Abstract
RS-Predictor is a tool for creating pathway-independent, isozyme-specific site of metabolism (SOM) prediction models using any set of known cytochrome P450 substrates and metabolites. Until now, the RS-Predictor method was only trained and validated on CYP 3A4 data, but in the present study we report on the versatility the RS-Predictor modeling paradigm by creating and testing regioselectivity models for substrates of the nine most important CYP isozymes. Through curation of source literature, we have assembled 680 substrates distributed among CYPs 1A2, 2A6, 2B6, 2C19, 2C8, 2C9, 2D6, 2E1 and 3A4, which we believe is the largest publicly accessible collection of P450 ligands and metabolites ever released. A comprehensive investigation into the importance of different descriptor classes for predicting the regioselectivity of each isozyme is made through the generation of multiple independent RS-Predictor models for each set of isozyme substrates. Two of these models include a DFT reactivity descriptor derived from SMARTCyp. Optimal combinations of RS-Predictor and SMARTCyp are shown to have stronger performance than either method alone, while also exceeding the accuracy of the commercial regioselectivity prediction methods distributed by StarDrop and Schrödinger, correctly identifying a large proportion of the metabolites in each substrate set within the top two rank-positions: 1A2(83.0%), 2A6(85.7%), 2B6(82.1%), 2C19(86.2%), 2C8(83.8%), 2C9(84.5%), 2D6(85.9%), 2E1(82.8%), 3A4(82.3%) and merged(86.0%). Comprehensive datamining of each substrate set and careful statistical analyses of the predictions made by the different models revealed new insights into molecular features that control metabolic regioselectivity and enable accurate prospective prediction of likely SOMs.
Introduction
The cytochrome P450 (CYP) enzymes are a family of ubiquitous heme-thiolate proteins that catalyze the metabolism of a large number of xenobiotic and endobiotic compounds. Members of three CYP families alone, CYP1, CYP2, and CYP3, account for the metabolism of over 90% of clinical drugs.1,2 Nine of the most prevalent isoforms within these families are CYPs 1A2, 2A6, 2B6, 2C19, 2C8, 2C9, 2D6, 2E1 and 3A4; the percentage of drugs metabolized by each isozyme are: 1A2(15%), 2A6(3%), 2B6(8%), 2C19(12%), 2C8(8%), 2C9(20%), 2D6(25%), 2E1(4%) and 3A4(50%), respectively.2–9 The P450 phase I mediated metabolism of orally administered drugs occurs primarily in the human liver, where CYP isozymes are expressed at different levels: 1A2(13%), 2A6(4%), 2B6(10%), 2C19(unk.), 2C8(7%), 2C9(20%), 2D6(2%), 2E1(7%), 3A4(30%).6,10 Importantly, P450-mediated metabolism may not be limited to the liver, as the genes that express specific CYP isoforms have been found in many different tissues throughout the human body, allowing for tissue-specific pharmacokinetic profiles to emerge that may vary between individuals.5,6,8,10–17
It is clear that lead compound PK and ADME/Tox optimization efforts would benefit greatly through knowledge of the regiochemical location where any given compound would be susceptible to metabolism by specific CYP isozymes. For example, such information would allow susceptible regions of a lead molecule to be modified in ways that retain potentcy, while optimizing PK properties and moderating hepatotoxicity. Unfortunately, using experimental means to determine the exact location that a xenobiotic compound may be susceptible to P450-mediated metabolism is both time and resource intensive. A solution to this problem lies in the availability of reliable in silico models of CYP-mediated metabolic regioselectivity that are applicable at all stages of the drug-discovery process. A number of such models have been developed in recent years, but most have not yet achieved a desirable level of predictive accuracy.
In the current state of the art, P450-regioselectivity models fall into two broad categories: ligand-based, where putative sites of metabolism (SOMs) on the substrate are ranked according to their respective reactivity scores, and enzyme structure-based, where the reactivity of potential SOMs are weighted by the quality and number of poses that place those SOMs close to the oxidizing heme of the CYP isozyme. Hybrid methods that utilize signal from both sources have also been proposed. Some of regioselectivity models are isoform-specific while others are not, but all are data-driven in that they require training from existing substrate sets to form predictive models. Of the few 1A2, 2B6, 2C8, and 2E1 regioselectivity models proposed in the literature, all but two of them are purely based on docking methods that were validated using fewer than than 20 substrates.18–23 One of the exceptions is the commercial application MetaSite, a hybrid method which was successfully applied in 2005 to 135 substrates of 1A2 and 125 substrates of 2C19, as well as substrates sets of CYPs 2C9, 2D6 and 3A4.24 These last three are the most promiscuous P450 isoforms, and each has had a number of regioselectivity models proposed in the literature. The majority of solutions for 2C924–29 and 2D624,30–37 are docking-based, while 3A4 models are primarily ligand-based,27,38,39 with some reactivity/docking hybrids,24,40,41 and one docking-based solution.42 The proposed rationale for why reactivity is more relevant for 3A4 regioselectivity prediction is that the binding pocket is relatively large, allowing substrates a high degree of orientational freedom, thereby giving each site equal access to the catalytic heme.38 This theory may hold for other CYPs as well, as recently disclosed crystal structures of other isozymes indicate that the large binding pocket of 3A4 is not unique — other isozymes also posess binding sites with significant volumes: 1A2(375Å3), 2A6(260Å3), 2B6(no xtal), 2C19(no xtal), 2C8(1438Å3), 2C9(1667Å3), 2D6(540Å3), 2E1(190Å3), 3A4(1386Å3).6 Another explanation for why reactivity plays a greater role in 3A4-mediated metabolism is that the backbone of 3A4 is much more flexible than those of other P450 isozymes;43 this provides explanations for both the high number of drugs metabolized by 3A4 and why only one purely docking-based 3A4 regioselectivity solution has been proposed. The electronic environment around each potential SOM of a substrate, and how that environment reacts in proximity to a CYP catalytic heme are extremely important factors that have been used successfully by multiple isozyme non-specific regioselectivity prediction methods.44–47 The majority of these models utilize AM1-derived descriptors to approximate SOM reactivity, which other groups have shown to be useful in estimating CYP regioselectivity.38,48 One of the more innovative approaches, published by the Sheridan group at Merck in 2007, quantified putative SOMs with local substructure information derived from 2D molecular topology, and then used random-forest machine learning to determine which oxidative pathways were most likely to be utilized for a given set of substrates.27 The benefit of this algorithm described by Sheridan et al. is that isozyme-specific models could be trained from a large set of isozyme substrates and quickly be applied to new substrates without explicitly calculating either the local SOM electronic reactivity or the orientation of each ligand within the enzyme binding pocket. Another valuable contribution of this work was the release of structures and metabolites of 324 substrates of 3A4, 134 substrates of 2D6 and 101 substrates of 2C9 — the largest public release of P450 metabolites made up to that time.
Our recently published method, RegioSelectivity-Predictor (RS-Predictor), expands on the work of Sheridan et al. by encoding putative SOMs with 148 topological descriptors that capture local structural information, while characterizing local electronic reactivity through 392 quantum chemical descriptors derived by MOPAC49 2007 AM1 calculations on the candidate substrate.50 Regioselectivity models are then created using MIRank, a customized implementation of support vector machines (SVMs) technology that was specifically designed to optimized the ranking of observed sites of P450-mediated oxidation over non-observed sites on a substrate by substrate basis.51,52 RS-Predictor models may be built from any sufficiently diverse set of substrates, such as those released by Sheridan et al., and were proven to be highly effective at identifying the regioselectivity of a set of 394 substrates of 3A4. This set of substrates was originally used as the validation set for SMARTCyp, an isozyme-blind method that differs from other reactivity-based methods in that high quality DFT calculations are used to estimate the SOM reactivity of specific substructures rather than using theoretically less sound AM1-derived charges.47 In SMARTCyp, the transition state energies between a generic CYP heme and individual molecular fragments were encoded as a SMARTS reactivity look-up table which collectively represents a large range of known CYP-mediated reactions.53 The recorded reactivity of each putative SOM is weighted slightly by the site’s span, a quantity that reflects its relative location to the middle or end of the molecule. The strength of SMARTCyp lies in the encoding of a high quality, first-order reactivity signal that is quickly accessible, while its weakness is the lack of isozyme-specific signal that represents the different propensities of each P450 isozyme to mediate specific pathways. RS-Predictor was specifically designed to capture this type of information from any set of isozyme-specific substrates, but was originally dependent upon a suboptimal reactivity signal from AM1-derived quantum chemical descriptors. The weakness of each method is the corresponding strength of the other, which suggested that a synthesis of the two methods would yield highly predictive isozyme-specific regioselectivity models. This supposition proved to be correct, resulting in enhanced prediction quality of RS-Predictor models described later in this report.
Prior to this publication, RS-Predictor results were only reported using 3A4 data, where it was shown to outperform the state of the art methods that existed at the time. The main focus of this work then is to establish the versatility of the RS-Predictor modeling paradigm through the creation of robust SOM prediction models for all nine important CYP isozymes. A fundamental part of this endeavor was the gathering and curation of CYP data from public sources. In total 680 unique substrates were collated and then distributed into newly assembled substrate sets for CYPs 1A2, 2A6, 2B6, 2C19, 2C8, 2C9, 2D6, 2E1 and 3A4. The structures and metabolites within each of these sets may be found within the Supporting Information and represent the largest collection of P450 substrates ever made publicly available. A comprehensive investigation into the utility of different classes of descriptors towards accurate identification of CYP-mediated metabolism is made, which includes the enhancement of the RS-Predictor framework through the incorporation of SMARTCyp reactivities as a new descriptor. Optimal combinations RS-Predictor and SMARTCyp are shown to outperform both methods individually, as well as the commercial methods available through StarDrop54 and Schrödinger55 for 2C9, 2D6 and 3A4 substrate sets. Meanwhile, regioselectivity QSARs representing the first ligand-based SOM prediction models ever created for substrates of 1A2, 2A6, 2B6, 2C8, 2C19, and 2E1 were prepared and shown to have high levels of predictive accuracy. To ensure that the RS-Predictor method was producing valid results, another set of models were trained using only the substrates of 2C9, 2D6 and 3A4 that had previously been released by Sheridan et. al. The models created from these older datasets were used to make blind predictions on the newly assembled sets of substrates for each isozyme, while treating the new compounds as blind external sets. This approach allowed us to make fair comparisons of the predictive accuracies of RS-Predictor, SMARTCyp, StarDrop and Schrodinger on identical external datasets, providing a way for medicinal chemists to identify which method might be best for use on their in-house compounds.
Results and Discussion
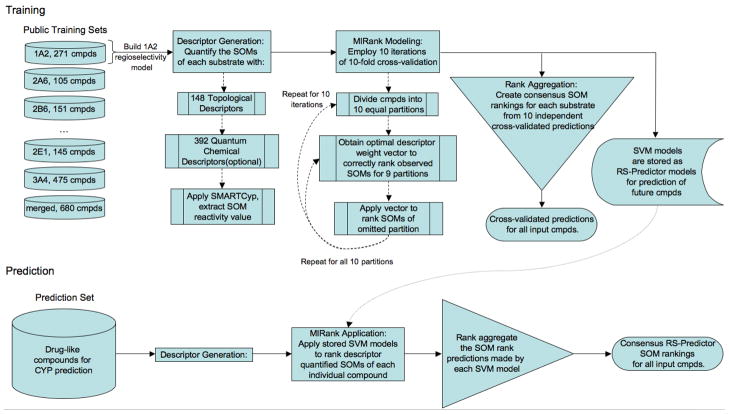
Complete details of the RS-Predictor algorithm have been described in our earlier published work; a high level flowchart of the algorithm is given in Figure 1.50 To train an isozyme-specific regioselectivity model, a set of known isozyme substrates are represented as individual competitions between candidate SOMs, each of which is characterized by a specified set of descriptors. SOM prediction models are then trained through the application of MIRank to 10 independent substrate set partitions using a 10-fold cross-validation scheme. Each compound then has 10 separate SOM rankings obtained from a collection of models, each trained on 90% of the compounds in the dataset. Rank aggregation is then employed to merge these independent rank predictions into a single consensus ranked list of putative SOMs.
Figure 1.
RS-Predictor Flowchart. Stored SVM models from the Training schema are optimized for all training set substrates, using the optimal model parameter values obtained from each iteration of cross-validation. Cross-validated results in Table 1 and Calibration set results in Table 2 were obtained from predictions made using the Training schema. External set results in Table 2 and proprietary set results in Table 3 were obtained from predictions made using the Prediction schema.
During the course of this investigation multiple RS-Predictor models were created for each isozyme substrate set using different sets of descriptors. In this way we were able to investigate the relative importance and contribution of each descriptor type for predicting susceptable SOMs. The descriptor sets that were chosen represent an exhaustive combination of 148 topological descriptors (TOP), 392 quantum chemical descriptors (QC), and the SMARTCyp reactivity descriptor (SCR). One combination of descriptor types was omitted: QC SCR. The complete definitions of the descriptors in each set may be found in our prior work; some examples of TOP descriptors include whether a given SOM is in a ring, the size and nature of the ring (aromatic or not), and the distribution of different atom-types (C, O, N, etc.) up to four bond-lengths away from the given SOM. Examples of QC descriptions include the AM1 partial charge, nucleophilicity, and σ – σ, σ –π, and π – π bond orders.
To simplify overall presentation, only results for the stand-alone TOP QC RS-Predictor model and the two incorporations of the SCR descriptor as TOP SCR and TOP QC SCR models are given in this section. Results for TOP and QC models may be found in Supporting Information. Predictions accuracies for independent RS-Predictor models are compared with those of SMART-Cyp(V1.5), and the commercial methods available from StarDrop(V4.3) and Schrödinger(V1.0) in Table 1. At the time of submission, results for RS-Predictor and SMARTCyp had only been reported for substrates of 3A4. SMARTCyp(V1.5) has not been calibrated towards any specific isoform, though post-submission a separate 2D6 SMARTCyp model has been released.37 At the time of publication, neither StarDrop nor Schrödinger possess 1A2, 2A6, 2B6, 2C19, 2C8 or 2E1 regioselectivity models.
Table 1.
The percentage of each substrate set with an experimentally observed SOM predicted in the top two rank-positions by the given methoda
| Isozyme | 1A2 | 2A6 | 2B6 | 2C19 | 2C8 | 2C9 | 2D6 | 2E1 | 3A4 | merged |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of Substrates | 271 | 105 | 151 | 218 | 142 | 226 | 270 | 145 | 475 | 680 |
| RS-Predictor (TOP SCR)b | 82.3 | 85.7 | 76.8 | 86.2 | 83.8 | 84.1 | 83.7 | 80.7 | 82.1 | 86.0 |
| RS-Predictor (TOP QC SCR)b | 83.0 | 81.0 | 82.1 | 86.2 | 83.8 | 84.5 | 85.9 | 82.8 | 82.3 | 84.1 |
| RS-Predictor (TOP QC)b | 79.7 | 79.0 | 80.1 | 82.6 | 77.5 | 80.5 | 83.3 | 80.7 | 77.7 | 81.6 |
| SMARTCyp | 78.9 | 83.3 | 73.6 | 73.7 | 73.2 | 67.3 | 58.4 | 81.0 | 74.4 | 74.8 |
| StarDrop | — | — | — | — | — | 78.0 | 75.3 | — | 74.1 | — |
| Schrödinger | — | — | — | — | — | 72.1 | 68.1 | — | 76.4 | — |
|
| ||||||||||
| Random Model | 26.0 | 31.9 | 24.8 | 20.2 | 22.6 | 22.2 | 21.1 | 36.5 | 21.0 | 26.3 |
For each CYP, the optimal model is shown in bold, as are all other models found not to be statistically different using Fisher’s exact test of independence.
Independent RS-Predictor models encode putative SOMs with different combinations of 148 topological descriptors (TOP), 392 quantum chemical descriptors (QC), and the SMARTCyp reactivity descriptor (SCR).
Cross-validated RS-Predictor results were obtained from predictions made using the Training schema described in Figure 1.
Consistent with previous regioselectivity investigations, each substrate was evaluated on the basis of whether one or more of the experimentally observed SOM(s) were predicted within the top two predicted rank-positions of the given method.24,27,38–40,44–47,50 This metric is used because the source experimental data comes from a large number of different sources and groups with potentially different motivations and techniques for the determination of CYP-mediated metabolism, and so each putative SOM is given a binary response value, metabolized or not-metabolized by the given CYP. Each entry in Table 1 gives the percentage of known substrates of the column CYP that had an experimental SOM identified within the top two predicted rank-positions by the given row method, with the performance of the optimal model being shown in bold. If no evidence was found that the proportion of correctly to incorrectly predicted compounds by the optimal method is greater (using a right-tailed alternative hypothesis) than the proportion of another non-optimal method (i.e. accepting the null hypothesis at a 5% significance level) then the performance of that non-optimal method is also bolded. Fisher’s exact test of independence was invoked for this purpose.73
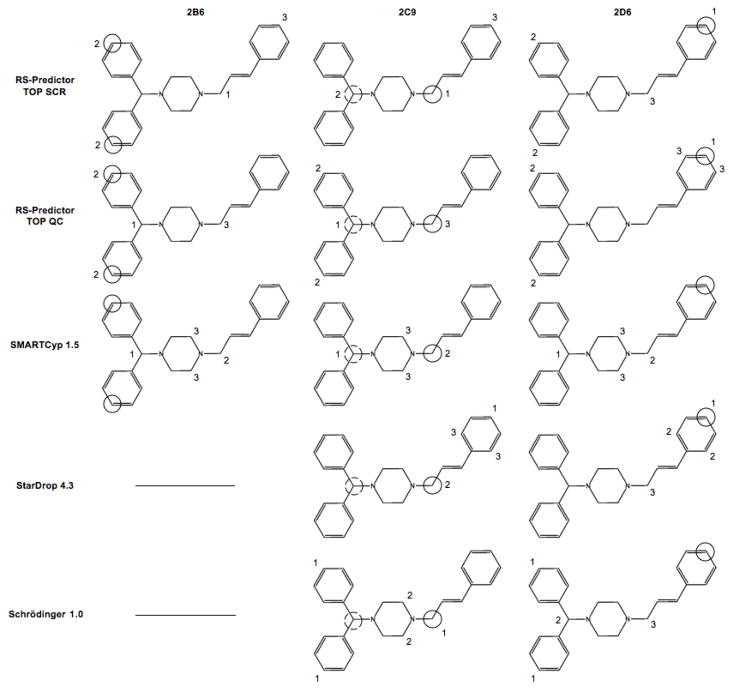
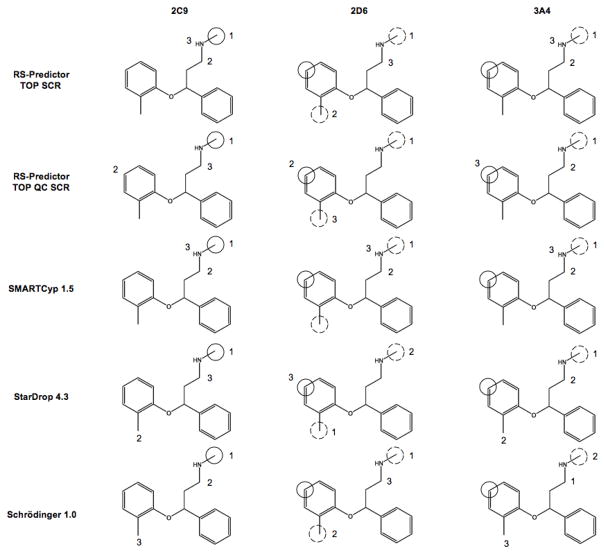
The overall results clearly demonstrate the ability of RS-Predictor to capture the regioselectivity propensities of any set of substrates regardless of the metabolizing isozyme, provided that the dataset is large enough. The models that were produced were in fact isozyme-specific, despite the fact that no enzyme structure was used during model construction. An example of this is shown in Figure 2 through the metabolism of cinnarizine, one of the few substrates that undergoes completely different 2B6, 2C9 and 2D6-mediated pathways. Isozyme-specific RS-Predictor models identify the observed sites of oxidation for each isozyme, and the predictions were found to change from CYP model to CYP model, just as the metabolism of the ligand changes from isozyme to isozyme. SMARTCyp was able to identify the both sites of 2C9-mediated N-deaklylation, but neither the 2B6 nor 2D6-mediated sites of aromatic hydroxylation. Both StarDrop and the Schrödinger method identify the primary site of 2C9-mediated N-dealkylation, but miss the secondary oxidation site, while only StarDrop was found to be able to identify the observed site of 2D6-mediated metabolism. Neither of the commercial methods have 2B6 models, though the 2C9 and 2D6 Schrödinger models both identify the observed 2B6-mediated reaction in the primary predicted rank-position.
Figure 2.
Different CYP-mediated metabolic pathways and model predictions of cinnarizine; solid circles, coarse hashed circles and finely hashed circles designate primary, secondary, and tertiary experimentally observed SOMs for the given isozyme. The numerals near each site designate the primary, secondary and tertiary predicted sites of the given isozyme model of the given method upon the substrate. This molecule does not posses any observed tertiary SOMs, but later graphics that use this labeling scheme do.
As hypothesized, the incorporation of SMARTCyp reactivities into the RS-Predictor modeling framework yields a set of highly predictive regioselectivity models. On average, TOP SCR and TOP QC SCR success rates surpass those of TOP QC models by 2.8% and 3.3% respectively, thereby illustrating the high quality signal of SMARTCyp reactivities and the complementary information contained within different SCR and QC representations of electronic reactivity. Further analysis revealed that the relative proportion of correctly to incorrectly predicted compounds by SCR models were not statistically different to the proportions of non-SCR models for a majority of the substrate sets. This disparity is likely due to the small sizes of the majority of the assembled sets. Indeed for the two largest sets, 3A4 and merged, prediction rates between TOP QC models and optimal SCR models differ by ~4.5%, and SCR models were found to return a statistically higher proportion of correctly-predicted compounds than non-SCR models. But for the smaller sets, such as 2A6 and 2C8 where optimal SCR model rates are respectively 6.7% and 6.3% higher than non-SCR model rates, the proportions of correctly to incorrectly predicted compounds between the SCR and non-SCR models were found to be statistically equivalent. We hypothesize that as more and more metabolite data becomes available for each isozyme, the performance of SCR models will not just be greater than those of non-SCR models, but greater by a statistically significant measure.
Another explanation for statistical equivalence across multiple RS-Predictor models is that the main strength of the algorithm lies not in electronic descriptors, but in the isozyme-specific reaction pathway propensities elucidated through robust modeling of topological descriptors. Consequently TOP SCR and TOP QC SCR models offer a significant boost in performance over the isozyme non-specific SMARTCyp model, having higher average rates of 9.3% and 9.7%, respectively. The prediction rates of RS-Predictor models with SMARTCyp reactivities surpass those of StarDrop by ~6%, ~9% and ~8% and those of Schrödinger by ~12%, ~16.5% and ~6% for 2C9, 2D6 and 3A4 sets respectively. In addition, the relative proportions of correctly to incorrectly predicted substrates for optimal RS-Predictor models are statistically greater than those of both StarDrop and Schrödinger. While the SCR descriptor improves model performances regardless of isozyme, the same cannot be said of AM1-derived QC descriptors. For the majority of the sets, TOP QC SCR models are optimal, though TOP SCR rates are within 2% of TOP QC SCR rates for all sets but 2B6. This highlights an interesting phenomena, in that AM1-derived descriptors demonstrate significant signal towards the identification of 2B6-mediated metabolism; prediction rates for the set of 2B6 substrates represent the only case where TOP QC models outperform TOP SCR models. In other sets however, QC descriptors contribute no signal or even decrease overall accuracy, as demonstrated by 2A6, 2C19, 2C8 and merged results. Differences between TOP and TOP QC models, available within the Supporting Information, show that TOP QC rates actually fall below TOP rates for 2A6 and 2C8 sets. To further investigate the utility of QC descriptors, as well as make additional comparisons between different methods, we have performed pathway-based analysis of each method and model using specialized techniques developed in our prior work.
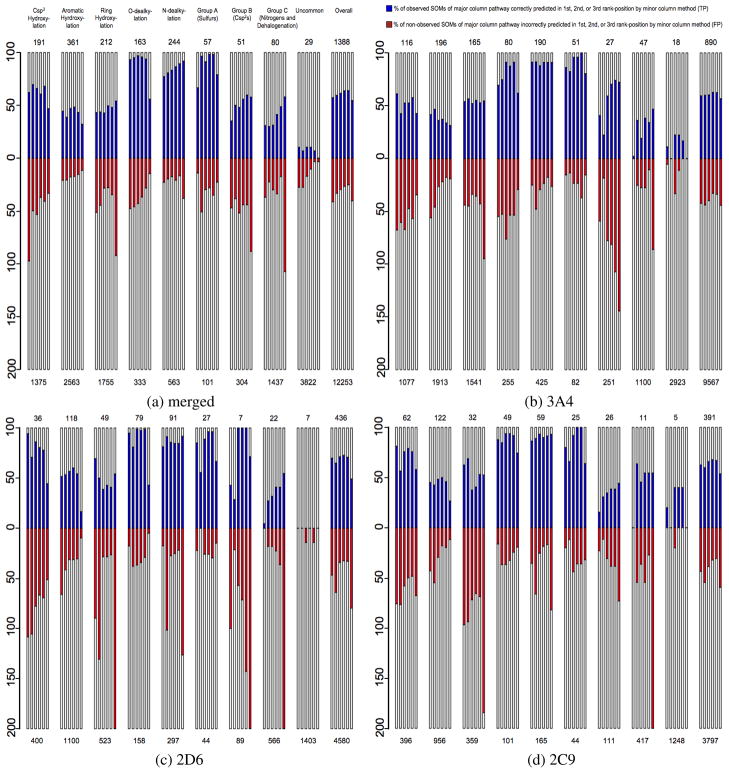
An alternative way to access the regioselectivity performance of a given prediction method is to analyze the numbers of true-positive (TP) and false-positive (FP) predictions made by a given model on a pathway-by-pathway, molecule-by-molecule basis. To accomplish this, each SOM is placed into a set based upon the CYP-mediated reaction pathway that they have the potential to follow. Each set is then composed of a certain number of observed and non-observed SOMs; the exact ratios of SOMs for each pathway and substrate set may be found in Table 6 (see Methods). Due to the unbalanced nature of CYP-mediated metabolism, there are usually many more non-observed SOMs than observed SOMs in each pathway set, though there are exceptions such as with sulfur-based oxidation reactions. The number of observed SOMs in each set that are correctly predicted in the top three rank-positions (TP), as well as the number of non-observed SOMs that are incorrectly predicted in the top three rank-positions (FP) are shown for individual methods and models for merged, 3A4, 2D6 and 2C9 substrate sets in Figure 3. Comparisons were only made between SOMs that occur within the same substrate. This evaluation technique has some inherent ambiguity in that the TPs and FPs of each set were obtained by being ranked above members of a different set, ie. a correctly predicted N-dealkylation reaction was ranked above a non-observed aromatic ring hydroxylation. Despite this limitation, this evaluation technique is a powerful way to compare a large amount of disparate information on a single informative graphic. Using this approach, users who wish to evaluate the effectiveness of a particular prediction method may quickly identify which pathways or substrate sets are most accurately predicted using each method. More importantly, end-users with access to limited MS/MS data that identifies only the region but not the exact location of a CYP-mediated reaction site can objectively decide which model is best suited to predict the regioselectivity profile of a specific compound. As an important note, these pathway-based findings necessarily represent only the chemical information contained within the 680 substrates curated for this work; when additional information becomes available, the relative performance of each method on specific pathways may change.
Table 6.
CYP-mediated pathway propensities (%) according to the of known substrates of each isozyme. Similar pathways, or those with low relative populations such as Csp2-based reactions (B), are grouped together in order to simplify pathway-based performance analysis in Figure 3
| Pathway | 1A2 | 2A6 | 2B6 | 2C19 | 2C8 | 2C9 | 2D6 | 2E1 | 3A4 | merged |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Csp3 Hydroxylation | 13.0 | 11.3 | 9.9 | 10.9 | 10.8 | 15.7 | 9.0 | 14.5 | 10.8 | 13.9 |
| Aromatic Hydroxylation | 16.1 | 11.0 | 12.3 | 7.6 | 10.9 | 12.8 | 10.7 | 18.2 | 10.3 | 14.1 |
| Ring Hydroxylation | 14.1 | 20.0 | 8.4 | 10.4 | 11.8 | 9.0 | 9.1 | 14.2 | 10.5 | 11.9 |
| O-dealkylation | 48.1 | 48.5 | 39.3 | 51.8 | 34.9 | 48.5 | 50.0 | 45.8 | 31.0 | 48.9 |
| N-dealkylation | 43.1 | 48.5 | 44.0 | 38.7 | 46.7 | 35.8 | 29.6 | 39.2 | 44.7 | 43.4 |
| Sulfur(II) Oxidation (A) | 50.0 | 73.3 | 66.7 | 53.3 | 47.0 | 61.1 | 57.9 | 58.8 | 59.7 | 54.4 |
| Sulfur(IV) Oxidation (A) | 100 | 0.0 | 100 | 42.9 | 0 | 33.3 | 100 | None | 100 | 90.9 |
| Desulfuration (A) | 66.7 | 0.0 | 50.0 | 57.1 | 42.9 | 40.0 | 75.0 | 100 | 40.0 | 36.4 |
| Csp2 Oxidation (B) | 17.4 | 20.0 | 18.2 | 15.7 | 19.4 | 20.9 | 5.4 | 17.8 | 8.5 | 20.4 |
| Aldehyde Oxidation (B) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 75.0 | 71.4 | 70.0 |
| Alcohol Oxidation (B) | 29.4 | 66.7 | 14.3 | 13.6 | 13.6 | 23.8 | 9.7 | 14.3 | 9.6 | 9.2 |
| N-hydroxylation (C) | 9.0 | 15.2 | 7.1 | 7.0 | 7.6 | 5.6 | 7.0 | 13.0 | 5.4 | 8.7 |
| N-oxide Formation (C) | 3.0 | 2.6 | 0 | 1.9 | 3.8 | 2.4 | 3.4 | 5.3 | 4.2 | 4.5 |
| Nitro-group Reduction (C) | 21.4 | 0 | 0 | 0 | 0 | 0 | 0 | 14.3 | 5.9 | 18.2 |
| Dehalogenation (C) | 3.6 | 8.1 | 2.8 | 0 | 1.6 | 0 | 1.1 | 17.6 | 1.3 | 3.3 |
| Uncommon | 0.6 | 1.0 | 0.8 | 0.6 | 1.2 | 0.4 | 0.6 | 1.0 | 0.6 | 0.8 |
| Overall | 12.1 | 12.5 | 10.6 | 9.2 | 10.2 | 10.3 | 9.5 | 13.6 | 9.2 | 11.3 |
Figure 3.
True-positive (TP) and false-positive (FP) prediction rates of individual methods broken down by CYP-mediated pathway. Each major column represents a specific pathway set, identified at the top of 3a, that is composed of all SOMs from the given substrate set that have the potential to undergo that pathway. Similar pathways, or those with low relative populations, were grouped together as detailed in Table 6 (see Methods) to simplify overall presentation. Each pathway column is composed of multiple mini-columns that represent the number of predicted TPs/FPs by (from left to right) QC, TOP, TOP QC, TOP QC SCR, TOP SCR and SMARTCyp models for 3a and StarDrop, Schrödinger, TOP QC SCR, TOP SCR and SMARTCyp models for 3b, 3c, and 3d. SMARTCyp was not specifically calibrated for 2C9 or 2D6, but is a measure of the importance of reactivity for specific sets of substrates and isozymes. The y-column of each pathway column represents the percentage of observed SOMs from the given pathway of the given substrate set, with the actual number being shown at the top of each column. This scaling was chosen to ensure that visualization of true-negatives would not overshadow more interesting results. The number of potential (observed and non-observed) SOMs of each pathway for each substrate set is given at the bottom of each column.
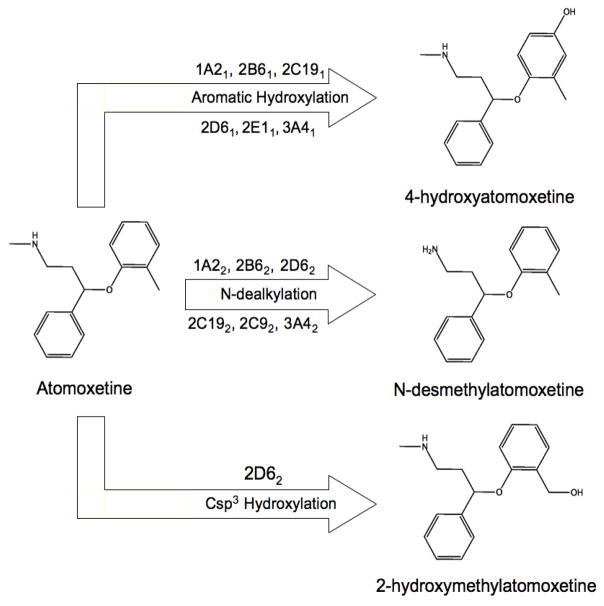
Using the comparative method described above, it is clear that in general, StarDrop has a greater preference to predict aliphatic Csp3 hydroxylation than other methods, regardless of isoform. This means that StarDrop predicts a greater number of both TPs and FPs than other methods for reactions of this type. Schrödinger predicts a similar number of FPs as StarDrop, but fewer TPs. SMARTCyp identifies a similar number of TPs as Schrödinger, but has much fewer FPs than other methods, with the exception of RS-Predictor models for the 2C9 set. The majority of RS-Predictor models have greater TP rates than all methods save StarDrop and lower FP rates than all methods save SMARTCyp. In this case, QC descriptors did not contribute to the accurate identification of aliphatic hydroxylation sites. An example case where the inclusion of QC descriptors increases RS-Predictor accuracy is in the case of hydroxylation of ellipticine, as shown in Figure 4. In a different example (Figure 5), TOP QC SCR models identified the secondary observed site of 2D6-mediated hydroxylation of atomoxetine in the third rank-position, but TOP SCR models predict the same site in the second rank position.
Figure 4.
Different CYP-mediated metabolic pathways and model predictions of ellipticine (labeling scheme detailed in Figure 2). For this substrate the inclusion of QC descriptors improves the RS-Predictor identification of observed sites of Csp3 hydroxylation. The signal provided by QC descriptors enhances the identification of aromatic ring reactions, while SMARTCyp reactivities assist in the identification of nitrogen-based reactions. This is likely why only the TOP QC SCR model is able to identify the primary observed site of 3A4-mediated aromatic ring N-oxidation.
Figure 5.
Different CYP-mediated metabolic pathways and model predictions of atomoxetine (labeling scheme detailed in Figure 2). RS-Predictor models with QC descriptors are shown to increase the prediction accuracy of the primary observed 2D6 and 3A4-mediated aromatic ring metabolite relative to models without them. The SMARTCyp model is shown to have high preference to predict nitrogen-based reactions, identifying the primary 2C9, and secondary 2D6,3A4-mediated N-dealkylation metabolite in the first rank-position and non-observed sites of N-hydroxylation and N-dealkylation in the second and third rank-positions respectively.
One of our most surprising findings was that QC descriptors add significant information towards the correct identification of CYP-mediated aromatic ring hydroxylation sites. This signal was identified through the visible increase in TP rates and occasional decrease in FP rates that QC models have relative to non-QC models. The effect is most easily seen in the second major column of Figure 3a, as well as the aromatic ring pathway analysis in the Supporting Information. Additional analysis of the top three predicted rank-positions of different RS-Predictor models shows that TOP QC models identify 8, 9, 12 and 30 more TPs and −3, 2, −6 and 10 fewer FPs than TOP models for 2C9, 2D6, 3A4 and merged sets, respectively. Respective increases in TP rates of TOP QC SCR models relative to TOP SCR rates are somewhat smaller, 5, 7, 7 and 19, while the number of TOP QC SCR FPs actually increases by −2, 1, 8, 7 relative to TOP SCR rates. Still, with the exception of the 3A4 set, the addition of QC descriptors to TOP SCR descriptors gives RS-Predictor a net improvement in the identification of aromatic hydroxylations. This also indicates that the SMARTCyp reactivities can be improved for aromatic hydroxylations. The sets for which QC models have the least effect are 1A2 and 2E1. Looking at Table 6, these are the two sets that have the highest ratios of aromatic hydroxylations; perhaps better ratios allow RS-Predictor models to elucidate a greater discriminatory signal for this reaction type solely through topological descriptors and quantum chemical descriptors offer no additional signal. The 2B6 substrate set has the greatest proportional increase in QC TP rate and decrease in FP rate relative to non-QC models. Both QC models identify five more observed reactions than equivalent non-QC models, while TOP QC models have 15 fewer FPs than TOP models, and TOP QC SCR models have 9 fewer FPs fewer than TOP SCR models. These numbers are relatively close to those mentioned for the other substrate sets, which is significant considering that 2B6 contains approximately and of the number of observed and potential aromatic hydroxylations as 2C9, 2D6, 3A4 and merged sets respectively. This provides further justification for the importance of QC descriptors towards the identification of 2B6-mediated regioselectivity. SMARTCyp was found to identify fewer TPs and FPs than other methods. Both StarDrop and Schrödinger have significantly higher numbers of FPs than other methods, while their TP rates only surpass those of RS-Predictor for the 3A4 set.
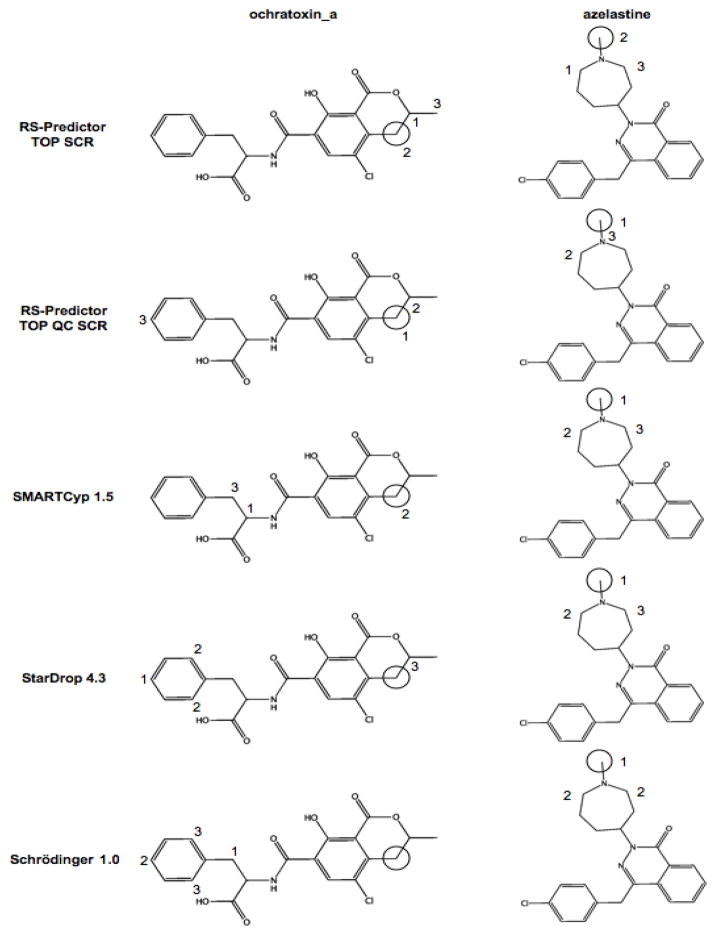
While QC descriptors do not have as much effect on the TP prediction rates for non-aromatic ring hydroxylation, they do lessen the number of predicted FP sites. TOP QC models identify 7, 7, 31 and 34 fewer non-observed ring hydroxylations than TOP models for 2C9, 2D6, 3A4 and merged sets respectively. As with aromatic hydroxylation, the relative difference between different SCR models is less pronounced, likely reflecting the additional signal represented by the SMARTCyp reactivity descriptor. The FP rates of TOP QC SCR models are 1, −1, 12 and 14 lower than corresponding TOP SCR models for respective 2C9, 2D6, 3A4 and merged sets. Predictions of ochratoix_a and azelastine in Figure 6 are two cases where TOP QC SCR models have improved TP predictions due to a lowered rank-prediction of a non-aromatic ring false-positive incorrectly identified by a TOP SCR model. Based upon this evidence we believe that if available mass-spectrometry data indicates that an unknown substrate is undergoing ring-based metabolism, QC models should be applied to determine the exact site of P450-mediated metabolism. With regards to other methods, SMARTCyp identifies a few more observed ring hydroxylations than RS-Predictor, but significantly more FPs for this reaction type. The only cases where RS-Predictor TP rates match SMARTCyp rates are for TOP SCR model for the 2C9 set, and for all models for the 3A4 set, where every method has very similar TP rates. Both Schrödinger and StarDrop have higher ring hydroxylation TP and FP rates than RS-Predictor for 2C9 and 2D6 sets. These two methods have higher TP rates and lower FP rates than SMARTCyp, except when Schrödinger TP rates match those of SMARTCyp for 2D6. Schrödinger and StarDrop have similar rates for 2C9 and 3A4 sets, but StarDrop has higher ring hydroxylation TP rates and lower FP rats than Schrödinger for the 2D6 set.
Figure 6.
Different 3A4-mediated metabolic pathways and model predictions of Ochratoxin_a and Azelastine (labeling scheme detailed in Figure 2). For each ligand the inclusion of QC descriptors decreases the predicted rank-position of non-observed sites of non-aromatic ring hydroxylation while simultaneously increasing the rank-position of the observed site of metabolism.
SMARTCyp and Schrödinger have similar TP prediction rates of sulfur-based reactions, while Schrödinger has lower FP rates. StarDrop has similar FP rates to those of SMARTCyp, but higher TP rates. RS-Predictor has a greater preference to predict sulfur-based reactions than other methods, having higher TP and FP rates for all substrate sets. Analysis of different RS-Predictor model predictions for the merged set indicate that QC models alone are quite poor at identifying observed sulfur-based reactions, though they also have a very low FP rate. The situation is reversed with TOP models, which have high TP and FP prediction rates for sulfur-based reactions. When models are trained from both descriptor sets the TP and FP rates fall in-between those of both models separately, with a better overall ratio than either. It appears to be a general trend across the different substrate sets that the addition of SCR descriptors to TOP QC models improves TP prediction rates of sulfur-based reactions while leaving the FP rates unchanged. Removal of QC descriptors from these models increases FP prediction rates, while giving 3A4 models a slight improvement in TP rates and improving the rank-positions of certain TP sulfur predictions for other sets.
Of the three Csp2-based reactions, Csp2 oxidation, aldehyde oxidation and alcohol oxidation, StarDrop is only parametrized to predict alcohol oxidation. While Schrödinger is parametrized for Csp2 oxidation and aldehyde oxidation, only very rarely does it predict them. Consequently both methods have lower TP and FP rates than RS-Predictor and SMARTCyp for this reaction class. SMARTCyp has greater TP and FP rates than RS-Predictor, while SCR models have better ratios than non-SCR models.
The nitrogen-based reaction set does not include N-dealkylation reactions, which occur with much greater frequency than N-hydroxylation, N-oxidation or Nitro-group reduction. StarDrop is not parameterized for any of these reactions, its TP and FP predictions for this category all represent dehalogenation reactions; these reactions were placed into this set because they represent a known reaction type with low overall population, and graphic space was limited. The most important conclusion to be drawn from the analysis of this pathway-set is the strong propensity for SMARTCyp to predict nitrogen-based reactions. RS-Predictor is able to successfully incorporate this signal; SCR models have significantly higher TP rates than non-SCR models. At the same time the FP rates of TOP QC SCR models are quite similar to those of TOP QC models, which was not guaranteed to occur given the high FP rates of SMARTCyp. Even more interesting, the removal of QC descriptors from TOP QC SCR models results in decreased FP rates, which is the opposite situation of what occurs for sulfur-based reactions, where removal of QC descriptors gives TOP SCR models higher FP rates. The FP rates of Schrödinger are similar to those of QC models, and have more, less and equal TP rates to SCR models for 2C9, 2D6 and 3A4 sets respectively. The signal of QC descriptors towards the identification of aromatic ring reactions, and the signal of SCR descriptors towards the identification of nitrogren-based reactions, is likely why TOP QC SCR is the only model able to successfully identify the primary observed site of 3A4-mediated aromatic ring N-oxidation of ellipticine, illustrated in Figure 4.
In addition to having high prediction propensity for the low population N-based reactions, SMARTCyp also identifies a large number of N-dealkylations. The TP rates of SMARTCyp are greater than or equal to those of other methods, and its FP rates are as well, except for the 3A4 set where Schrödinger FP rates surpass those of SMARTCyp. Schrödinger N-dealkylation FPs are significantly higher than either StarDrop or RS-Predictor, which have comparable FP rates. As with the other N-reaction set, incorporation of SCR descriptors and subsequent removal of QC descriptors each decreases the number of predicted FPs. While SMARTCyp has a strong propensity to predict N-dealkylations it has an equally low propensity to predict O-dealkylation. RS-Predictor has the highest O-deaklylation TP rates of all the methods, with decreasing FP rates upon the addition of the SCR descriptor and subsequent removal of QC descriptors. StarDrop TP rates are slightly higher than those of Schrödinger for 2C9, much higher for 2D6 and slightly lower for 3A4. Schrödinger FP rates are similar to those of TOP QC models, while StarDrop FP rates are much lower than both RS-Predictor and Schrödinger for 2C9 and 2D6.
The sets of SOMs undergoing O-dealkylation and N-dealkylation have higher ratios of reactions than most other pathway sets. Even more interesting, these catalytic propensities have a high degree of variance between isozymes. The average ratio of O-dealkyation for both 2C9 and 2D6 datasets is 49.3, whereas it is 32.7 for N-dealkylation. In contrast, the catalytic propensities of 3A4 substrates is 31.0 for O-dealkylation and 44.7 for N-dealkylation. The previously observed biases of SMARTCyp to favor the prediction of N-based reactions and disfavor the prediction of O-dealkation reactions provide a chemical rational for observed SMARTCyp performances on distinct substrate sets. SMARTCyp has an average prediction rate of 62.9% for 2C9, 2D6 datasets, compared with 74.4% for 3A4 and an average of 74.1% across all nine substrate sets. In contrast, SMARTCyp has an average performance rate of 82.2% for 2A6 and 2E1 sets, the two sets with the smallest average substrate size. These are two of the three sets where the proportion of correct SMARTCyp predictions are found to be statistically equivalent to those of RS-Predictor. We hypothesize the high quality reactivities of SMARTCyp are best suited to identify the CYP-mediated regioselectivity of smaller substrates because each putative SOM has similar likelihood of reaching the oxidative heme, making the transition state energy of each fragment the most discriminatory factor between SOMs. For a similar reason SMARTCyp is likely to perform better on isozymes with a flexible backbone, which are better able to expand and accommodate substrates in any orientation. The strong SMARTCyp performance upon 1A2(78.9%) substantiates this views, because 1A2 the only CYP isoform beyond 3A4 that has been reported to accommodate multiple ligands within its binding site at the same time.56 Investigations by Cruciani et al. also found that local SOM reactivity was a more important factor for 1A2 and 3A4 isozymes than it was for 2C9, 2D6 or 2C19 isozymes.24 The 1A2 set is the third case where the proportion of correct SMARTCyp predictions are found to be statistically equivalent to those of RS-Predictor.
Isozyme specific RS-Predictor models, which elucidate pathway-based regioselectivity trends through topological descriptors, have relatively consistent performance regardless of whether or not SMARTCyp reactivities are incorporated into model generation. Respective TOP QC and TOP SCR RS-Predictor models have average prediction rates of 79.9% and 83.2% for 2A6 and 2E1 substrate sets, 81.9% and 83.9% for 2C9 and 2D6 datasets, and 80.3% and 83.2% for all nine substrate sets. To ensure that these results are truly representative of how RS-Predictor will perform upon novel ligands, further validation experiments were employed.
Model Validity
Each individual substrate prediction made by RS-Predictor was performed by a model trained on 90% of the compounds within the substrate set for each isozyme in order to allow for bootstrapping and model validation. Subsequent to validation, production-level models were created using 100% of the available substrates in order to obtain the best possible RS-Predictor models for making blind predictions. This means that predictions made using these models will best reflect real-world situations. However, it may be that the in-house compounds to which these models are one day applied may occupy a different region of chemical space than those represented by our collated substrate sets, resulting in lower accuracy until models are extended to that portion of chemical space as well. As a separate issue, the substantial degree to which RS-Predictor outperforms SMARTCyp, StarDrop and Schrödinger methods may stem from the fact that it alone has training access to the large amount of experimental metabolite information that is only now being released in this work.
To address these issues we have performed additional validation tests by training RS-Predictor models on updated versions of the 98 2C9, 134 2D6 and 321 3A4 substrates previously released by Sheridan et al. in 2007. These Calibration models were then applied to the 128 2C9, 136 2D6 and 154 3A4 substrates that were newly collated in this work. These new data were treated as blind External sets using the Prediction schema in Figure 1. The Calibration and External sets of each isozyme represent independent samples of the available literature regioselectivity information; despite the fact that each set of is composed of substrates of the same isozyme, each sample may in fact represent very different regioselectivity spaces, just as the proprietary compounds within a given pharmaceutical company may represent a unique regioselectivity space. The differences in the SOMs of different oxidative pathways from the substrates of each Calibration and External set are given in the Supporting Information and help to illustrate these differences. Further analysis of similarity between the Calibration and External sets of each isozyme were made through fingerprint similarity analysis. MACCS fingerprints as implemented in MOE were calculated for the all of the substrates of each isozyme dataset.57 It was determined that using MACCS fingerprints and a 95% Tanimoto similarity that Calibration and External sets of 3A4, 2D6 and 2C9 respectively contained 16, 2 and 7 similar compounds. This suggests that the Calibration and External sets of each isozyme are significantly different in composition.
An additional benefit to performing Calibration and External set investigations is that results of RS-Predictor, SMARTCyp, StarDrop and Schrödinger may be compared with those previously released for Merck and MetaSite methods. These are not entirely one-to-one comparisons, as the Calibration sets of this work have been slightly updated based upon new literature information (see Methods), but they are still informative. We have also applied the 3A4 reactivity model of Schrödinger, which represents half of the Schrödinger 2C9 and 2D6 models, to both 2C9 and 2D6 sets. This additional experiment lets us to gauge the relative benefits of running the time-intentsive Glide docking component of Schrödinger 2C9 and 2D6 models, while also giving us a second purely reactivity-based set of results for each set from a source independent of SMARTCyp.
Overall results in Table 2 show that External RS-Predictor rates fall below those of Calibration rates; the drops in performance vary significantly between isozymes and descriptor sets, reflecting both differences in regioselectivity space, as well as the applicability of different descriptors towards those spaces. At the same time the Calibration models of RS-Predictor, even those that do not incorporate SMARTCyp reactivities, outperform all other methods for all sets, with the exception of SMARTCyp for the 3A4 External set. Since External 3A4 substrates contain on average four fewer potential SOMs than Calibration substrates, the fact that SMARTCyp External rates are higher than Calibration rates corroborates the hypothesis that SMARTCyp is best suited to identify the regioselectivity of smaller substrates. It also gives evidence beyond SMARTCyp results upon other isozyme substrate sets that the first-order principle represented through time-intensive DFT calculations may be viably applied to additional compounds for which they were not exclusively calibrated. The question then becomes: Why do other methods fare so poorly upon the 3A4 External set as compared to how well they perform upon the 3A4 Calibration set?
Table 2.
The percentage of each substrate set with an experimentally observed SOM predicted in the top two rank-positions by the given method for Calibration (Cal.)a and External (Ext.)b setsc
| Isozyme Substrate Set | 2C9 Cal. | 2C9 Ext. | 2D6 Cal. | 2D6 Ext. | 3A4 Cal. | 3A4 Ext. |
|---|---|---|---|---|---|---|
| Number of Substrates | 98 | 128 | 134 | 136 | 321 | 154 |
| RS-Predictor (TOP SCR) | 84.7 | 80.5 | 85.8 | 79.4 | 81.9 | 79.2 |
| RS-Predictor (TOP QC SCR) | 81.6 | 79.7 | 86.6 | 78.7 | 85.7 | 72.7 |
| RS-Predictor (TOP QC) | 78.6 | 78.9 | 84.3 | 77.2 | 81.0 | 68.8 |
| SMARTCyp | 67.7 | 66.9 | 48.5 | 68.1 | 73.1 | 77.2 |
| StarDrop | 77.4 | 78.4 | 81.5 | 69.2 | 77.5 | 66.9 |
| Schrödinger | 69.6 | 74.0 | 66.2 | 70.1 | 80.2 | 68.2 |
|
| ||||||
| Schrödinger (3A4 Model) | 73.5 | 71.9 | 58.5 | 68.1 | 80.2 | 68.2 |
| Merckd (Sheridan et al.) | 72.4 | — | 71.9 | — | 77.4 | — |
| MetaSited (2.7.5) | 68.8 | — | 65.4 | — | 61.8 | — |
|
| ||||||
| Random Model | 22.5 | 22.0 | 20.2 | 22.0 | 19.4 | 24.5 |
| Avg. # Observed SOMs | 1.9 | 1.6 | 1.6 | 1.6 | 1.8 | 2.0 |
| Avg. # Potential SOMs | 17.0 | 16.5 | 17.3 | 16.5 | 21.3 | 17.5 |
Cross-validated RS-Predictor results for the Calibration sets were obtained from predictions made using the Training schema described in Figure 1.
Blind RS-Predictor results for the External sets were obtained from predictions made using the Prediction schema described in Figure 1.
For each CYP, the optimal model is shown in bold, as are all other models found not to be statistically different using Fisher’s exact test of independence.
Since Merck and MetaSite models are not made public, results for these methods could not be obtained for the External sets.
Our findings indicate that the empirical modeling of electronic reactivity through 392 quantum chemical descriptors derived through AM1 semiempirical Hamiltonian result in overdetermined RS-Predictor models for the 3A4 Calibration set. Prediction rates of RS-Predictor models trained with QC descriptors on the External 3A4 set fall on average 11.6% below Calibration rates. However models trained using just topological descriptors, or topological descriptors with SMARTCyp reactivities, have External rates that fall just 2.7% below those of equivalent Calibration rates. The reason for this appears to be that AM1-derived signal has greater relevance for the 3A4 Calibration set than for the 3A4 External set. Performance rates of TOP QC and TOP QC SCR models for the 3A4 Calibration set exceed those of TOP and TOP SCR models by 6.2% and 3.8% respectively. These results indicate that the QC descriptors have significant signal for the 3A4 Calibration set, just as they do for the set of 2B6 substrates, but that the elucidated signal for QC models is not guaranteed to be applicable to a different set of 3A4 substrates that encompasses a different area of regioselectivity space. Respective performances drops of 12% and 10.6% for Schrödinger and StarDrop show that this phenomena of different 3A4 regioselectivity spaces is not indicated by RS-Predictor alone. As Schrödinger and StarDrop represent fundamentally different modeling algorithms than RS-Predictor, drops in performances are more likely to stem from different domains of applicability between the small set of substrates each method was calibrated upon (unreleased by either method) and the External sets of this work, rather than the particulars of a given modeling algorithm.
The signal relevance of QC descriptors for the largest set of 3A4 substrates available prior to our literature collation efforts helps to explain the large number of 3A4 and isozyme non-specific AM1-depedent regioselectivity methods that have been proposed, and raises questions about their overall utility.39,40,44,45 Another group investigating CYP hydrogen-abstraction models using PM3, SAM1 and AM1-derived regression models found results to be internally consistent within the given training set, but could only be applied to predict external DFT-derived activation energies for alkane reactions, and no other reaction types.58 They suggest, and we agree, that semiempirical-derived models should be interpreted and applied with a certain degree of caution. Still the small 0.2% difference between TOP SCR and TOP QC SCR rates for the complete set of 475 substrates of 3A4 suggests that robust MIRank modeling of the entire 3A4 regioselectivity space is able elucidate QC and non-QC signal with similar degrees of overall effectiveness.
This was proven when select 3A4 RS-Predictor models were applied using the Prediction schema in Figure 1 to a set of 38 in-house 3A4 substrates provided by a major pharmaceutical company that we are currently collaborating with. TOP QC, TOP QC SCR and TOP SCR models were respectively able to identify the observed SOMs in the top two rank-positions 71%, 71% and 76% of the time. StarDrop also performed well with an accuracy of 74%; in contrast SMART-Cyp and Schrödinger faired poorly upon this set, with respective prediction accuracies of 55% and 58%. The higher accuracy of RS-Predictor and StarDrop models, while encouraging, do not imply that they are the only methods that should be applied to compounds under development, merely that on on average they are more accurate than the other two methods. In Table 3 we see that each individual method and model is able to identify the 3A4-mediated metabolism of at least one compound that is not correctly identified by another method or model. The SMARTCyp reactivity descriptor has less overall effect on the base TOP QC RS-Predictor model accuracy for this set (TOP QC models have the highest top three rank-prediction accuracy of all the methods) than for the public 3A4 substrate set. Meanwhile the QC models of 3A4 models are shown to be just as robust as non-QC models for this set of substrates. A point made in our prior work was that the base 3A4 TOP-QC RS-Predictor model is less accurate than other methods when considering only the first predicted rank-position, but more accurate than other methods when the second and third positions are taken into account.50 Results for this set of compounds corroborate this finding, while showing the incorporation of the SMARTCyp reactivity descriptor improves the accuracy of the first rank-position while leaving second and third rank-position accuracy relatively unchanged.
Table 3.
The number of 38 proprietary 3A4 substrates correctly predicted in the top two rank-positions by the column method but not the row method. When row and column designate the same model the number of substrates correctly predicted in the top 1, 2 and 3 rank-positions are given
| Model | TOP QC | TOP QC SCR | TOP SCR | SMARTCyp | StarDrop | Schrödinger |
|---|---|---|---|---|---|---|
| TOP QC | 16,27,34 | 4 | 4 | 6 | 7 | 4 |
| TOP QC SCR | 3 | 19,27,33 | 4 | 4 | 6 | 3 |
| TOP SCR | 3 | 2 | 22,29,34 | 1 | 5 | 2 |
| SMARTCyp | 11 | 9 | 8 | 16,22,26 | 10 | 5 |
| StarDrop | 6 | 5 | 6 | 4 | 21,28,30 | 4 |
| Schrödinger | 9 | 8 | 9 | 5 | 10 | 16,22,27 |
Quantum chemical descriptors have less overall significance towards the identification of 2D6- mediated regioselectivity. The prediction rates of TOP SCR and TOP QC SCR models are within 1% of each other for both Calibration and External 2D6 sets. The DFT-derived transition state energies encoded within SMARTCyp are not equally effective at the identification of 2D6-mediated metabolism, with Calibration and External rates that differ by almost 20%. SMARTCyp prediction rates for the 2D6 Calibration set are astonishingly low at 48.5%, but at 68.1% almost equal the performances of StarDrop and Schrödinger for the External set. Such discrepancies cannot be explained by overall substrate size; propensities of Csp3 hydroxylation, O-, N-dealkylation and S(II)-oxidation have some variance between sets, though not enough to justify such significant differences in SMARTCyp performance. Instead these findings indicate that electronic reactivity is much more important for the External 2D6 set than it is for the Calibration set. Indeed, the Schrödinger 3A4 model, a reactivity-based method developed completely independently from SMARTCyp has a 9.6% performance increase for the External 2D6 set relative to Calibration set rates. When Glide docking is combined with the 3A4 model to create the Schrödinger 2D6 model, Calibration rates rise by 7.7%, corroborating the importance of isozyme structure towards 2D6-mediated regioselectivity. The discrepancy between Schrödinger 2D6 and 3A4 model performance is much more pronounced for the Calibration set (7.7%) than for the External set (2.0%), likely due to the greater signal of the Schrödinger reactivity model for the External set. Schrödinger Calibration rates are significantly lower (15.3%) than those of StarDrop, but when the third predicted rank-position is taken into consideration, Schrödinger rates fall only 3.5% lower than those of StarDrop. RS-Predictor does approximately 5% better upon the 2D6 Calibration than StarDrop, but 10% better for the External set. Where SMARTCyp and Schrödinger External rates improve relative to Calibration rates by 19.6% and 3.9% respectively, StarDrop and RS-Predictor External rates respectively fall by 12.3% and ~7%. Differences between RS-Predictor Calibration and External 2D6 rates are relatively constant, regardless of the descriptor set used. Few details are known about the orientation and steric descriptors used within StarDrop, and we are therefore unsure why External rates should drop so significantly, beyond the fact that Calibration and External 2D6 sets appear to represent significantly different regioselectivity spaces.
Whereas each method has significant differences in performance for the Calibration and External sets of 3A4 and 2D6, the same cannot be said 2C9. Performances for each method and model are relatively stable across all 2C9 substrates. This is somewhat surprising given that this is the only case where the External set is substantially larger than the Calibration set, and there are significant differences in pathway propensities of Csp3 and Csp2 oxidation. One explanation could be the relative similarity in observed propensities of O-, N-dealkylation and S(II)-oxidation between Calibration and External sets; 2C9 is the only isozyme for which these mechanisms all have similar propensities between sets. It is also the only isozyme for which both TOP QC and StarDrop External rates rise relative to the Calibration rates. Schrödinger rates also rise, but only for the 2C9 model, the 3A4 reactivity-based model rates fall slightly. While Glide docking makes the Schrödinger 2C9 model more robust for the External set, the 3A4 model actually does better for the Calibration set, raising questions about which Schrödinger model should be applied to an unknown 2C9 substrate. 2C9 is also the only isozyme where SMARTCyp External rates fall below Calibration rates, and though the fall is by only by a small amount (0.8%), it helps to explain the lack of robustness of TOP SCR models relative to TOP QC SCR models.
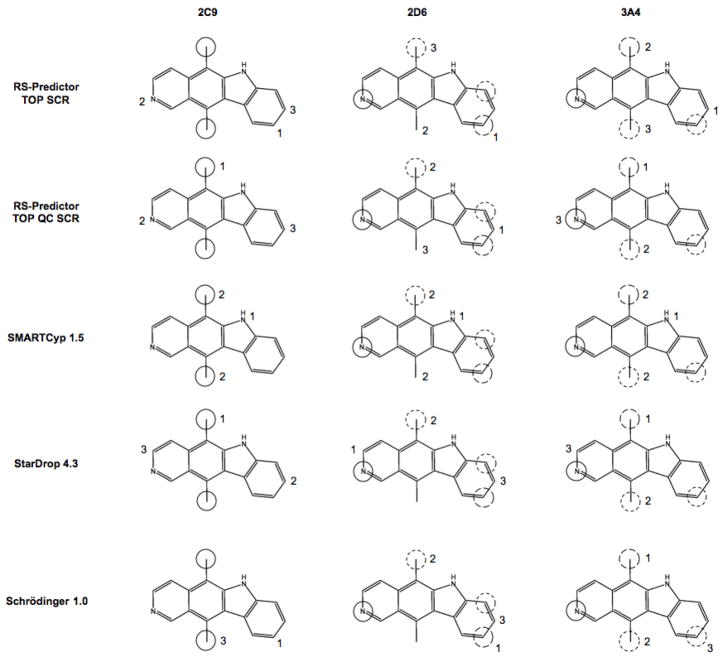
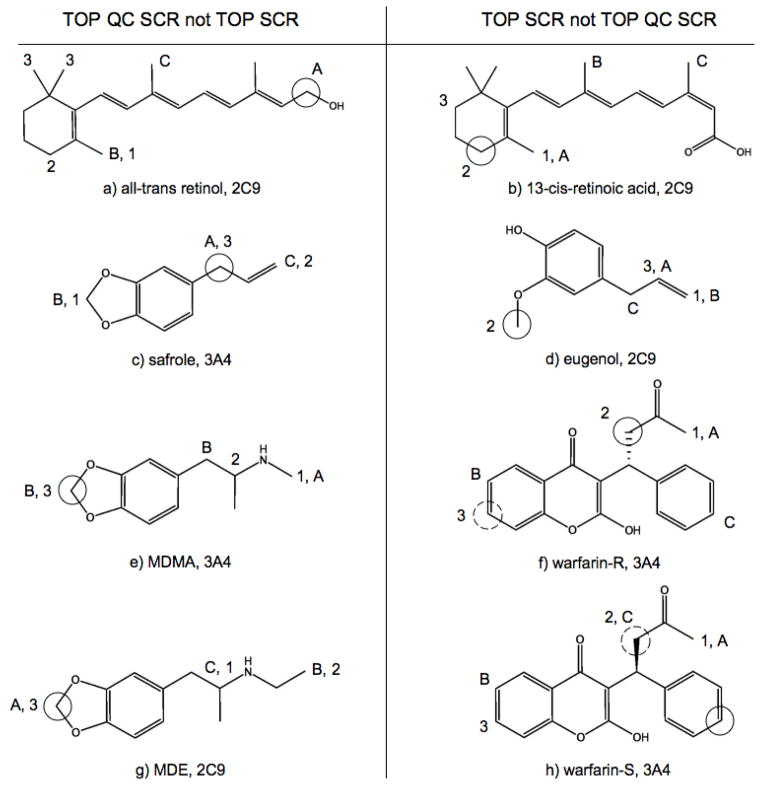
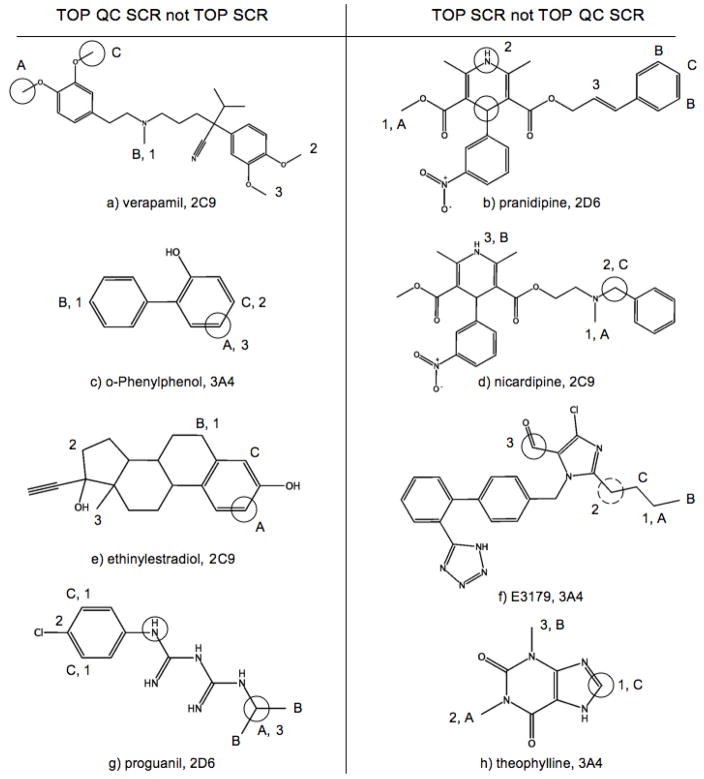
While TOP SCR rates are optimal for the three External sets, TOP QC SCR models only predict one substrate less than TOP SCR models for 2C9 and 2D6 sets. Meanwhile the QC-regioselectivity space of 3A4 Calibration and External sets are significantly different, as evidenced by Calibration/External performance rates of 74.8%/72.1% and 81.0%/68.8% for TOP and TOP QC models respectively. As shown earlier TOP QC, TOP QC SCR and TOP SCR models have equivalent overall performances for a different set of 38 proprietary substrates of 3A4. While the number of accurately predicted substrates between TOP SCR and TOP QC SCR models only differ by one for 2C9 and 2D6 External sets, this does not mean that TOP SCR models correctly predict one additional substrate that was missed by TOP QC SCR models. In fact TOP SCR models identified 9 2C9, 8 2D6 and 16 3A4 External substrates missed by TOP QC SCR models, while TOP QC SCR models identified 8 2C9, 7 2D6 and 6 3A4 External substrates missed by TOP SCR models. This then raises the question which model, TOP SCR or TOP QC SCR, should be applied to predict the regioselectivity of an unknown compound. To help future users of RS-Predictor answer this question, as well to aid the endeavors of future modelers, we have shown example substrates from the External sets that were predicted by one of these models but not the other in Figure 7, Figure 8 and Figure 9.
Figure 7.
Substrates with an observed SOM predicted in the top two rank-positions by TOP QC SCR models and not TOP SCR models in the left panel, and by TOP SCR models but not TOP QC SCR models in the right panel. The first, second and third predicted SOMs are designated by 1, 2 and 3 for TOP SCR models and A, B and C for TOP QC SCR models. Primary and secondary observed sites of oxidation are designated by solid circles and coarsely hashed circles respectively.
Figure 8.
Substrates correctly predicted by either TOP SCR or TOP QC SCR models, but not both. Labeling scheme detailed in Figure 7.
Figure 9.
Substrates correctly predicted by either TOP SCR or TOP QC SCR models, but not both. Labeling scheme detailed in Figure 7.
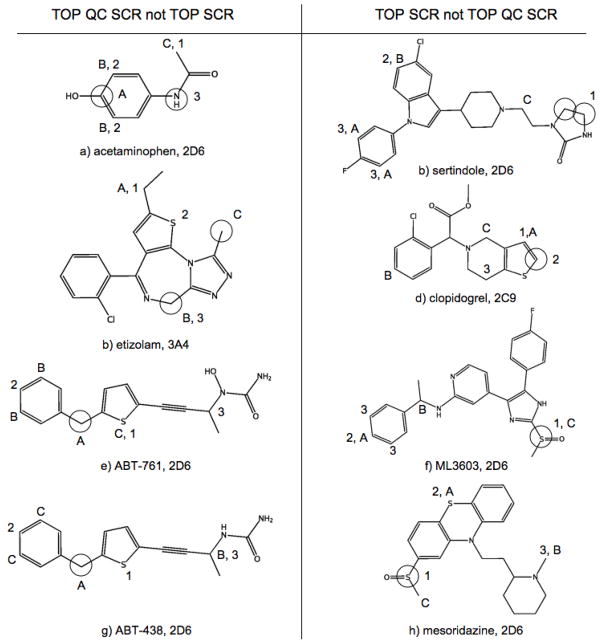
In the oxidation of all-trans retinol(6a) by 2C9, RS-Predictor models created with topological and SMARTCyp descriptors alone erroneously predicted that the preferred metabolic sites would be on the allylic methyl and methylene groups on the cyclohexene ring, and the third site involving the geminal methyl groups; the addition of QC descriptors allowed the TOP QC SCR models to properly identify the observed Csp3 oxidation site in the first rank-position. However, in 13-cis-retinoic acid(6b), the terminal carbon bearing the hydroxyl group is no longer available as a potential Csp3 reaction site, and the predictions made without QC descriptors are more accurate. A similar situation occurs in the naturally occurring compound eugenol(6d), an extensively used flavoring agent and fragrance whose CYP-mediated O-dealkylation results in the toxic product hydroxychavicol that can damage DNA. In this case, the inclusion of QC descriptors bias the outcome and predict that potential reactions at the allylic or vinylic positions are more favorable than at the actual observed SOM, whereas TOP SCR models identify the observed O-deakylation in the second rank-position. Safrole is another natural product that only differs from eugenol in that the two ortho oxygens on the benzene ring are joined as a 1,3-dioxolane. This region of the fused bicyclic molecule is then more stable to CYP-mediated oxygen dealkylation, and the reaction outcome changes from O-demethylation for eugenol to Csp3 hydroxylation at the benzylic/allylic position for safrole, making TOP QC SCR models more accurate in this case. Replacement of the vinyl moiety of safrole with two different saturated secondary amines give the structures of the widely abused amphetamine derivatives MDMA(6e) and MDE(6g). The addition of QC descriptors to TOP SCR models was found to increase the predicted rank-position of the observed SOMs of these two compounds. CYPs 1A2, 2B6 and 3A4 also mediate the N-deethylation of MDE as a secondary reaction; TOP SCR and TOP QC SCR models both identify the observed SOMs in the first and second rank-positions, but only QC models predict them in the correct order. These findings all indicate that QC descriptors are better suited to predict compounds with 1,3-dioxolane moiety fused to an phenyl ring.
The 3A4-mediated pathways of the R(6f) and S(6h) enantiomers of warfarin are better predicted by TOP SCR models than those involving QC descriptors. This represents additional evidence that the inclusion of QC descriptors tends to overestimate the reactivity of certain aromatic and enolic sites. Further analysis of the reactivity patterns of both warfarin enantiomers shows that both of the aromatic ring hydroxylations mediated by 3A4 are also mediated by 2C9, as is an additional aromatic ring reaction that was properly identified by the second predicted SOM of 3A4 TOP QC SCR models. For these 2C9-mediated reactions, TOP QC SCR models were better at predicting the observed reactions, in contrast to what was observed in 3A4 warfarin modeling. Since QC descriptors were not designed to encode enantiospecific information, some loss of accuracy may be expected, and suggests that the incorporation of chiral descriptors into RS-Predictor modeling would be a viable path for future research.
With verapamil(7a), a calcium channel blocking agent used in the treatment of cariovascular disorders, models trained with quantum chemical descriptors alone identify the two observed SOMs in the first two rank-positions, and the non-observed para-oxygen dealkylation in the third rank-position. The addition of topological descriptors changes changes that last prediction to the non-observed N-deaklylation, while the addition of SCR descriptor places moves the N-deaklylation to the second predicted position, while the observed meta O-deakylation moves to the third rank-position. A conclusion to be drawn from this observation is that for relatively symmetric molecules, QC descriptors will be more informative than the 2D-based atom distributions represented through topological descriptors that do not explicitly represent information about the local electronic environment around a putative site. Another relatively symmetric molecules that is more accurately predicted by QC models than non-QC models is o-Phenylphenol(7c), a fungicide used on harvested citrus fruits with a toxic CYP-mediated metabolite. The CYP-mediated site of benzenol hydroxylation was identified by TOP SCR models in the third rank-position, but by TOP QC SCR models in the first rank-position.
Pranidipine(7b) and nicardipine(7d) are calcium channel blockers with significantly different scaffolds than verapamil; each substrate is composed of a Hantzsch dihydropyridine ring with an attached meta-nitrobenzene, but differ by having slightly different ester chains. SMARTCyp highly favors the prediction involving Hantzsch pyridine hydroxylation, with the two observed sites of 2D6-mediated pranidipine metabolism in the first and second rank-positions. These same sites are predicted by SMARTCyp in the same positions for nicardipine, barnidipine and nifedipine - these molecules are metabolized in those positions by some CYP isoforms and not others. Neither topological or quantum chemical descriptors were able to identify this reaction class. Therefore, it is possible to suggest that TOP SCR models, which do not contain the large number of QC descriptors that have the potential to dilute encoded SMARTCyp reactivity information, should be used to for predicting the sites of CYP metabolism on molecules with a Hantzsch pyridine scaffold. Results for E3179(7f) and theophylline(7h) indicate tha substrates witht imidazole moieties should also be predicted by TOP SCR models, or by SMARTCyp alone. For both of these cases, TOP, QC and TOP QC models were unable to identify the observed SOMs on or adjacent an imidazole, whereas SMARTCYP identifies the observed SOMs in the 1st and 2nd rank-positions. Another case where QC models make less accurate predictions near an nitrogen-containing aromatic ring is the atypical serotonin-dopamine antagonist antipsychotic drug sertindole(8b). In this case, however, the observed 2D6-mediated reaction is an imidazolidine hydroxylation on a different region of the molecule, which TOP SCR models identify in the first rank-position.
As stated earlier QC models tend to have higher aromatic ring TP rates, and lower non-aromatic ring FP rates. One example of this is the greater accuracy TOP QC SCR models upon the steroid hormone ethinylestradiol(7e). TOP SCR models identify two non-observed non-aromatic ring hydroxylations, but QC signal let RS-Predictor identify the observed aromatic ring hydroxylation in the first rank-position, and a different position on the same ring in the third rank-position. Another case of improved QC prediction quality is the 2D6-mediated ring formation of the prodrug proguanil(7g). Topological descriptors alone identify the aromatic ring region of the molecule as being the active site in the first and second predicted rank-positions and the addition of SMARTCyp reactivities place the observed SOM in the third rank-position. On the hand, Quantum chemical descriptors alone identify the isopropane group as being reactive in the second and third rank-posisionts, while the addition of topological descriptors places the observed site of oxidation in the first rank-position.
The mechanism through which the widely used analgesic and antipyretic agent acetaminophen(8a) is metabolized is not completely understood. It is therefore fitting that TOP SCR and TOP QC SCR models would each identify only one of the reaction sites that have the potential to yield the known product NAPQI, a toxic compound that causes severe hepatocellular damage. In this case, SOM predictions of TOP and TOP QC models are identical to their SCR counterparts, which is favorable considering that SMARTCyp alone is not able to identify either of the observed reaction sites. Unfortunately, lack of change in SCR predictions relative to non-SCR predictions is not always a good situation. Etizolam(8b) is a short-acting benzodiazepine derivative prescribed for the treatment of insomnia and related sleep disturbances that undergoes 3A4-mediated Csp3 hydroxylation and 2-hydroxylation of a 1,4 diazapine. Both these sites are identified by SMARTCyp in the top two rank-positions, while QC models only identify the Csp3 hydroxylation site and TOP models only identify the ring hydroxylation site. TOP QC models were able to identify both reaction sites, but only in the second and third rank positions. The incorporation of SMARTCyp reactivities leaves the results unchanged, despite optimal SMARTCyp predictions. Etizolam also provides an example of how QC descriptors can lessen sulfur oxidation FPs, specifically on a thiophene ring.
TOP SCR models for thiophene sulfur oxidation made false-positive predictions on two 5-lipoxygenaze inhibitors released by Abbott Laboratories: ABT-761(8g) and its N-hydroxylated metabolite ABT-438(8e). The 2D6-mediated oxidation at the benzylic methano bridge are only predicted by TOP QC models, but not by TOP or QC models individually; the incorporation of SMARTCyp reactivities appear to have little effect. For the antithrombotic prodrug clopidogrel(8d) however, the observed 2C9-mediated monooxygenation of the fused thiophene ring is only predicted by TOP SCR models. It appears that non-QC models tend to predict thiophene reactions more often than QC models do, regardless of whether CYP-mediated metabolism actually occurs on that moiety for a particular substrate. This could explain why both the TP and FP rates of TOP SCR models are higher than those of TOP QC SCR models for sulfur-based oxidation reactions. TOP SCR models were also found to perform better in the prediction of S(IV) oxidations. The observed S(IV) oxidation of ML3603(8f), a metabolite of the potent inhibitor of p38 MAP kinase ML3403 is better predicted by non-QC models, another case of a poor quality QC prediction for a site near an imidazole moiety. Removal of QC descriptors lowers sulfur FP rates while simultaneously identifying sites of 2D6-mediated S(IV) oxidation of the phenothiazine neuroleptic mesorisazine(8h), a blocker of dopaminergic D2 and noradrenergic α1 receptors.
The main conclusions to be drawn from the above cases about how and when QC descriptors and models are most appropriate can be summarized as follows:
QC descriptors are often better suited to the identification of sites on or near aromatic rings
QC models should be used when making predictions on compounds with a benzene-fused 1,3-dioxolane moiety
QC descriptors are not sufficient to discriminate between enantiospecific CYP-mediated pathways in chiral compounds.
QC models have difficulties identifying observed SOMs on or near nitrogen containing rings, specifically Hantzsch dihydropyridine, imidazole and imidazolidine moieties.
Non-QC models have a greater ability to correctly prioritize sulfur-based reactions, and have greater numbers of TPs and FPs than QC models
It is important to remember that the cases described above represent comparisons between two different RS-Predictor models as applied to specific External sets which represent a relatively small sampling of the assembled P450-substrates. Conclusions drawn from such a sampling should only be considered suggestive, and not be taken as canon.
Conclusion
The incorporation of fragment DFT transition state energies from SMARTCyp into the RS-Predictor modeling framework enabled the creation of robust isozyme-specific CYP regioselectivity QSARs trained using the largest set of P450 substrate data released to date. Optimal combinations of RS-Predictor and SMARTCyp were able to identify experimentally observed sites of metabolism within the top two rank-positions for substrate sets for each CYP isozyme with the a high level of cross-validated accuracy: CYP(number of substrates, accuracy), 1A2(271, 83.0%), 2A6(105, 85.7%), 2B6(151, 82.1%), 2C19(218, 86.2%), 2C8(142, 83.8%), 2C9(226, 84.5%), 2D6(270, 85.9%), 2E1(145, 82.8%), 3A4(475, 82.3%) and merged(680, 86.0%). Performance of these models were demonstrated to be higher than those made by commercially available regioselectivity models from StarDrop(78.0%, 75.3%, 74.1%) and Schrödinger(72.1%, 68.1%, 76.4%) for the 2C9, 2D6 and 3A4 sets, respectively. Combined models were also shown to have higher respective performances than RS-Predictor models trained without SMARTCyp reactivities (80.5%, 83.3%, 77.7%), or SMARTCyp alone (67.3%, 58.4%, 74.4%). To validate RS-Predictor performance rates, models were created using the largest sets of 2C9(98), 2D6(134) and 3A4(321) substrates available prior to this work which were then applied to the newly assembled datasets of substrates of each isozyme, which were used as blind external sets (2C9:128, 2D6:136, 3A4:154). Calibration and external rates were found to be statistically equivalent for 2C9 models, and the same was found for 3A4 models that were created without quantum chemical descriptors. Quantum chemical descriptors were found to have significantly better results when identifying the regioselectivity of the 3A4 calibration set alone, but the added effectiveness did not translate to the 3A4 external set. External rates of the 2D6 models fell roughly 7% below those of equivalent calibration rates, while all other methods except the Schrödinger method showed even greater performance discrepancies between sets. Performance differences for the majority of methods indicate that 2D6 calibration and external sets represent significantly different areas of regioselectivity space. Meanwhile the prediction rates of optimal combinations of RS-Predictor and SMARTCyp surpassed those previously reported by Merck (12.3%, 14.7% and 8.2% more accurate) and by MetaSite (15.9%, 21.2% and 23.9% more accurate) for the 2C9, 2D6 and 3A4 calibration sets, respectively. In addition to performing well on 2C9, 2D6 and 3A4 substrates, this work describes the first ligand-based regioselectivity models ever released for CYPs 1A2, 2A6, 2B6, 2C19, 2C8 and 2E1.
What makes RS-Predictor modeling different from the majority of regioselectivity prediction methods is that no explicit modeling of a particular substrate within an isozyme binding pocket is made. Instead, regioselectivity signal is elucidated from three sources: 1) topological descriptors, which represent the relative propensities of different oxidative pathways to occur for a particular isozyme, 2) quantum chemical descriptors, which represent the local electronic environment of putative sites with respect to the global electronic environment of the substrate, and 3) SMARTCyp transition state energies, which represent the local energy barrier that each unique fragment must overcome to be oxidized by a CYP heme. The benefit of this strategy is that once catalytic trends have been obtained, the encoding of a candidate ligand with these descriptors and application of a previously generated QSAR may be accomplished in approximately 2 s for a non-quantum chemical model and 9 seconds for a quantum chemical model. These runtimes are shorter than the one to ten minute runtime of StarDrop required by MOPAC calculation of substrate abstraction energies, and significantly faster than the two to two and a half hour runtime of Schrödinger requires for induced-fit docking. In contrast to this, the reactivity-only models from both SMARTCyp and Schrödinger may be applied in less than a second, but both lack the isozyme-specific signal provided by RS-Predictor models. While the implicit representation of the catalytic trends of a particular isozyme using topological descriptors is quick and yields impressive results, incorporation of data from the explicit docking of candidate substrates will likely produce even better overall performance. Investigation into the development of quickly calculable docking-based descriptors compatible with the RS-Predictor modeling framework is currently underway.
The main weakness of RS-Predictor and the majority of other regioselectivity prediction models is the implicit assumption that any compound submitted to the model is assumed to be a substrate of the isoform for which the given model was calibrated. This, of course, may not be the case. Isoform specificity models would address this issue by determining through molecule-specific descriptors whether or not a given compound is a substrate of a specific isozyme. The rationale behind these models is relatively straightforward — if there is an accurate method to identify which, if any, CYP will metabolize an unknown compound, then the actual CYP-mediated regioselectivity of that compound is more likely to be identified by the isoform-specific model of that CYP. In recent years, a few isoform specificity models have been published, but more work is needed in that area.59–62 The available CYP selection models cover only 1A2, 2C19, 2C9, 2D6 and 3A4 isozymes, and were trained from smaller substrate sets (226, 379) than those used in this work. Consequently, the 680 substrates collated in this work serve as a strong foundation upon which improved isozyme specificity models may be built. It is important to note however, that the datasets used in this work were compiled specifically for isoform regioselectivity modeling, not for isoform specificity selection. The fact that a given substrate is not a member of a particular isozyme substrate set does not necessarily mean that it is not metabolized by that isoform; it may just mean that the investigating group did not test for the regioselectivity of that isoform. Further analysis and curation of the literature would facilitate the development of isozyme specificity models.
Specificity models function in a different fashion than regioselectivity models in that descriptors are calculated for the entire compound, not for the potential oxidation sites (SOMs) of that compound. This makes isoform specificity modeling and subsequent regioselectivity modeling a subclass of multi-task modeling, a process by which multiple related tasks may be modeled simultaneously.63 In the case of CYP-mediated metabolism, this covers not only isoform specific mediated pathways, but differences in the pathways mediated upon different types of compound. The differences in performances of multiple methods upon different sets of 2D6 and 3A4 substrates illustrate how substrates of the same isozyme may encompass different regioselectivity spaces. In a similar fashion substrates identified by models trained either with quantum chemical descriptors or without them, but not by both, demonstrate the relative utility of different descriptors and models towards identifying the regioselectivity of the same set of substrates.
The end-goal of regioselectivity modeling is to identify how a human being will metabolize a specific compound without actually administering that compound. Accurate regioselectivity modeling is an important component of early-state drug discovery efforts. Continued advancements in the creation of robust regioselectivity models require that future investigators be as adaptable as the CYP isozymes that they seek to model. The combination of RS-Predictor and SMARTCyp results reported here as applied to the largest open-source set of P450 substrates ever released provides a state of the art benchmark by which future efforts should be judged.
Methods
Substrate Sets
Prior to this work, the most extensive publicly available collection of P450 substrates with identified SOMs and source papers was released by Merck in the form of six datasets for three isozymes. Substrates for 2C9, 2D6 and 3A4 were respectively released in the form of calibration sets of sizes 91, 124 and 305, and external sets of sizes 10, 10 and 19.27 These sets serve as the foundation from which we have compiled nine isozyme-specific substrate sets, as well as a conglomerate merged set of P450 substrates. Substrates and metabolites from source papers identified by Sheridan et al., review papers of Rendic and Brown,64,65 the SuperCYP database,66 as well those found within the public literature were curated as SDF files serving as the starting point for RS-Predictor model generation. These files are part of the the Supporting Information associated with this publication.
Curation of the primary literature has resulted in new substrate sets for CYPs 1A2, 2A6, 2B6, 2C19, 2C8, and 2E1 of respective sizes 271, 105, 151, 218, 142, and 145. Meanwhile the original 98 2C9, 134 2D6 and 321 3A4 substrates released by Merck have been updated based on new source information, as well as being respectively extended by 128, 136, and 154 substrates. Literature curation is a manual, time intensive process that involves the collation of reported substrate oxidations from a number of different sources. The techniques used to determine oxidation sites vary between reporting groups. Two of the most common techniques are the incubation of the “unknown” substrate with isolated microsomes of a specific CYP isozyme or with multiple samples of human liver combined with known inhibitors of specific CYP isozymes. Mass-spectroscopy is used to determine the structure of observed metabolites, while differences in the inhibition levels each CYP with and without the given substrate are used to gauge rate constants for the given isoform. The reactant concentrations, tools used and motivations can vary significantly between studies; occasionally the use of smaller concentrations can yield a greater rate constant, while different studies often report different rate constants or even different metabolites for the same substrate and isozyme. Examine the CYP-mediated metabolism of atomoxetine in Figure 10. One study tested for two different metabolites for nine CYP isozymes, while another did an in-depth analysis of atomoxetine metabolism by CYP 2D6. The 2D6 study identified a 2D6-mediated Csp3 hydroxylation in addition too the aromatic ring hydroxylation and N-dealkylation investigated in the other work. It is possible that other isozymes also metabolize the Csp3 hydroxylation but the investigating group did not test for it. For the purposes of this work any metabolite reported from any paper with a clearance ( ) value above 0.05 for a specific human isoform was included in the substrate set of that isozyme. When possible, differentiation between primary, secondary and tertiary observed metabolites were also recorded. The atom identified by the author to receive the oxidation radical from the CYP heme was the one marked, though in more cases than not the mechanism of reaction was not a part of the study. Human error in terms of correct identification of the mechanism of metabolism and mislabeling of sites are also possible. Errors in source data is an unfortunate, but common issue with retrospective regioselectivity modeling; this is one of the main reasons why the standard metrics of gauging regioselectivity prediction accuracy treat a given substrate as being correctly predicted as long as any observed metabolite, regardless of the source, is identified within the top two predicted rank-positions.
Figure 10.
The CYP-mediated metabolism of atomoxetine. The primary and secondary metabolites, in terms of observed reaction rates, are indicated by the subscript after each isozyme.
In addition to mining the public literature for new works that had not been curated by Sheridan et. al, a large amount of our efforts involved looking up the papers originally identified by them and spreading the structures and response information to 1A2, 2A6, 2B6, 2C19, 2C8, and 2E1 isozyme sets. During this process, errors within the original Merck substrate sets were identified. For the 3A4 set, the compound H_259_31 occurred twice, while SSR97193 and lu25_109 were removed due to low reported oxidation rates.67,68 Reports of low or non-existent oxidation of SSR97193, BPU and luciferin resulted in their removal from the 2C9 dataset.67,69,70 Errors in structure were identified and fixed for aflatoxin_b1 and promazine for 3A4 and Δ3-carene for 2D6. Meanwhile 143, 59 and 41 compounds from respective 3A4, 2D6 and 2C9 datasets had their observed metabolites updated based upon source literature. The majority of these fixes involved the switching of rank-positions of sites that were incorrectly identified as primary observed site of oxidation versus a secondary observed site of oxidation, which would not effect comparison between the results of Merck and MetaSite within that paper and our results presented in this paper. Occasionally however an observed site of metabolism was only substantiated by non-human CYP studies, in which case that site was removed. Other times the marked site of oxidation did not correspond to the reported metabolite, in which case the correct site of oxidation was recorded. There is an additional field in the 2C9, 2D6 and 3A4 substrate sets within the Supporting Information that indicates whether a given substrate or the observed metabolites of that substrate have been altered from their original release by Sheridan et al... Unfortunately the Supporting Information of Sheridan et al. only provides the structures and experimentally observed SOMs of each substrate, not the predictions of each individual method upon that substrate. Only the overall prediction rate of each method was given in the main paper; also 3A4 results were presented for 335 compounds but only structures for 324 compounds were released. One structure for 2C9 was withheld as well. Given these facts it is important to realize comparisons between Merck and MetaSite results and those of RS-Predictor, SMARTCyp, StarDrop and Schrödinger are not truly one-to-one. In addition the results reported for MetaSite were for version 2.7.5 a significantly older version of the software. While we have access to MetaSite version 3.1, their legal policy prohibits the public benchmarking of MetaSite against other technologies and so were unable to calculate the up-to-date performance of this technology for our new substrate sets. This is unfortunate given that MetaSite is the only method to date(with the exception of RS-Predictor) with reported 1A2 or 2C19 regioselectivity models that have been validated on large-scale substrate sets.24
Given the importance of P450-mediated Phase I metabolism to biological survival it is unsurprising that functional redundancy between isozymes has arisen, whereby the same substrate is often metabolized by multiple isoforms. The number of substrates that overlap between different isozyme sets are shown in Table 4. Further analysis, made available to the interested reader in the Supporting Information, shows that when the same substrate is metabolized by different isozymes, the resultant metabolites have a high degree of overlap; only a small percentage of the time does the same substrate undergo completely different P450-mediated reactions. Observed metabolites are often the same, but relative reaction rates vary between isoforms, resulting in situation where the primary metabolite of one isoform is sometimes the secondary or tertiary metabolite of another. Differences in metabolic function between isoforms likely reflects evolutionary specialization of different P450 isozymes. The most extreme of cases contradictory regioselectivity signals are found in the 192 compounds metabolized by both 2D6 and 3A4. Of these, 107 have identical observed SOMs, while 130 share the same primary site of oxidation and only 21 have completely different 3A4 and 2D6-mediated pathways. In total there are 680 unique compounds, which have been merged together into an isozyme non-specific P450 substrate set. All observed metabolites, regardless of isozyme, are represented in the merged dataset.
Table 4.
Substrate set sizes and the number of overlapping substrates between sets
| Isozyme | 1A2 | 2A6 | 2B6 | 2C19 | 2C8 | 2C9 | 2D6 | 2E1 | 3A4 |
|---|---|---|---|---|---|---|---|---|---|
| 1A2 | 271 | 63 | 100 | 139 | 88 | 129 | 162 | 98 | 192 |
| 2A6 | 105 | 57 | 51 | 42 | 62 | 63 | 63 | 71 | |
| 2B6 | 151 | 96 | 68 | 89 | 89 | 65 | 122 | ||
| 2C19 | 218 | 90 | 141 | 147 | 62 | 167 | |||
| 2C8 | 142 | 106 | 83 | 51 | 125 | ||||
| 2C9 | 226 | 125 | 76 | 169 | |||||
| 2D6 | 270 | 79 | 192 | ||||||
| 2E1 | 145 | 94 | |||||||
| 3A4 | 475 |
The regioselectivity signal of each set is shown in Table 5 as the average number of observed and potential SOMs per substrate within each set. A listing of the sets in order of decreasing regioselectivity signal are 2E1, 2A6, 1A2, merged, 2B6, 2C9, 2C8, 2C19, 3A4 and 2D6. Ordering of the sets by increasing substrate size yields a similar, though not identical list: 2E1, 2A6, 2B6, 1A2, 2C19, 2C9, 2D6, 2C8, merged and 3A4. The pathway-based catalytic trends of different isozymes are shown in Table 6. Pathway propensities are determined by placing the SOMs of all substrates of an isozyme set into the CYP-mediated reaction pathway set that they have the potential to undergo. Differences in pathway-mediated trends between CYP isoforms are used to highlight relative strengths and weaknesses of different methods and models for accurately identifying the substrate metabolites of specific isozymes. To simplify pathway-based performance analysis made in the Results section, similar pathways, or those with low relative populations such as sulfur-based reactions, are grouped together into the same pathway-analysis set.
Table 5.
Available regioselectivity signal (%) from the average number of observed and potential SOMs per substrate per dataset
| 1A2 | 2A6 | 2B6 | 2C19 | 2C8 | 2C9 | 2D6 | 2E1 | 3A4 | merged | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| observed | 1.9 | 1.5 | 1.5 | 1.6 | 1.7 | 1.7 | 1.6 | 1.7 | 1.9 | 2.0 | |
| potential | 16.0 | 12.3 | 14.5 | 16.7 | 17.2 | 16.8 | 17.0 | 12.2 | 20.1 | 18.0 | |
|
|
11.9% | 12.2% | 10.3% | 9.6% | 9.9% | 10.1% | 9.4% | 13.9% | 9.5% | 11.1% |
Method Details
One difference between the RS-Predictor algorithm of our prior work and of this work is that models are created using 10-fold cross-validation, instead of 5-fold cross-validation. This decision was made because for some of the smaller substrate sets (2A6, 2E1), results would occasionally suffer when models were trained upon 80% instead of 90% of the metabolite data. The only other change is the incorporation of SMARTCyp reactivity values as an additional descriptor, which has been shown throughout this work to substantially increase overall accuracy of RS-Predictor models.
Descriptor generation takes on average 1 s per substrate for non-QC models and 3 s for QC models. The rate-limiting step of RS-Predictor is the MIRank employment of 10 iterations of 10-fold cross-validation. MIRank models were calibrated upon multiple 2.6 GHz Opteron Linux workstations with 8 CPUs, often with three independent runs being created at the same time by each machine. Overall runtime is highly influenced the number of dimensions that are being optimized; the number of dimensions are directly dependent upon the number of substrates in the training set, and the number of descriptors being used to quantify the SOMs of each substrate. The greater the number of either, the greater the runtime. The time taken to create models (TOP SCR, TOP QC SCR), for the 105 substrates of 2A6(9.0 h, 13.7 h), 226 substrates of 2C9(1.1 d, 1.71 d), 475 substrates of 3A4(2.95 d, 4.55 d), and all 680 substrates of the merged(3.86 d, 6.23 d) set illustrate the broad run-time trends. It is important to remember that these runtimes reflect 10 iterations of 10-fold cross-validation; using only one iteration would require a tenth of the runtime, but would not give the additional signal bump obtained through use of rank aggregation (see Supporting Information). Once models have been trained however the only steps necessary to make a prediction are to encode an input compound with the relevant descriptors and apply the previously trained models to that encoding. It takes approximately 1.8 s to apply a TOP model and 9 s to apply a TOP QC model to a given substrate. SMARTCyp takes approximately .5 s to make a prediction on an unknown compound. Consequently, the incorporation of SMARTCyp reactivities into the RS-Predictor modeling paradigm does not greatly impact overall runtime; it takes approximately 9 s per TOP QC SCR prediction and 2 s per TOP SCR prediction.
Details of SMARTCyp(V1.5) may be found here.47,53 StarDrop(V4.3) determines SOM reaction barriers through on-the-fly AM1 calculations with a modified version MOPAC97, a process that can take one to ten minutes per compound on a modern workstation.54 These barriers are combined with orientation and steric accessibility descriptors to evaluate potential reaction pathways.71 The Schrödinger P450 SOM Prediction 1.0 module assigns an intrinsic reactivity to each SOM using values that were derived from a Hammett-Taft approach with parameters optimized from the reactivity profiles of selected substrates of 3A4.55,72 This 3A4 model from the Schrödinger suite is combined with the Induced Fit Docking (IFD) Glide protocol when predicting sites of 2C9 and 2D6-mediated metabolism. The use of IFD reduces the number of predicted false-positives of the module on the Schrödinger 2C9 and 2D6 test sets by 16% and 17% respectively, at cost of a 2–2.5 h run-time per ligand on a 2-CPU machine. The Schrödinger 3A4 model alone is quite fast, with a runtime of approximately .5 s per substrate.
The random model used in this work is equivalent to the classic probability of having a bag filled with multiple marbles that are one of two colors, and one must pick a specific color combination of marbles within two guesses and no replacements. For a given substrate i composed of S SOMs (marbles), of which M undergo CYP-mediated metabolism (colored blue), and NM (NM == S − M) do not undergo CYP-mediated metabolism (colored red), the likelihood of an accurate random prediction of an observed SOM in the top two rank-positions is predicted as follows
The converse of this event,
may be calculated as shown above with the hypergeometric distribution, resulting in the expression
| (1) |
The R2 values for all substrates within a given set are then averaged to determine the random model accuracy for that particular set.
Supplementary Material
Abbreviations
- RS-Predictor
Regioselectivity-Predictor
- CYP
Cytochrome P450
- SOM
site of metabolism
- MIRank
multiple instance ranking
- SVM
support vector machines
- IFD
induced fit docking
- TP
true-positive
- FP
false-positive
Footnotes
Supporting Information Available
Structure and metabolite data for all 680 compounds, including the top three predicted SOMs of each method and model are made available. Additional graphics, tables, and discussion are also provided. This information is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 2.Guengerich FP. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 2006;8:E101–E111. doi: 10.1208/aapsj080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou S, Chan E, Zhou Z, Xue CC, Lai X, Duan W. Insights into the structure, function, and regulation of human cytochrome P450 1A2. Curr Drug Metab. 2009;10:713–729. doi: 10.2174/138920009789895552. [DOI] [PubMed] [Google Scholar]
- 4.Di YM, Chow VD, Yang L, Zhou S. Structure, function, regulation and polymorphism of human cytochrome P450 2A6. Curr Drug Metab. 2009;10:754–780. doi: 10.2174/138920009789895507. [DOI] [PubMed] [Google Scholar]
- 5.Mo S, Liu Y, Duan W, Wei MQ, Kanwar JR, Zhou S. Substrate specificity, regulation, and polymorphism of human cytochrome P450 2B6. Curr Drug Metab. 2009;10:730–753. doi: 10.2174/138920009789895534. [DOI] [PubMed] [Google Scholar]
- 6.Lai X, Yang L, Li X, Liu J, Zhou Z, Zhou S. Human CYP2C8: Structure, substrate specificity, inhibitor selectivity, inducers and polymorphisms. Curr Drug Metab. 2009;10:1009–1047. doi: 10.2174/138920009790711832. [DOI] [PubMed] [Google Scholar]
- 7.Mo S, Zhou Z, Yang L, Wei MQ, Zhou S. New insights into the structural features and functional relevance of human cytochrome P450 2C9. Part II. Curr Drug Metab. 2009;10:1127–1150. doi: 10.2174/138920009790820101. [DOI] [PubMed] [Google Scholar]
- 8.Zhou S, Liu J, Lai X. Substrate specificity, inhibitors and regulation of human cytochrome P450 2D6 and implications in drug development. Curr Med Chem. 2009;16:2661–2805. doi: 10.2174/092986709788681985. [DOI] [PubMed] [Google Scholar]
- 9.Czodrowski P, Kriegl JM, Scheuerer S, Fox T. Computational approaches to predict drug metabolism. Expert Opin Drug Meta. 2009;5:15–27. doi: 10.1517/17425250802568009. [DOI] [PubMed] [Google Scholar]
- 10.Mo S, Zhou Z, Yang L, Wei MQ, Zhou S. New insights into the structural features and functional relevance of human cytochrome P450 2C9. Part I. Curr Drug Metab. 2009;10:1075–1126. doi: 10.2174/138920009790820129. [DOI] [PubMed] [Google Scholar]
- 11.Meyer RP, Gehlhaus M, Knoth R, Volk B. Expression and function of cytochrome P450 in brain drug metabolism. Curr Drug Metab. 2007;8:297–306. doi: 10.2174/138920007780655478. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh C, Gonzalez-Martinez J, Hossain M, Cucullo L, Fazio V, Janigro D, Marchi N. Pattern of P450 expression at the human blood-brain barrier: Roles of epileptic condition and laminar flow. Epilepsia. 2010;51:1408–1417. doi: 10.1111/j.1528-1167.2009.02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Rayes BF, Ali S, Heilbrun LK, Lababidi S, Bouwman D, Visscher D, Philip PA. Cytochrome P450 and glutathione transferase expression in human breast cancer. Clin Cancer Res. 2003;9:1705–1709. [PubMed] [Google Scholar]
- 14.Modugno F, Knoll C, Kanbour-Shakir A, Romkes M. A potential role for the estrogen-metabolizing cytochrome P450 enzymes in human breast carcinogenesis. Breast Cancer Res Treat. 2003;82:191–197. doi: 10.1023/B:BREA.0000004376.21491.44. [DOI] [PubMed] [Google Scholar]
- 15.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: Function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 16.Bergheim I, Bode C, Parlesak A. Distribution of cytochrome P450 2C, 2E1, 3A4, and 3A5 in human colon mucosa. BMC Clin Pharmaco. 2005;5:4. doi: 10.1186/1472-6904-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Totah RA, Rettie AE. Cytochrome P450 2C8: Substrates, inhibitors, pharmacogenetics, and clinical relevance. Clin Pharmaco Ther. 2005;77:341–352. doi: 10.1016/j.clpt.2004.12.267. [DOI] [PubMed] [Google Scholar]
- 18.Vasanthanathan P, Hritz J, Taboureau O, Olsen L, Jørgensen FS, Vermeulen NPE, Oostenbrink C. Virtual screening and prediction of site of metabolism for cytochrome P450 1A2 ligands. J Chem Inf Model. 2009;49:43–52. doi: 10.1021/ci800371f. [DOI] [PubMed] [Google Scholar]
- 19.Ekins S, Bravi G, Ring BJ, Gillespie TA, Gillespie JS, Vandenbranden M, Wrighton SA, Wikel JH. Three-dimensional quantitative structure activity relationship analyses of substrates for CYP2B6. J Pharmacol Exp Ther. 1999;288:21–29. [PubMed] [Google Scholar]
- 20.Wang B, Zhou S. Synthetic and natural compounds that interact with human cytochrome P450 1A2 and implications in drug development. Curr Med Chem. 2009;16:4066–4218. doi: 10.2174/092986709789378198. [DOI] [PubMed] [Google Scholar]
- 21.Melet A, Marques-Soares C, Schoch GA, Macherey A, Jaouen M, Dansette PM, Sari M, Johnson EF, Mansuy D. Analysis of human cytochrome P450 2C8 substrate specificity using a substrate pharmacophore and site-directed mutants. Biochemistry. 2004;43:15379–15392. doi: 10.1021/bi0489309. [DOI] [PubMed] [Google Scholar]
- 22.Park J, Harris D. Construction and assessment of models of CYP2E1: Predictions of metabolism from docking, molecular dynamics, and density functional theoretical calculations. J Med Chem. 2003;46:1645–1660. doi: 10.1021/jm020538a. [DOI] [PubMed] [Google Scholar]
- 23.Rydberg P, Vasanthanathan P, Oostenbrink C, Olsen L. Fast Prediction of Cytochrome P450 Mediated Drug Metabolism. ChemMedChem. 2009;4:2070–2079. doi: 10.1002/cmdc.200900363. [DOI] [PubMed] [Google Scholar]
- 24.Cruciani G, Carosati E, Boeck BD, Ethirajulu K, Mackie C, Howe T, Vianello R. MetaSite: Understanding metabolism in human cytochromes from the perspective of the chemist. J Med Chem. 2005;48:6970–6979. doi: 10.1021/jm050529c. [DOI] [PubMed] [Google Scholar]
- 25.de Groot MJ, Alex AA, Jones BC. Development of a combined protein and pharmacophore model for cytochrome P450 2C9. J Med Chem. 2002;45:1983–1993. doi: 10.1021/jm0110791. [DOI] [PubMed] [Google Scholar]
- 26.Zamora I, Afzelius L, Cruciani G. Predicting drug metabolism: A site of metabolism prediction tool applied to the cytochrome P450 2C9. J Med Chem. 2003;46:2313–2324. doi: 10.1021/jm021104i. [DOI] [PubMed] [Google Scholar]
- 27.Sheridan RP, Korzekwa KR, Torres RA, Walker MJ. Empirical regioselectivity models for human cytochromes P450 3A4, 2D6, and 2C9. J Med Chem. 2007;50:3173–3184. doi: 10.1021/jm0613471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sykes MJ, McKinnon RA, Miners JO. Prediction of metabolism by cytochrome P450 2C9: Alignment and docking studies of a validated database of substrates. J Med Chem. 2008;51:780–791. doi: 10.1021/jm7009793. [DOI] [PubMed] [Google Scholar]
- 29.Tarcsay A, Kiss R, Keseru GM. Site of metabolism prediction on cytochrome P450 2C9: A knowledge-based docking approach. J Comput-Aided Mol Des. 2010;24:399–408. doi: 10.1007/s10822-010-9347-3. [DOI] [PubMed] [Google Scholar]
- 30.Vermeulen NPE. Prediction of drug metabolism: The case of cytochrome P450 2D6. Curr Top Med Chem. 2003;3:1227–1239. doi: 10.2174/1568026033451998. [DOI] [PubMed] [Google Scholar]
- 31.de Graaf C, Oostenbrink C, Keizers PHJ, van der Wijst T, Jongejan A, Vermeulen NPE. Catalytic site prediction and virtual screening of cytochrome P450 2D6 substrates by consideration of water and rescoring in automated docking. J Med Chem. 2006;49:2417–2430. doi: 10.1021/jm0508538. [DOI] [PubMed] [Google Scholar]
- 32.Hritz J, de Ruiter A, Oostenbrink C. Impact of plasticity and flexibility on docking results for cytochrome P450 2D6: A combined approach of molecular dynamics and ligand docking. J Med Chem. 2008;51:7469–7477. doi: 10.1021/jm801005m. [DOI] [PubMed] [Google Scholar]
- 33.Unwalla RJ, Cross JB, Salaniwal S, Shilling AD, Leung L, Kao J, Humblet C. Using a homology model of cytochrome P450 2D6 to predict substrate site of metabolism. J Comput-Aided Mol Des. 2010;24:237–256. doi: 10.1007/s10822-010-9336-6. [DOI] [PubMed] [Google Scholar]
- 34.Santos R, Hritz J, Oostenbrink C. Role of water in molecular docking simulations of cytochrome P450 2D6. J Chem Inf Model. 2010;50:146–154. doi: 10.1021/ci900293e. [DOI] [PubMed] [Google Scholar]
- 35.Moors SLC, Vos AM, Cummings MD, Van Vlijmen H, Ceulemans A. Structure-Based Site of Metabolism Prediction for Cytochrome P450 2D6. J Med Chem. 2011;54:6098–6105. doi: 10.1021/jm2006468. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Schneebeli ST, Bylund J, Farid R, Friesner RA. IDSite: An Accurate Approach to Predict P450-Mediated Drug Metabolism. J Chem Theory Comput. 2011;7:3829–3845. doi: 10.1021/ct200462q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rydberg P, Olsen L. Ligand-Based Site of Metabolism Prediction for Cytochrome P450 2D6. ACS Med Chem Lett. 2012;3:69–73. doi: 10.1021/ml200246f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh SB, Shen LQ, Walker MJ, Sheridan RP. A model for predicting likely sites of CYP3A4-mediated metabolism on drug-like molecules. J Med Chem. 2003;46:1330–6. doi: 10.1021/jm020400s. [DOI] [PubMed] [Google Scholar]
- 39.Kim DN, Cho K, Oh WS, Lee CJ, Lee SK, Jung J, No KT. EaMEAD: Activation energy prediction of cytochrome P450 mediated metabolism with effective atomic descriptors. J Chem Inf Model. 2009;49:1643–1654. doi: 10.1021/ci900011g. [DOI] [PubMed] [Google Scholar]
- 40.Oh WS, Kim DN, Jung J, Cho K, No KT. New combined model for the prediction of regioselectivity in cytochrome P450/3A4 mediated metabolism. J Chem Inf Model. 2008;48:591–601. doi: 10.1021/ci7003576. [DOI] [PubMed] [Google Scholar]
- 41.Hasegawa K, Koyama M, Funatsu K. Quantitative prediction of regioselectivity toward cytochrome P450/3A4 using machine learning approaches. Mol Inform. 2010;29:243–249. doi: 10.1002/minf.200900086. [DOI] [PubMed] [Google Scholar]
- 42.Prusis P, Afzelius L. Improvement of Site of Metabolism Predictions for CYP3A4 by Using Discriminant Analysis of Compound Preference of CYP3A4 X-Ray Structural Conformers and Subsequent Docking. QSAR Comb Sci. 2009;28:865–868. [Google Scholar]
- 43.Skopalík J, Anzenbacher P, Otyepka M. Flexibility of Human Cytochromes P450: Molecular Dynamics Reveals Differences between CYPs 3A4, 2C9, and 2A6, which Correlate with Their Substrate Preferences. J Phys Chem B. 2008;112:8165–8173. doi: 10.1021/jp800311c. [DOI] [PubMed] [Google Scholar]
- 44.Zheng M, Luo X, Shen Q, Wang Y, Du Y, Zhu W, Jiang H. Site of metabolism prediction for six biotransformations mediated by cytochromes P450. Bioinformatics. 2009;25:1251–1258. doi: 10.1093/bioinformatics/btp140. [DOI] [PubMed] [Google Scholar]
- 45.Hennemann M, Friedl A, Lobell M, Keldenich J, Hillisch A, Clark T, Göller AH. CypScore: Quantitative prediction of reactivity toward cytochromes P450 based on semiempirical molecular orbital theory. ChemMedChem. 2009;4:657–669. doi: 10.1002/cmdc.200800384. [DOI] [PubMed] [Google Scholar]
- 46.Smith J, Stein V. SPORCalc: A development of a database analysis that provides putative metabolic enzyme reactions for ligand-based drug design. Comput Biol and Chem. 2009;33:149–159. doi: 10.1016/j.compbiolchem.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Rydberg P, Gloriam DE, Zaretzki J, Breneman C, Olsen L. SMARTCyp: A 2D method for prediction of cytochrome P450-mediated drug metabolism. ACS Med Chem Lett. 2010;1:96–100. doi: 10.1021/ml100016x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones JP, Mysinger M, Korzekwa KR. Computational models for cytochrome P450: a predictive electronic model for aromatic oxidation and hydrogen atom abstraction. Drug Metab Dispos. 2002;30:7–12. doi: 10.1124/dmd.30.1.7. [DOI] [PubMed] [Google Scholar]
- 49.Stewart JJP. MOPAC: A semiempirical molecular orbital program. J Comput-Aided Mol Des. 1990;4:1–103. doi: 10.1007/BF00128336. [DOI] [PubMed] [Google Scholar]
- 50.Zaretzki J, Bergeron C, Rydberg P, Huang T-w, Bennett KP, Breneman CM. RS-Predictor: A New Tool for Predicting Sites of Cytochrome P450-Mediated Metabolism Applied to CYP 3A4. J Chem Inf Model. 2011;51:1667–1689. doi: 10.1021/ci2000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergeron C, Zaretzki J, Breneman C, Bennett KP. Multiple instance ranking. Proceedings of the 25th International Conference on Machine Learning; Helsinki, Finland. 2008; New York, New York: ACM; 2008. pp. 48–55. [Google Scholar]
- 52.Bergeron C, Moore G, Zaretzki J, Breneman CM, Bennett KP. Fast bundle algorithm for multiple instance learning. [Accepted 2011.];IEEE T Pattern Anal. doi: 10.1109/TPAMI.2011.194. [DOI] [PubMed] [Google Scholar]
- 53.Rydberg P, Gloriam DE, Olsen L. The SMARTCyp cytochrome P450 metabolism prediction server. Bioinformatics. 2010;26:2988–2989. doi: 10.1093/bioinformatics/btq584. [DOI] [PubMed] [Google Scholar]
- 54.StarDrop, version 4.3. Optibrium Ltd; Cambridge, United Kingdom: 2009. [Google Scholar]
- 55.P450 SOM Prediction, version 1.0. Schrödinger LLC; New York, NY: 2011. [Google Scholar]
- 56.Isin EM, Sohl CD, Eoff RL, Guengerich FP. Cooperativity of cytochrome P450 1A2: Interactions of 1, 4-phenylene diisocyanide and 1-isopropoxy-4-nitrobenzene. Arch Biochem Biophys. 2008;473:69–75. doi: 10.1016/j.abb.2008.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MOE, version 2009.10. Chemical Computing Group; Montreal, Canada: 2009. [Google Scholar]
- 58.Mayeno AN, Robinson JL, Yang RSH, Reisfeld B. Predicting Activation Enthalpies of Cytochrome-P450-Mediated Hydrogen Abstractions. 2. Comparison of Semiempirical PM3, SAM1, and AM1 with a Density Functional Theory Method. J Chem Inf Model. 2009;49:1692–1703. doi: 10.1021/ci8003946. [DOI] [PubMed] [Google Scholar]
- 59.Yap CW, Chen YZ. Prediction of cytochrome P450 3A4, 2D6, and 2C9 inhibitors and substrates by using support vector machines. J Chem Inf Model. 2005;45:982–992. doi: 10.1021/ci0500536. [DOI] [PubMed] [Google Scholar]
- 60.de Graaf C, Vermeulen NPE, Feenstra KA. Cytochrome P450 in silico: An integrative modeling approach. J Med Chem. 2005;48:2725–2755. doi: 10.1021/jm040180d. [DOI] [PubMed] [Google Scholar]
- 61.Terfloth L, Bienfait B, Gasteiger J. Ligand-based models for the isoform specificity of cytochrome P450 3A4, 2D6, and 2C9 substrates. J Chem Inf Model. 2007;47:1688–1701. doi: 10.1021/ci700010t. [DOI] [PubMed] [Google Scholar]
- 62.Mishra N, Agarwal S, Raghava G. Prediction of cytochrome P450 isoform responsible for metabolizing a drug molecule. BMC Pharmaco. 2010;10:8. doi: 10.1186/1471-2210-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caruana R. Multitask learning. Machine Learn. 1997;28:41–75. [Google Scholar]
- 64.Rendic S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab Rev. 2002;34:83–448. doi: 10.1081/dmr-120001392. [DOI] [PubMed] [Google Scholar]
- 65.Brown CM, Reisfeld B, Mayeno AN. Cytochromes P450: A structure-based summary of biotransformations using representative substrates. Drug Metab Rev. 2008;40:1–100. doi: 10.1080/03602530802309742. [DOI] [PubMed] [Google Scholar]
- 66.Preissner S, Kroll K, Dunkel M, Senger C, Goldsobel G, Kuzman D, Guenther S, Winnenburg R, Schroeder M, Preissner R. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res. 2009;38:D237–D243. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daher W, Pelinski L, Klieber S, Sadoun F, Meunier V, Bourrié M, Biot C, Guillou F, Fabre G, Brocard J, Fraisse L, Maffrand J, Khalife J, Dive D. In vitro metabolism of ferroquine (SSR97193) in animal and human hapatic models and antimalarial activity of major metabolites on plasmodium falciparum. Drug Meta Dispos. 2006;34:667–682. doi: 10.1124/dmd.104.003202. [DOI] [PubMed] [Google Scholar]
- 68.Jensen KG, Dalgaard L. In vitro metabolism of the M1-muscarinic agonist 5-(2-ethyl-2H-tetrazol-5-yl)-1-methyl-1,2,3,6-tetrahydropyridine by human hepatic cytochromes P-450 determined at pH 7.4 and 8.5. Drug Metab Dispos. 1999;27:125–132. [PubMed] [Google Scholar]
- 69.Rudek MA, Zhao M, Smith NF, Robey RW, He P, Hallur G, Khan S, Hidalgo M, Jimeno A, Colevas AD, Messersmith WA, Wolff AC, Baker SD. In vitro and in vivo clinical pharmacology of dimethyl benzoylphenylurea, a novel oral tubulin-interactive agent. Clin Cancer Res. 2005;11:8503–8511. doi: 10.1158/1078-0432.CCR-05-1037. [DOI] [PubMed] [Google Scholar]
- 70.Guo Y, Wang Y, Si D, Fawcett PJ, Zhong D, Zhou H. Catalytic activities of human cytochrome P450 2C9*1, 2C9*3 and 2C9*13. Xenobiotica. 2005;35:853–861. doi: 10.1080/00498250500256367. [DOI] [PubMed] [Google Scholar]
- 71. [accessed October 21, 2011];Optibrium FAQs: P450 Models. http://www.optibrium.com/community/faq/p450-models.
- 72. [accessed October 21, 2011];Schrödinger Support Center - Documentation. http://www.schrodinger.com/supportdocs/18/other13/
- 73.McDonald JH. Handbook of Biological Statistics. 2. Sparky House Publishing; Baltimore, Maryland: 2009. pp. 70–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.