Abstract
A method has been designed for patterning supported phospholipid bilayers (SLBs) on planar substrates and inside microfluidic channels. To do this, bovine serum albumin (BSA) monolayers were formed via adsorption at the liquid/solid interface. Next, this interfacial protein film was selectively patterned by using deep UV lithography. Subsequently, SLBs could be deposited in the patterned locations by vesicle fusion. By cycling through this process several times, spatially addressed bilayer arrays could be formed with intervening protein molecules serving as two-dimensional corrals. By employing this method, phospholipid bilayers containing various concentrations of ganglioside GM1 were addressed along the length of individual microfluidic channels. Therefore, the binding of GM1 with pentameric cholera toxin B (CTB) subunits could be probed. A seven-channel microfluidic device was fabricated for this purpose. Each channel was simultaneously patterned with four chemically distinct SLBs containing 0, 0.2, 0.5, and 2.0 mol % GM1, respectively. Varying concentrations of CTB were then introduced into each of the channels. With the use of total internal reflection fluorescence microscopy, it was possible to simultaneously abstract multiple equilibrium dissociation constants as a function of ligand density for the CTB-GM1 system in a single shot.
Multivalent ligand–receptor interactions are ubiquitous on cell surfaces. They have a wide variety of consequences including the modulation of equilibrium dissociation constants, high receptor selectivity, and receptor clustering.1 Multivalency can also play a direct role in signal transduction processes.2 Examination of the underlying thermodynamics of ligand–receptor binding may lead to a greater understanding of its biological role and could provide insight into biomedical applications involving inhibitory drug design.1,3,4 Unfortunately, high-throughput, low-protein consumption assays are not presently well enough developed in this field to afford rapid, systematic studies of binding data at two-dimensionally fluid bilayer interfaces.5
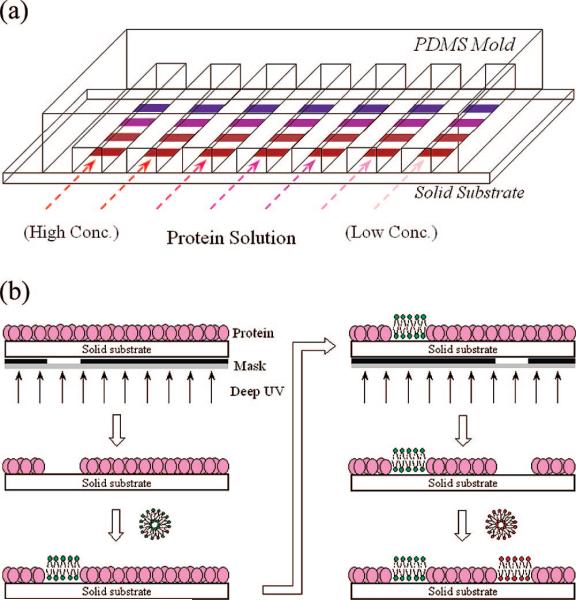
In previous work it has been demonstrated that microfluidic devices can be designed for measuring binding affinities in multivalent systems.5–14 In our previous setup, ligands were incorporated into supported lipid bilayers (SLBs) coated on the walls and floors of polydimethylsiloxane/glass microchannels. Linear arrays of channels were then monitored by total internal reflection fluorescence microscopy (TIRFM)15 to obtain equilibrium dissociation constants in one-shot assays. This could be done by using the same surface chemistry in each microchannel while varying the solution concentration of the aqueous proteins. Such a setup made these assays relatively rapid, afforded high accuracy, and used only a few microliters of protein solution. Nevertheless, such platforms could be markedly improved by measuring multiple binding constants with various membrane chemistries simultaneously. This could be achieved by addressing distinct lipid bilayer chemistries along individual microfluidic channels while varying the solution concentration of aqueous proteins introduced above the entire array (Figure 1a). This would essentially amount to a two-dimensional assay whereby both the surface chemistry and aqueous solution conditions are varied on a single chip. Such assays would be extremely useful for probing multivalent binding as a function of ligand density, cholesterol content, membrane charge, and related variables in one-shot experiments. Such designs are also reminiscent of other on-chip two-dimensional assays.12,16–18
Figure 1.
(a) Schematic representation of a seven-channel microfluidic device. Each channel is addressed with four distinct lipid bilayers designated by colored patches. Aqueous solutions with various protein concentrations are arrayed from left to right. (b) Schematic diagram of the sequential patterning technique employed for creating spatially addressed bilayer arrays.
The key to achieving two-dimensional binding assays is to create spatially addressed arrays of bilayers along individual microchannels. There have already been a number of techniques developed to array different bilayer chemistries at each address. For example, we first showed the use of pulled microcapillaries to deliver unique vesicle solutions to an array of hydrophilic patches patterned on a substrate with hydrophobic barriers.19 Other methods for creating spatially addressed bilayer arrays were also developed. These included microcontact printing,20–23 mechanical writing and erasing,24 laminar flow vesicle deposition,25 robotic pin printing,26 scanning probe lithography,27 and dip-pen nanolithography.28 Although each of these techniques has its merits, it would be most convenient to develop a procedure that allows bilayers to be arrayed inside enclosed microchannels with a well characterized and predetermined chemistry at each location. The most attractive way of obtaining such a result would be to develop a light directed patterning method analogous to the ones we have already developed for arraying immobilized ligands inside microfluidic devices.29,30
Pioneering work has been done by the Parikh laboratory to pattern solid supported lipid bilayers using deep UV radiation.31–34 Additionally, deep UV radiation had been employed to pattern organosilane monolayers,35,36 polysaccharides,37 and S-layer proteins.38 In this paper, we extend this idea to patterning sacrificial adsorbed protein layers at the liquid/solid interface. We show that it is possible to deposit films made from bovine serum albumin (BSA). Areas containing the protein layer resist the fusion of phospholipid vesicles, while irradiated regions allow supported bilayers to form by the fusion of vesicles from bulk solution (Figure 1b). We exploit this ability to pattern linear arrays of supported phospholipid membranes containing four different concentrations of ganglioside GM1. Each channel in a seven-channel device contained the identical membrane chemistry. Therefore, it was possible to simultaneously obtain 28 unique data points for CTB-GM1 interactions by flowing a unique concentration of the protein into each channel. This afforded three distinct equilibrium binding curves plus one set of control data for the nonspecific adsorption of CTB on bilayers containing no GM1 ligands.
EXPERIMENTAL SECTION
Materials
Bovine serum albumin (BSA, catalog number A-7638, lyophilized powder) was purchased from Sigma (St. Louis, MO). The protein should contain bound fatty acids. Texas Red-conjugated BSA, Rabbit IgG, and Texas Red-labeled 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (Texas Red-DHPE) were purchased from Molecular Probes, Inc. (Eugene, OR). CTB from Vibrio cholerae was obtained from Sigma-Aldrich. Ganglioside GM1 (brain, ovine-ammonium salt), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (ammonium salt) (NBD-PE) were purchased from Avanti Polar Lipids (Alabaster, AL). Polydimethylsiloxane (PDMS) was obtained from Dow Corning (Sylgard, silicone elastomer-184).
CTB was labeled with Alexa Fluor-594 dye using a standard protein-labeling kit (A10239, Molecular Probes, Eugene, OR). The degree of labeling was about 0.8 fluorophores per protein as determined by UV–vis absorption spectroscopy. The dye-labeled protein was stored in a phosphate buffered saline (PBS) solution which consisted of 10 mM sodium phosphate, 150 mM NaCl, and 0.2 mM sodium azide. The pH of the PBS solution was set to 7.2 by the dropwise addition of 2.0 M NaOH. This buffer was also used for vesicle preparation and the successive dilution of protein solutions. Purified water for these experiments came from a NANOpure Ultrapure Water System (≥18.2 MΩ cm, Barnstead, Dubuque, IA). Quartz coverslips (1 in. × 1 in., 0.2 mm thick, Electron Microscopy Sciences, Hatfield, PA) were used as substrates for lipid bilayer formation. Electron microscopy grids (EM-grids, Gilder Grids from Electron Microscopy Sciences, Hatfield, PA) were employed as photomasks for deep UV lithography.
Small Unilamellar Vesicles Preparation
Small unilamellar vesicles (SUVs) were prepared by vesicle extrusion as reported previously.6,39,40 Briefly, the appropriate lipid mixtures were combined in organic solvent. Chloroform was used to dissolve most lipids; however, methanol was used as the solvent for GM1 as this glycolipid is not soluble in pure chloroform. The chloroform/methanol solvent mixture was evaporated away under a stream of nitrogen followed by vacuum desiccation for 4 h. This left the desired lipid mixture behind, which was then rehydrated in PBS. The concentration of lipids in solution was 2.0 mg/mL. After five freeze–thaw cycles, the solutions were then extruded more than seven times through a polycarbonate filter (Whatman) with 50 nm pores. SUVs prepared by this method were characterized by dynamic light scattering (90Plus particle size analyzer, Brookhaven Instruments Corp.) and showed an average diameter of 70 ± 10 nm.
Microfluidic Device Fabrication
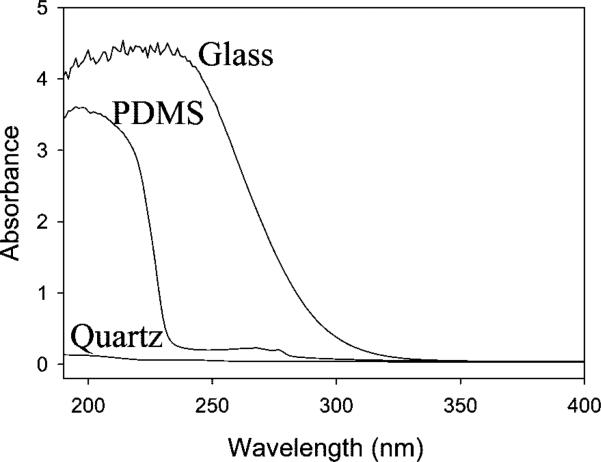
Seven-channel PDMS/quartz microfluidic devices were fabricated largely according to our previously published methods.13 The key difference herein is that we employed a 0.2 mm thick piece of square fused quartz as the substrate rather than borosilicate glass. Fused quartz was used because it passes light deep into the UV. For reference, UV–vis spectra (Lambda 35, UV–vis spectrometer, PerkinElmer, Inc.) for 1 mm thick quartz, PDMS, and borosilicate slides are provided in Figure 2. To make a device, a seven-channel pattern was transferred from a photomask onto a clean soda lime microscope slide by standard photoresist and buffered oxide etchant (BOE) techniques. Degassed PDMS was then poured over this glass master and cured in a convection oven at 55 °C overnight. The elastomeric mold was carefully peeled off, washed with ethanol and purified water, and dried under a stream of nitrogen. Inlets were reamed at the channel termini using a hollow flat-tipped syringe needle. Finally, the PDMS mold and a clean planar quartz coverslip were placed in an oxygen plasma for 30 s (Basic Plasma Cleaner, Harrick Scientific). The two were immediately brought into contact to create the finished seven-channel microfluidic device. It should be noted that the quartz substrate was cleaned in a boiling 1:3 solution of ICN 7X detergent and purified water. It was then rinsed with copious amounts of purified water, dried with nitrogen, and annealed in a kiln at 500 °C for 5 h before use.
Figure 2.
UV–vis spectra for 1 mm thick quartz, PDMS, and borosilicate glass substrates.
Epifluorescence and TIRFM
Epifluorescence images of protein patterns and SLBs were obtained using a Nikon E800 fluorescence microscope with a Roper Scientific MicroMAX 1024B charge-coupled device (CCD) camera (Princeton Instruments). Fluorescence recovery after photobleaching (FRAP)41,42 experiments were conducted to check the quality and fluidity of the SLBs. This was done using an inverted epifluorescence Nikon Eclipse TE2000-U microscope equipped with a 10× objective. Laser radiation from a 2.5 W mixed gas Ar+/K+ laser (Stabilite 2018, Spectra Physics) was used to bleach the lipid bilayer samples. FRAP images were captured with a Photometrics SenSys CCD camera.
TIRFM15 was employed in CTB-GM1 binding studies to discriminate surface-bound CTB molecules from those in the bulk solution. To perform a TIRFM experiment, a 594 nm helium–neon laser beam (4 mW, Uniphase, Manteca, CA) was passed through a dove prism that was optically coupled to the quartz substrate of the microfluidic device by index matching immersion oil. The laser beam was telescoped out by a line generator lens (BK7 for 30°, Edmund Optics, Barrington, NJ) to create a uniform intensity profile across the microchannel array. The laser illuminated the quartz/water interface from the quartz side at an angle of 78° with respect to the surface normal and was totally internally reflected. This created an evanescent wave above the interface, which exponentially decayed to its 1/e value over ~70 nm.6,43 This allowed the proteins bound to the SLBs to be studied with high specificity. All images were collected using Metamorph software (Universal Imaging Corp.) and transferred to Microsoft Excel and Sigma Plot for further processing.
RESULTS
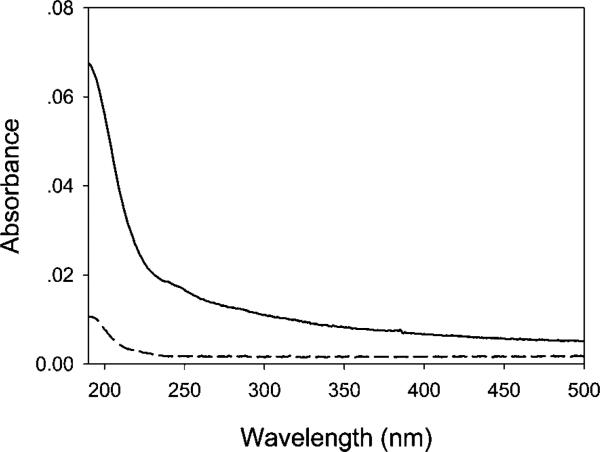
In a first set of experiments, the use of deep UV radiation to degrade an adsorbed protein monolayer was tested. To do this, a 10 mg/mL solution of BSA was introduced above a planar quartz surface and allowed to incubate for 20 min. This should form a continuous monolayer of BSA at the quartz/water interface.44,45 Excess protein molecules were washed away with copious amounts of purified water. Next, the substrate was exposed to deep UV radiation from a mercury Pen-Ray lamp (UVP Inc., Upland, CA) in a quartz envelope. Approximately 2.8 W of radiation with peaks at 190 and 254 nm were introduced to the sample surface for 2 min over ~2.5 cm2 area. The surface was then washed with copious amounts of purified water. UV-vis spectra of the BSA-coated quartz substrate were taken before and after exposure to the Pen-Ray lamp radiation. These are shown in Figure 3, and the data clearly demonstrate that the protein layer is highly degraded upon treatment with UV radiation. Such a result is consistent with previous reports that deep UV radiation (~190 nm) can be used to degrade organosliane monolayers,35,36 polysaccharides,37 and S-layer proteins.38 It should be noted that the small peak near 280 nm typically found in spectra from bulk BSA solutions does not appear to be present in Figure 3. This is probably due to the very small number of molecules measured at the surface compared with bulk assays. The protein adsorption and rearrangement processes at the surface may also alter the UV–vis spectrum.
Figure 3.
UV–vis spectra for BSA-coated quartz substrates before (–) and after (- - -) exposure to deep UV radiation.
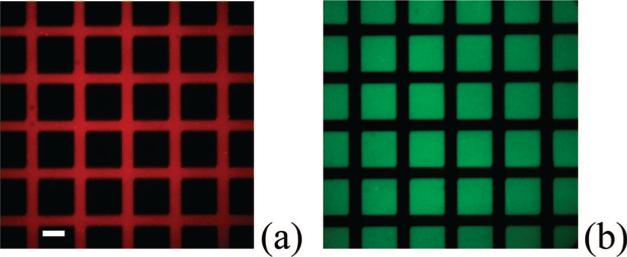
In a next set of experiments, an EM-grid was used as a photomask for patterning a BSA monolayer under conditions that were similar to Figure 3. In this case, however, protein molecules were conjugated with a Texas Red fluorophore so that the resulting pattern could be visualized under a fluorescence microscope (Figure 4a). As can be seen, ~90 μm × 90 μm square nonfluorescent regions were created that were separated by ~35 μm wide fluorescent lines. This was identical to the dimensions of the 200 mesh EM-grid used to make the pattern. At this point, POPC vesicle solutions containing 2.0 mol % NBD-PE were introduced above the patterned surface and allowed to create supported bilayers by the vesicle fusion method (Figure 4b).46 Significantly, supported POPC membranes were only deposited in the areas where the BSA monolayer was exposed to UV radiation.
Figure 4.
(a) An epifluorescence image of a Texas Red-conjugated BSA film on a quartz substrate after deep UV patterning. (b) An epifluorescence image of a 2.0 mol % NBD-PE/POPC bilayer array deposited over the same protein patterned film shown in part a. The scale bar is 50 μm.
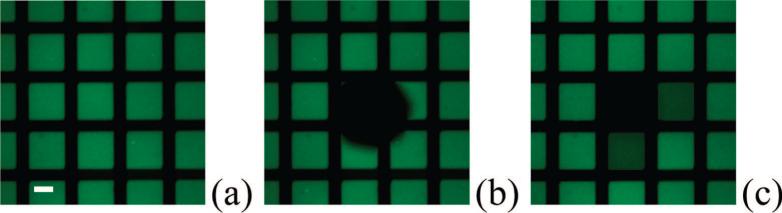
It should be noted that the deep UV illumination time is a key parameter for creating high quality supported bilayers. For example, no bilayers were deposited on a BSA-coated substrate without deep UV exposure (Figure 5a). Moreover, POPC vesicle fusion was incomplete when the illumination time was substantially shorter than 2 min. An example of this is given in Figure 5b where uneven supported bilayers are observed when the exposure time was limited to 30 s. Next, Figure 5c shows high quality bilayers with an illumination time of 2 min. Finally, we found that very long exposure times appeared to degrade the BSA barriers between the corralled bilayers. To demonstrate this, a 300 s exposure time is shown in Figure 5d.
Figure 5.
Epifluorescence images of supported POPC bilayers containing 2.0 mol % NBD-PE. The bilayers have been deposited on BSA-coated quartz substrates after deep UV patterning for (a) 0, (b) 30, (c) 120, and (d) 300 s. The scale bar is 50 μm.
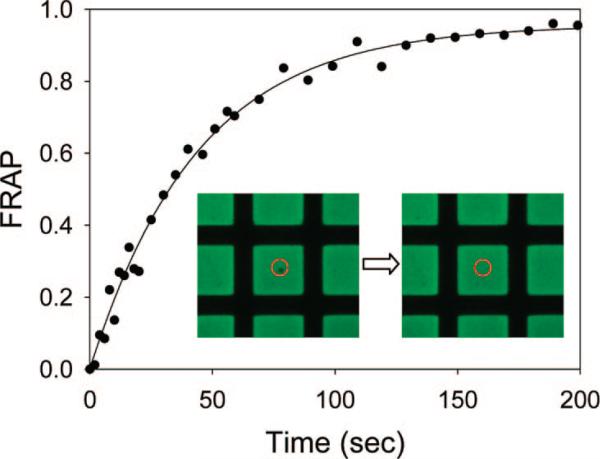
The two-dimensional fluidity of the supported bilayers inside individual two-dimensional protein corrals was probed by fluorescence recovery after photobleaching (Figure 6).41,42 The diffusion constant for the NBD-PE lipid was 3.4 ± 0.8 μm2/s and the mobile fraction was ~95 ± 2%. Moreover, individual SLBs were completely confined within their protein boundaries.47 To demonstrate this, the fluorescence recovery of a large bleach spot is shown in Figure 7. Taken together, the results in Figures 6 and 7 are strong evidence that high-quality supported bilayers can be patterned on BSA-coated substrates that are selectively illuminated by deep UV radiation.
Figure 6.
FRAP recovery curve for a 2.0 mol % NBD-PE/POPC bilayer on quartz formed by vesicle fusion after deep UV patterning of the BSA-coated substrate. The inset images were taken immediately after photobleaching with a 14 μm diameter laser spot as well as 200 s later (left and right image, respectively). The red circles designate the position of the bleach spot. The size of a bilayer square is ~90 μm × 90 μm.
Figure 7.
Photobleaching of a 2.0 mol % NBD-PE/POPC bilayer inside protein corrals. The bilayer array is shown (a) before bleaching, (b) immediately after bleaching, and (c) 15 min later. The scale bar is 50 μm.
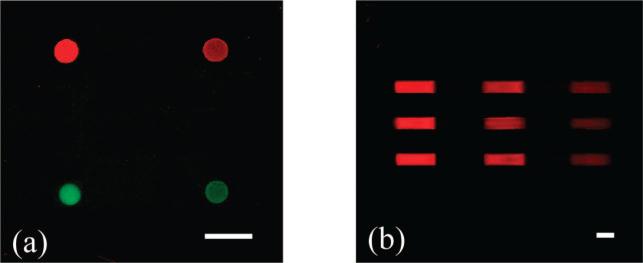
Next, it was necessary to demonstrate that this bilayer patterning method could be performed in sequential steps to spatially address distinct bilayer chemistries at the surface. The general idea is illustrated in Figure 1b. First, a fresh BSA monolayer was formed at the liquid/solid interface on a quartz substrate. Protein molecules from specific regions were selectively removed by UV illumination through a hole with a diameter of 100 μm. A supported lipid bilayer was then addressed over the light exposed region via vesicle fusion. In a next step, the photomask was aligned to a new location and the process was repeated with a fresh vesicle solution. Figure 8a shows a false color epifluorescence image of four circles addressed with POPC lipids contain 0.5 mol % Texas Red-DHPE (upper left), 0.1 mol % Texas Red-DHPE (upper right), 2.0 mol % NBD-PE (lower left), and 0.5 mol % NBD-PE (lower right). The circular supported bilayers had an average diameter of 99 ± 4 μm. It should be noted that control experiments demonstrated that sequential deposition of bilayers does not result in any significant mixing of lipids between a previously formed supported bilayer patch and nascently introduced vesicle solutions. It is well-known that individual lipids can exchange between the two membranes;48 however, this exchange process is slow compared to the time scale used for vesicle fusion to the substrate (less than 5 min).
Figure 8.
(a) A false color epifluorescence image of four spatially addressed bilayers. Top left, 0.5 mol % Texas Red-DHPE/POPC; top right, 0.1 mol % Texas Red-DHPE/POPC; bottom left, 2.0 mol % NBD-PE/POPC; and bottom right, 0.5 mol % NBD-PE/POPC. (b) An epifluorescence image of spatially addressed supported bilayers inside a linear array of microfluidic channels. The long access of each channel goes from left to right across the image. The three POPC bilayer patches along each channel contain 0.5, 0.2, and 0.05 mol % Texas Red-DHPE from left to right. The scale bar is 200 μm.
With the use of the strategy outlined in Figure 8a, SLBs with a variety of different components could be spatially addressed along a line inside microfluidic channels. To demonstrate this, protein solutions were individually injected into three parallel microchannels and allowed to incubate for 20 min to form a thin protein film coating the quartz surface. Protein molecules should also coat and passivate the PDMS walls.30 After incubation, purified water was flushed through the channels to remove excess proteins. Next, deep UV radiation was exposed to the channels via a single ~450 μm wide slit that was aligned normal to the direction of the channel flow. This photomask was placed beneath the microfluidic device to allow UV radiation to pass through the quartz substrate as shown in Figure 1b. After selectively radiating and rinsing away protein molecules, a vesicle solution consisting of 0.5 mol % Texas Red-DHPE in POPC was injected into each microchannel, allowed to incubate for 5 min, and rinsed away with buffer. This should form a continuous supported bilayer on the PDMS walls as well as the quartz substrate.49 The 450 μm wide slit was then translated ~1.0 mm along the channels and the process was repeated with POPC vesicles containing 0.2 mol % Texas Red-DHPE. Finally, the process was repeated one last time with POPC vesicles containing 0.05 mol % Texas Red-DHPE. An epifluorescence image of the resulting patterned surface is shown in Figure 8b.
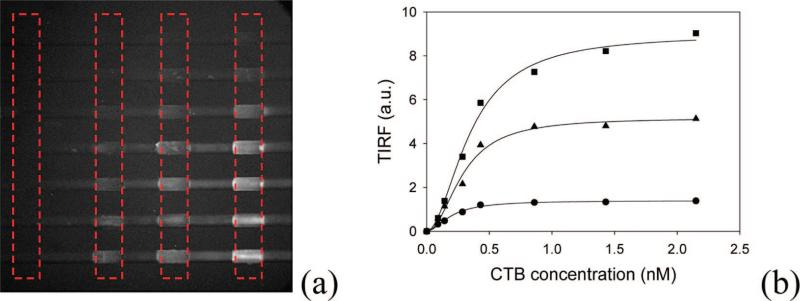
In a final set of experiments, we wished to exploit this spatial addressing process to obtain equilibrium dissociation constant data from the CTB-GM1 system as a function of ligand density. To do this, a seven-channel microfluidic device was used. Each channel was addressed with ~350 μm wide POPC bilayer patches containing 0, 0.2, 0.5, and 2.0 mol % GM1 from left to right. TIRF microscopy was applied to study the pentavalent CTB-GM1 interactions as a function of ligand density. Before the injection of CTB solutions, the bilayer-coated microchannels were incubated for 30 min with a 0.5 mg/mL rabbit IgG antibody solution to suppress nonspecific adsorption. After rinsing, various concentrations of Alexa 594-labeled CTB solutions were flowed simultaneously through each channel at a rate of 0.2 μL/min. The surface binding process was monitored by TIRF microscopy as a function of time until the fluorescence intensity remained constant. A TIRF image of this system is shown in Figure 9a. The bulk CTB concentration ranged from 0.09 to 2.15 nM (top to bottom). The column on the far left, which consisted of bilayers without GM1, was used to judge nonspecific CTB adsorption at each bulk protein concentration. It was found that the background fluorescence level was below the detection limit under all circumstances. This finding is in agreement with previous measurements.13,50 By contrast, some nonspecific CTB adsorption was found outside the bilayer patches on the BSA-coated regions, although this does not affect the binding measurements. Intensity profiles across the channels (dashed red rectangles shown in Figure 9a) were employed to obtain quantitative binding data for each of the three GM1 densities. A plot of the fluorescence signal as a function of bulk protein concentration for each ligand density is provided in Figure 9b.
Figure 9.
(a) TIRF image of bilayer-arrayed microchannels containing various concentrations of dye-labeled CTB. The four sets of bilayer patches designated with the dashed red boxes represent identical membrane chemistries. From left to right, the bilayer patches contain 0.0, 0.2, 0.5, and 2.0 mol % GM1 in POPC, respectively. The long access of each channel is from left to right across the image. (b) TIRF signal vs bulk CTB concentration for each of the three GM1 concentrations: 2.0 (■), 0.5 (▲), and 0.2 mol % (●). The solid lines are fits to the data with eq 1.
To abstract equilibrium dissociation constant data for the CTB-GM1 system, the curves were fit to the Hill–Waud (eq 1) binding model:8,13,51
| (1) |
where F is the fluorescence intensity from surface bound proteins, Fmax is the maximum fluorescence intensity when proteins completely saturated the bilayer surface, [P] is the bulk CTB concentration, Kd is the apparent equilibrium dissociation constant, and n is the Hill coefficient of cooperativity.51 In our previous work, this binding model was found to be more suitable to pentavalent CTB-GM1 interactions than the simple Langmuir adsorption model.13 The apparent equilibrium dissociation constants extracted from Figure 9b are 0.20, 0.29, and 0.35 nM for 0.2, 0.5, and 2.0 mol % GM1, respectively. These results are in good agreement with our previous studies of this sytem.13 Indeed, the apparent equilibrium dissociation constant should weaken for this system with increased GM1 concentration because of ligand clustering in the membrane. It should also be noted that the measured dissociation constant values for the CTB-GM1 system have ranged from several picomolar to several hundred nanomolar depending upon specific conditions.8,52 Most results, however, have typically fallen within the 0.1–14 nM range.13,50,51,53–58 These differences can probably be attributed to the variety of buffer conditions, temperatures, ligand densities, and membrane chemistries employed in the various experiments. Differences in the diagnostics used to make measurements as well as the specific binding model used for fitting may also affect Kd values.
DISCUSSION
Patterning supported bilayers has shown considerable promise for addressing fundamental biophysical questions about cell membrane behavior and the creation of a new generation of biosensors.59–61 Moreover, various ideas for forming spatially addressed bilayer assays have been pursued over the past decade.19–21,25–28,62 A key innovation of the present work is to combine spatially addressed arraying technology with microfluidics strategies. This allows not only the bilayer chemistry to be varied but also the aqueous solutions above them. Such technology is quite powerful because arraying chemistry along one dimension and the solution conditions along the other opens up the door for studying ligand–receptor binding in a high throughput/low sample volume fashion whereby multiple Kd values can be abstracted simultaneously. Although we have demonstrated the ability to obtain dissociation constants as a function of ligand density, other variables such as cholesterol content, charge on the membrane, lipid tailgroup, and headgroup chemistry could also be varied. Other applications might include enzyme patterning and characterization,30,49 cell-supported bilayer interactions,63 as well as microfluidic drug discovery platforms.26
Now that spatially addressed bilayer arrays have been combined with microfluidics, there are three general goals that need to be pursued in association with this analytical methodology. First, the array size could be expanded by increasing the field of view of the fluorescence microscope. Indeed, the image in Figure 9 was captured with a 4× objective. This led to simultaneous data capture for four unique bilayer chemistries and seven bulk protein concentrations (28 unique data points). It should also be possible, however, to use a 1× objective from an epifluorescence macroscope to increase the field of view by a factor of 16.62 Moreover, a denser array of bilayer chemistries and/or channels could also be employed. Second, data from these systems could be taken in a label-free fashion by using techniques such as surface plasmon resonance imagining,16,64 imaging ellipsometry,65,66 or even interferometry.67 Care will need to be taken with such designs to ensure that they are compatible with the UV patterning techniques employed herein. Finally, new methods for simultaneously, rather than sequentially, addressing bilayers should be pursued. This will speed up the time required for array formation.
ACKNOWLEDGMENT
We thank the NIH (Grant R01 GM070622), the ARO (Grant W911NF-05-1-0494), and the ONR (Grant N00014-08-1-0467) for support.
References
- 1.Choi SK. Synthetic Multivalent Molecules. Wiley; Hoboken, NJ: 2004. [Google Scholar]
- 2.Clemons PA. Curr. Opin. Chem. Biol. 1999;3:112–115. doi: 10.1016/s1367-5931(99)80020-9. [DOI] [PubMed] [Google Scholar]
- 3.Mammen M, Choi SK, Whitesides GM. Angew. Chem., Int. Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Kiessling LL, Gestwicki JE, Strong LE. Angew. Chem., Int. Ed. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang TL, Baryshnikova OK, Mao HB, Holden MA, Cremer PS. J. Am. Chem. Soc. 2003;125:4779–4784. doi: 10.1021/ja029469f. [DOI] [PubMed] [Google Scholar]
- 6.Yang TL, Jung SY, Mao HB, Cremer PS. Anal. Chem. 2001;73:165–169. doi: 10.1021/ac000997o. [DOI] [PubMed] [Google Scholar]
- 7.Griffitts JS, Haslam SM, Yang TL, Garczynski SF, Mulloy B, Morris H, Cremer PS, Dell A, Adang MJ, Aroian RV. Science. 2005;307:922–925. doi: 10.1126/science.1104444. [DOI] [PubMed] [Google Scholar]
- 8.Moran-Mirabal JM, Edel JB, Meyer GD, Throckmorton D, Singh AK, Craighead HG. Biophys. J. 2005;89:296–305. doi: 10.1529/biophysj.104.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips KS, Cheng Q. Anal. Chem. 2005;77:327–334. doi: 10.1021/ac049356+. [DOI] [PubMed] [Google Scholar]
- 10.Cannon B, Weaver N, Pu QS, Thiagarajan V, Liu SR, Huang JY, Vaughn MW, Cheng KH. Langmuir. 2005;21:9666–9674. doi: 10.1021/la0502645. [DOI] [PubMed] [Google Scholar]
- 11.Kim P, Lee SE, Jung HS, Lee HY, Kawai T, Suh KY. Lab Chip. 2006;6:54–59. doi: 10.1039/b512593f. [DOI] [PubMed] [Google Scholar]
- 12.Taylor JD, Phillips KS, Cheng Q. Lab Chip. 2007;7:927–930. doi: 10.1039/b618940g. [DOI] [PubMed] [Google Scholar]
- 13.Shi JJ, Yang TL, Kataoka S, Zhang YJ, Diaz AJ, Cremer PS. J. Am. Chem. Soc. 2007;129:5954–5961. doi: 10.1021/ja069375w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung HS, Yang T, Lasagna MD, Shi JJ, Reinhart GD, Cremer PS. Biophys. J. 2008;94:3094–3103. doi: 10.1529/biophysj.107.115519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Axelrod D, Burghardt TP, Thompson NL. Annu. Rev. Biophys. Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- 16.Lee HJ, Goodrich TT, Corn RM. Anal. Chem. 2001;73:5525–5531. doi: 10.1021/ac010762s. [DOI] [PubMed] [Google Scholar]
- 17.Wegner GJ, Lee HJ, Corn RM. Anal. Chem. 2002;74:5161–5168. doi: 10.1021/ac025922u. [DOI] [PubMed] [Google Scholar]
- 18.Su J, Bringer MR, Ismagilov RF, Mrksich M. J. Am. Chem. Soc. 2005;127:7280–7281. doi: 10.1021/ja051371o. [DOI] [PubMed] [Google Scholar]
- 19.Cremer PS, Yang TL. J. Am. Chem. Soc. 1999;121:8130–8131. [Google Scholar]
- 20.Majd S, Mayer M. Angew. Chem., Int. Ed. 2005;44:6697–6700. doi: 10.1002/anie.200502189. [DOI] [PubMed] [Google Scholar]
- 21.Hovis JS, Boxer SG. Langmuir. 2001;17:3400–3405. [Google Scholar]
- 22.Sapuri AR, Baksh MM, Groves JT. Langmuir. 2003;19:1606–1610. [Google Scholar]
- 23.Jung SY, Holden MA, Cremer PS, Collier CP. ChemPhysChem. 2005;6:423–426. doi: 10.1002/cphc.200400540. [DOI] [PubMed] [Google Scholar]
- 24.Cremer PS, Groves JT, Kung LA, Boxer SG. Langmuir. 1999;15:3893–3896. [Google Scholar]
- 25.Kam L, Boxer SG. J. Am. Chem. Soc. 2000;122:12901–12902. [Google Scholar]
- 26.Yamazaki V, Sirenko O, Schafer RJ, Nguyen L, Gutsmann T, Brade L, Groves JT. BMC Biotechnol. 2005;5:18. doi: 10.1186/1472-6750-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson BL, Groves JT. J. Am. Chem. Soc. 2004;126:13878–13879. doi: 10.1021/ja046040a. [DOI] [PubMed] [Google Scholar]
- 28.Lenhert S, Sun P, Wang YH, Fuchs H, Mirkin CA. Small. 2007;3:71–75. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- 29.Holden MA, Cremer PS. J. Am. Chem. Soc. 2003;125:8074–8075. doi: 10.1021/ja035390e. [DOI] [PubMed] [Google Scholar]
- 30.Holden MA, Jung SY, Cremer PS. Anal. Chem. 2004;76:1838–1843. doi: 10.1021/ac035234q. [DOI] [PubMed] [Google Scholar]
- 31.Yee CK, Amweg ML, Parikh AN. Adv. Mater. 2004;16:1184–1189. [Google Scholar]
- 32.Yee CK, Amweg ML, Parikh AN. J. Am. Chem. Soc. 2004;126:13962–13972. doi: 10.1021/ja047714k. [DOI] [PubMed] [Google Scholar]
- 33.Howland MC, Sapuri-Butti AR, Dixit SS, Dattelbaum AM, Shreve AP, Parikh AN. J. Am. Chem. Soc. 2005;127:6752–6765. doi: 10.1021/ja043439q. [DOI] [PubMed] [Google Scholar]
- 34.Yu CH, Parikh AN, Groves JT. Adv. Mater. 2005;17:1477–1480. [Google Scholar]
- 35.Dulcey CS, Georger JH, Krauthamer V, Stenger DA, Fare TL, Calvert JM. Science. 1991;252:551–554. doi: 10.1126/science.2020853. [DOI] [PubMed] [Google Scholar]
- 36.Mooney JF, Hunt AJ, McIntosh JR, Liberko CA, Walba DM, Rogers CT. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12287–12291. doi: 10.1073/pnas.93.22.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka M, Wong AP, Rehfeldt F, Tutus M, Kaufmann S. J. Am. Chem. Soc. 2004;126:3257–3260. doi: 10.1021/ja038981d. [DOI] [PubMed] [Google Scholar]
- 38.Sleytr UB, Messner P, Pum D, Sara M. Angew. Chem., Int. Ed. 1999;38:1035–1054. doi: 10.1002/(SICI)1521-3773(19990419)38:8<1034::AID-ANIE1034>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 39.Kalb E, Frey S, Tamm LK. Biochim. Biophys. Acta. 1992;1103:307–316. doi: 10.1016/0005-2736(92)90101-q. [DOI] [PubMed] [Google Scholar]
- 40.Nollert P, Kiefer H, Jahnig F. Biophys. J. 1995;69:1447–1455. doi: 10.1016/S0006-3495(95)80014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albertorio F, Diaz AJ, Yang TL, Chapa VA, Kataoka S, Castellana ET, Cremer PS. Langmuir. 2005;21:7476–7482. doi: 10.1021/la050871s. [DOI] [PubMed] [Google Scholar]
- 43.Burmeister JS, Olivier LA, Reichert WM, Truskey GA. Biomaterials. 1998;19:307–325. doi: 10.1016/s0142-9612(97)00109-9. [DOI] [PubMed] [Google Scholar]
- 44.Wertz CF, Santore MM. Langmuir. 2001;17:3006–3016. [Google Scholar]
- 45.Sweryda-Krawiec B, Devaraj H, Jacob G, Hickman JJ. Langmuir. 2004;20:2054–2056. doi: 10.1021/la034870g. [DOI] [PubMed] [Google Scholar]
- 46.Sackmann E. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 47.Kung LA, Kam L, Hovis JS, Boxer SG. Langmuir. 2000;16:6773–6776. [Google Scholar]
- 48.Brown RF. Biochim. Biophys. Acta. 1992;1113:375–389. doi: 10.1016/0304-4157(92)90007-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao HB, Yang TL, Cremer PS. Anal. Chem. 2002;74:379–385. doi: 10.1021/ac010822u. [DOI] [PubMed] [Google Scholar]
- 50.Lauer S, Goldstein B, Nolan RL, Nolan JP. Biochemistry. 2002;41:1742–1751. doi: 10.1021/bi0112816. [DOI] [PubMed] [Google Scholar]
- 51.Lencer WI, Chu SHW, Walker WA. Infect. Immun. 1987;55:3126–3130. doi: 10.1128/iai.55.12.3126-3130.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuziemko GM, Stroh M, Stevens RC. Biochemistry. 1996;35:6375–6384. doi: 10.1021/bi952314i. [DOI] [PubMed] [Google Scholar]
- 53.Cuatrecasas P. Biochemistry. 1973;12:3547–3558. doi: 10.1021/bi00742a031. [DOI] [PubMed] [Google Scholar]
- 54.Cuatrecasas P. Biochemistry. 1973;12:3558–3566. doi: 10.1021/bi00742a032. [DOI] [PubMed] [Google Scholar]
- 55.MacKenzie CR, Hirama T, Lee KK, Altman E, Young NM. J. Biol. Chem. 1997;272:5533–5538. doi: 10.1074/jbc.272.9.5533. [DOI] [PubMed] [Google Scholar]
- 56.Cooper MA, Hansson A, Lofas S, Williams DH. Anal. Biochem. 2000;277:196–205. doi: 10.1006/abio.1999.4389. [DOI] [PubMed] [Google Scholar]
- 57.Janshoff A, Steinem C, Sieber M, elBaya A, Schmidt MA, Galla HJ. Eur. Biophys. J. 1997;26:261–270. doi: 10.1007/s002490050079. [DOI] [PubMed] [Google Scholar]
- 58.Masserini M, Freire E, Palestini P, Calappi E, Tettamanti G. Biochemistry. 1992;31:2422–2426. doi: 10.1021/bi00123a030. [DOI] [PubMed] [Google Scholar]
- 59.Groves JT, Ulman N, Boxer SG. Science. 1997;275:651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 60.Castellana ET, Cremer PS. Surf. Sci. Rep. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeMond AL, Groves JT. Curr. Opin. Immunol. 2007;19:722–727. doi: 10.1016/j.coi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Castellana ET, Cremer PS. Biointerphases. 2007;2:57–63. doi: 10.1116/1.2732312. [DOI] [PubMed] [Google Scholar]
- 63.Groves JT, Mahal LK, Bertozzi CR. Langmuir. 2001;17:5129–5133. [Google Scholar]
- 64.Phillips KS, Wilkop T, Wu JJ, Al-Kaysi RO, Cheng Q. J. Am. Chem. Soc. 2006;128:9590–9591. doi: 10.1021/ja0628102. [DOI] [PubMed] [Google Scholar]
- 65.Howland MC, Szmodis AW, Sanii B, Parikh AN. Biophys. J. 2007;92:1306–1317. doi: 10.1529/biophysj.106.097071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang ZH, Meng YH, Ying PQ, Qi C, Jin G. Electrophoresis. 2006;27:4078–4085. doi: 10.1002/elps.200500956. [DOI] [PubMed] [Google Scholar]
- 67.Markov DA, Swinney K, Bornhop DJ. J. Am. Chem. Soc. 2004;126:16659–16664. doi: 10.1021/ja047820m. [DOI] [PubMed] [Google Scholar]