) protein (TAC CCC TAC GAC GTC CCC GAC TAC GCC; peptide sequence: YPYDVPDYA) was inserted after the second methionine residue at the extracellular N-terminus of the A2A adenosine receptor gene.5 Oligonucleotides containing the HA tag sequence were designed and used to generate PCR fragments, which were then used to replace the homologous wild type pCDA2A sequences.
Transient Expression of Mutant Receptors in COS-7 Cells
COS-7 cells (2 × 106) were seeded into 100 mm culture dishes containing 10 mL of Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were transfected with plasmid DNA (4 μg of DNA/dish) by the DEAE-dextran method49 approximately 24 h later and grown for an additional 72 h at 37 °C.
Membrane Preparation and Radioligand Binding Assay
Cells were scraped into ice-cold lysis buffer (4 mL of 50 mM Tris, pH 6.8 at room temperature, containing 10 mM MgCl2). Harvested cells were homogenized using a Polytron homogenizer (Brinkmann Instruments Inc., Westbury, NY) and then spun at 27000g for 15 min. Cell membranes (pellet) were resuspended in the same buffer.
Competition binding with [3H]CGS21680,5,20 [3H]NECA,21 and [3H]XAC5 and saturation experiments with [3H]-CGS216805,20 were carried out as described previously. For typical saturation curves, 0.5–70 nM [3H]CGS21680 was used. Each tube contained 100 μL of membrane suspension (containing 2 U/mLadenosine deaminase), 50 μL of radioligand, and either 50 μL of buffer/competitor (50 mM Tris, pH 6.8, 10 mM MgCl2) or 50 μL of 160 μM CADO in buffer (to determine nonspecific binding). The mixtures were incubated at 25 °C for 120 min, filtered, and washed three times with approximately 5 mL of ice-cold buffer/wash using a Brandel cell harvester (Gaithersburg, MD). In competition experiments, Ki values were calculated from IC50 values according to the Cheng–Prusoff equation.50 Data analysis was performed using the KaleidaGraph program (Abelbeck Software, version 3.01).
ELISA
For indirect cellular ELISA measurements,5 cells were transferred to 96-well dishes (4–5 × 104 cells/well) 1 day after transfection. About 48 h after splitting, cells were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature. After washing with PBS three times and blocking with DMEM (containing 10% FBS), cells were incubated with HA specific monoclonal antibody (12CA5), 20 μg/mL, for 3 h at 37 °C. Plates were washed and incubated with a 1:2000 dilution of a peroxidase-conjugated goat anti-mouse IgG antibody for 1 h, at 37 °C. Hydrogen peroxide and o-phenylenediamine (2.5 mM each in 0.1 M phosphate–citrate buffer, pH 5.0) served as substrate and chromogen, respectively. The enzymatic reaction was stopped after 30 min at room temperature with 1 M H2SO4 solution containing 0.05 M Na2SO3, and the color development was measured bichromatically in the BioKinetics reader (EL 312, Bio Tek Instruments, Inc., Winooski, VT) at 490 and 630 nm (baseline).
Acknowledgments
We thank Dr. Marlene Jacobson (Merck Research Labs, West Point, PA) for providing the human A2A plasmid. A.M.vR. is grateful for support from the Cystic Fibrosis Foundation (Silver Spring, MD). M.G. and B.X.L. thank Gilead Sciences (Foster City, CA) for financial support.
Abbreviations
- BTH4
ethyl 3-(benzylthio)-4-oxo-4,5,6,7-tetrahydrobenzo[c]thiophene-1-carboxylate
- CADO
2-chloroadenosine
- CGS15943
9-chloro-2-furyl-1,2,4]triazolo[1,5-c]-quinazolin-5-amine
- CGS21680
2-[[[4-(2-carboxyethyl)phenyl]-ethyl]amino]-5′-(N-ethylcarbamoyl)adenosine
- CPX
8-cyclopentyl-1,3-dipropylxanthine
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethyl sulfoxide
- DPMA
N6-[2-(3,5-dimethoxyphenyl)-2-(2-methylphenyl)ethyl]adenosine
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- FITC-APEC
2-[[[4-[2-[2-[[[1,3-dihydro-1,1-bis(4-hydroxyphenyl)-3-oxo-5-isobenzofuranthioureidyl]ethyl]amino]carbonyl]ethyl]-phenyl]ethyl]amino]-5′-(N-ethylcarbamoyl)adenosine
- GPCR
G protein-coupled receptor
- HA
hemagglutinin
- IB-MECA
N6-(3-iodobenzyl)adenosine-5′-N-methyluronamide
- NECA
5′-(N-ethylcarbamoyl)adenosine
- NK
neurokinin
- PAPA-APEC
2-[[[4-[2-[[[2-[([4-aminophenyl)acetyl]amino]ethyl]amino]carbonyl]-ethyl]phenyl]ethyl]amino]-5′-(N-ethylcarbamoyl)adenosine
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- QNB
quinuclidinyl benzilate
- SAR
structure–activity relationships
- TM
(helical) transmembrane domain
- TRH
thyrotropin-releasing hormone
- Tris
tris(hydroxymethyl)aminomethane
- XAC
8-[[[[(2-aminoethyl)amino]carbonyl]methoxy]-phenyl]-1,3-dipropylxanthine
References
- 1.Fredholm BB, Abbracchio MP, Burnstock G, Daly JW, Harden KT, Jacobson KA, Leff P, Williams M. Nomenclature and classification of purinoceptors: a report from the IUPHAR subcommittee. Pharmacol Rev. 1994;46:143–156. [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobson KA, van Galen PJM, Williams M. Adenosine receptors - pharmacology, structure activity relationships, and therapeutic potential. J Med Chem. 1992;35:407–422. doi: 10.1021/jm00081a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Libert F, Parmentier M, Lefort A, Dinsart C, van Sande J, Maenhaut C, Simons MJ, Dumont JE, Vassart G. Selective amplification and cloning of four new members of the G protein-coupled receptor family. Science. 1989;244:569–572. doi: 10.1126/science.2541503. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson M. Molecular biology of adenosine receptors. In: Belardinelli L, Pelleg A, editors. Adenosine and Adenine Nucleotides: From Molecular Biology to Integrative Physiology. Kluver; Norwell, MA: 1995. pp. 5–14. [Google Scholar]
- 5.Kim JH, Wess J, van Rhee AM, Schöneberg T, Jacobson KA. Site-directed mutagenesis identifies residues involved in ligand recognition in the human A2a adenosine receptor. J Biol Chem. 1995;270:13987–13997. doi: 10.1074/jbc.270.23.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Q, van Rhee AM, Kim J, Yehle S, Wess J, Jacobson KA. Hydrophilic side chains in the third and seventh transmembrane helical domains of human A2A adenosine receptors are required for ligand recognition. Mol Pharmacol. 1996;50:512–521. [PMC free article] [PubMed] [Google Scholar]
- 7.IJzerman AP, von Frijtag Drabbe Künzel JK, Kim J, Jiang Q, Jacobson KA. Site-directed mutagenesis of the human A2a adenosine receptor. Critical involvement of Glu13 in agonist recognition. Eur J Pharmacol. 1996;310:269–272. doi: 10.1016/0014-2999(96)00495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olah ME, Ren HZ, Ostrowski J, Jacobson KA, Stiles GL. Cloning, expression, and characterization of the unique bovine-A1 adenosine receptor-studies on the ligand binding site by site-directed mutagenesis. J Biol Chem. 1992;267:10764–10770. [PMC free article] [PubMed] [Google Scholar]
- 9.Townsend-Nicholson A, Schofield PR. A threonine residue in the 7th transmembrane domain of the human A1-adenosine receptor mediates specific agonist binding. J Biol Chem. 1994;269:2373–2376. [PubMed] [Google Scholar]
- 10.Tucker A, Robeva AS, Taylor HE, Holeton D, Bockner M, Lynch KR, Linden J. A1 adenosine receptors-2 amino-acids are responsible for species-differences in ligand recognition. J Biol Chem. 1994;269:27900–27906. [PubMed] [Google Scholar]
- 11.Olah ME, Jacobson KA, Stiles GL. Identification of an adenosine receptor domain specifically involved in binding of 5′-substituted adenosine agonists. J Biol Chem. 1994;269:18016–18020. [PMC free article] [PubMed] [Google Scholar]
- 12.Rivkees SA, Lasbury ME, Barbhaiya H. Identification of domains of the human A1 adenosine receptor that are important for binding-receptor subtype-selective ligands using chimeric A1/A2a adenosine receptors. J Biol Chem. 1995;270:20485–20490. doi: 10.1074/jbc.270.35.20485. [DOI] [PubMed] [Google Scholar]
- 13.Barrington WW, Jacobson KA, Hutchison AJ, Williams M, Stiles GL. Identification of the A2 adenosine receptor binding subunit by photoaffinity crosslinking. Proc Natl Acad Sci US A. 1989;86:6572–6576. doi: 10.1073/pnas.86.17.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson KA, Pannell LK, Ji XD, Jarvis MF, Williams M, Hutchison AJ, Barrington WW, Stiles GL. Agonist-derived molecular probes for A2-adenosine receptors. J Mol Recognit. 1989;2:170–178. doi: 10.1002/jmr.300020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCabe RT, Skolnick P, Jacobson KA. FITC-APEC: A fluorescent ligand for A2-adenosine receptors. J Fluoresc. 1992;2:217–223. doi: 10.1007/BF00865279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piersen CE, True CD, Wells JN. I-125 2-[4-[2-[2-[(4-Azidophenyl)methylcarbonylamino]ethylaminocarbonyl]ethyl]-phenyl]ethylamino-5′-N-ethylcarboxamidoadenosine labels transmembrane span V of the A2A-adenosine receptor. Mol Pharmacol. 1994;45:871–877. [PubMed] [Google Scholar]
- 17.van Rhee AM, Jacobson KA. Molecular architecture of G protein-coupled receptors. Drug Dev Res. 1996;37:1–38. doi: 10.1002/(SICI)1098-2299(199601)37:1<1::AID-DDR1>3.0.CO;2-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Rhee AM, Fischer B, van Galen PJM, Jacobson KA. Modelling the P2Y purinoceptor using rhodopsin as template. Drug Des Discovery. 1995;13:133–154. [PMC free article] [PubMed] [Google Scholar]
- 19.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 20.Jarvis MF, Schulz R, Hutchison AJ, Do UH, Sills MA, Williams M. [3H]CGS 21680 a selective A2 adenosine receptor agonist directly labels A2 receptors in rat brain. J Pharmacol Exp Ther. 1989;251:888–893. [PubMed] [Google Scholar]
- 21.Bruns RF, Lu GH, Pugsley TA. Characterization of the A2 adenosine receptor labeled by [3H]NECA in rat striatal membranes. Mol Pharmacol. 1986;29:331–346. [PubMed] [Google Scholar]
- 22.Blüml K, Mutschler E, Wess J. Functional role in ligand binding and receptor activation of an asparagine residue present in the sixth transmembrane domain of all muscarinic acetylcholine receptors. J Biol Chem. 1994;269:18870–18876. [PubMed] [Google Scholar]
- 23.van Rhee AM, Jiang J-l, Melman N, Olah ME, Stiles GL, Jacobson KA. Interaction of 1,4-dihydropyridine and pyridine derivatives with adenosine receptors: selectivity for A3 receptors. J Med Chem. 1996;39:2980–2989. doi: 10.1021/jm9600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji XD, Melman N, Jacobson KA. Interactions of flavonoids and other phytochemicals with adenosine receptors. J Med Chem. 1996;39:781–788. doi: 10.1021/jm950661k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rhee AM, Siddiqi SM, Melman N, Shi D, Padgett WL, Daly JW, Jacobson KA. Tetrahydrobenzothiophenone derivatives as a novel class of adenosine receptor antagonists. J Med Chem. 1996;39:398–406. doi: 10.1021/jm9504823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji XD, Stiles GL, van Galen PJM, Jacobson KA. Characterization of human striatal A2-adenosine receptors using radioligand binding and photoaffinity labeling. J Recept Res. 1992;12:149–169. doi: 10.3109/10799899209074789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Askalan R, Richardson PJ. Role of histidine residues in the adenosine A2a receptor ligand binding site. J Neurochem. 1994;63:1477–1484. doi: 10.1046/j.1471-4159.1994.63041477.x. [DOI] [PubMed] [Google Scholar]
- 28.Garritsen A, IJzerman AP, Beukers MW, Cragoe EJ, Soudijn W. Interaction of amiloride and its analogues with adenosine A1 receptors in calf brain. Biochem Pharmacol. 1990;40:827–834. doi: 10.1016/0006-2952(90)90323-d. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson KA, van Rhee AM, Siddiqi SM, Ji XD, Jiang Q, Kim J, Kim HO. Molecular recognition in adenosine receptors. Perspect Recept Res. 1996:153–164. [Google Scholar]
- 30.Fong TM, Yu H, Cascieri MA, Underwood D, Swain CJ, Strader CD. The role of histidine 265 in antagonist binding to the neurokinin-1 receptor. J Biol Chem. 1994;269:2728–2732. [PubMed] [Google Scholar]
- 31.Ridge KD, Bhattacharya S, Nakayama TA, Khorana HG. Light-stable rhodopsin. II. An opsin mutant (Trp265Phe) and a retinal analog with a nonisomerizable 11-cis configuration form a photostable chromophore. J Biol Chem. 1992;267:6770–6775. [PubMed] [Google Scholar]
- 32.Perlman JH, Laakkonen L, Osman R, Gershengorn MC. A model of the thyrotropin-releasing hormone (TRH) receptor binding pocket. Evidence for a second direct interaction between transmembrane helix 3 and TRH. J Biol Chem. 1994;269:23383–23386. [PubMed] [Google Scholar]
- 33.Wang CD, Gallaher TK, Shih JC. Site-directed mutagenesis of the serotonin 5-hydroxytrypamine2 receptor: identification of amino acids necessary for ligand binding and receptor activation. Mol Pharmacol. 1993;43:931–940. [PubMed] [Google Scholar]
- 34.Ohta K, Hayashi H, Mizuguchi H, Kagamiyama H, Fujimoto K, Fukui H. Site-directed mutagenesis of the histamine H1 receptor: roles of aspartic acid107, asparagine198 and threonine194. Biochem Biophys Res Commun. 1994;203:1096–1101. doi: 10.1006/bbrc.1994.2295. [DOI] [PubMed] [Google Scholar]
- 35.Strader CD, Candelore MR, Hill WS, Dixon RA, Sigal IS. A single amino acid substitution in the beta-adrenergic receptor promotes partial agonist activity from antagonists. J Biol Chem. 1989;264:16470–16477. [PubMed] [Google Scholar]
- 36.Strader CD, Gaffney T, Sugg EE, Candelore MR, Keys R, Patchett AA, Dixon RA. Allele-specific activation of genetically engineered receptors. J Biol Chem. 1991;266:5–8. [PubMed] [Google Scholar]
- 37.Javitch JA, Fu D, Chen J, Karlin A. Mapping the binding-site crevice of the dopamine D2 receptor by the substituted-cysteine accessibility method. Neuron. 1995;14:825–831. doi: 10.1016/0896-6273(95)90226-0. [DOI] [PubMed] [Google Scholar]
- 38.Cho W, Taylor LP, Mansour A, Akil H. Hydrophobic residues of the D2 dopamine-receptor are important for binding and signal-transduction. J Neurochem. 1995;65:2105–2115. doi: 10.1046/j.1471-4159.1995.65052105.x. [DOI] [PubMed] [Google Scholar]
- 39.Strader CD, Candelore MR, Hill WS, Sigal IS, Dixon RA. Identification of two serine residues involved in agonist activation of the beta-adrenergic receptor. J Biol Chem. 1989;264:13572–13578. [PubMed] [Google Scholar]
- 40.Wess J, Gdula D, Brann MR. Site-directed mutagenesis of the m3 muscarinic receptor: identification of a series of threonine and tyrosine residues involved in agonist but not antagonist binding. EMBO J. 1991;10:3729–3734. doi: 10.1002/j.1460-2075.1991.tb04941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.IJzerman AP, van der Wenden EM, van Galen PJM, Jacobson KA. Molecular modeling of adenosine receptors-the ligand-binding site on the rat adenosine A2a receptor. Eur J Pharmacol Mol Pharmacol Sect. 1994;268:95–104. doi: 10.1016/0922-4106(94)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hibert MF, Trumpp-Kallmeyer S, Bruinvels A, Hoflack J. Three-dimensional models of neurotransmitter G-binding protein-coupled receptors. Mol Pharmacol. 1991;40:8–15. [PubMed] [Google Scholar]
- 43.Nordvall G, Hacksell U. Binding-site modeling of the muscarinic m1 receptor: a combination of homology-based and indirect approaches. J Med Chem. 1993;36:967–976. doi: 10.1021/jm00060a003. [DOI] [PubMed] [Google Scholar]
- 44.van der Wenden EM, Price SL, Apaya RP, IJzerman AP, Soudijn W. J Comput-Aided Mol Des. 1995;9:44–54. doi: 10.1007/BF00117277. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy AP, Mangum KC, Linden J, Wells JN. Covalent modification of transmembrane span III of the A1 adenosine receptor with an antagonist photoaffinity probe. Biochemistry. 1996;50:789–798. [PubMed] [Google Scholar]
- 46.Okayama H, Berg PA. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983;3:280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higuchi R. Using PCR to Engineer DNA. In: Ehrlich HA, editor. PCR Technology. Stockton Press; New York: 1989. pp. 61–70. [Google Scholar]
- 48.Sanger R, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci US A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cullen BR. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 50.Cheng YC, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 percent inhibition (IC50) of an enzyme reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]