Abstract
The disassembly of a core-satellite nanostructured substrate is presented as a colorimetric biosensor observable under dark field illumination. The fabrication method described herein utilizes thiol-mediated adsorption and streptavidin-biotin binding to self-assemble core-satellite nanostructures with a sacrificial linking peptide. Biosensing functionality is demonstrated with the protease trypsin and the optical properties of the nanoassemblies are characterized. A figure of merit is presented to determine the optimal core and satellite size for visual detection. Nanoassemblies with 50 nm cores and 30 nm or 50 nm satellites are superior as these structures achieve an orange to green color shift greater than 70 nm that is easily discernible by naked eye. This colorimetric substrate may prove to be a favorable alternative to liquid-based colloidal sensors and a useful visual readout mechanism for microfluidic diagnostic assays.
Keywords: nanoparticle, colorimetric, biosensor, plasmonic, nanoassembly, core-satellite, self-assembly
The simultaneous realization of self-assembled colloidal gold nanocrystals by Mirkin and Alivisatos established a platform technology for ‘bottom up’ fabrication that utilized fundamental principles from biology. The advent of this discovery has yielded a multitude of geometric nanoassembly variations such as lattice networks,1,2 dimer and trimer configurations,3,4,5 and core-satellite constructs.5–11 These noble metal nanoassemblies have enabled optical investigations of plasmon resonance coupling10,11 and the colorimetric detection of biological and chemical analytes such as single nucleotide polymorphisms (SNPs),12 cocaine, adenosine,13 lead,14 caspase,15 and rabbit IgG.16 Such technologies may bolster the groundwork for a new generation of point-of-care (POC) diagnostic devices capable of identifying diverse analyte species, an improvement upon the commercially available immunoassay-based lateral flow test.17 We propose an advancement upon the current suspension-based modality of colloidal biomolecular sensors that is better suited for on-chip diagnostic system integration while preserving the capability for diverse species detection. We present the fabrication of nanoassembles constructed on a two-dimensional glass substrate, demonstrate the colorimetric functionality of biomolecular sensing via disassembly, and systematically analyze the optical detection properties of the substrate. Nanoconstructs with 50 nm cores and 30 nm or 50 nm satellites maximized the signal-to-noise ratio during biosensing and enabled visible colorimetric shifts from orange (>630 nm) to green (~560 nm) via the observation of scattered light with dark field (DF) illumination.
The construction of colloidal gold biosensors using DNA, peptide, or aptamer linkers has enabled the detection of an assortment of species outside the typical antigen/antibody binding interactions observed in traditional immunoassays.12–17 This method is advantageous as plasmonic nanoparticles are ~500,000 times more luminous than a fluorophore, have an unlimited photon budget, and do not bleach, blink, or require expensive spectroscopic equipment.18 However, the technology to date has relied upon nanoconstructs suspended in solution. This assembly paradigm has five deficiencies that may be overcome by the use of disassembling nanostructures immobilized on a substrate. First, the sample analyte is diluted by three orders of magnitude when added to the suspension of nanostructures, diminishing detection limits. Second, impurities in the analyte such as salts and pH fluctuations might cause flocculation and subsequently a false positive readout. Third, the storage life is limited because suspended nanoparticles tend to aggregate and settle with time. Fourth, liquid-based assays are prone to containment issues such as spillage. Finally, fifth and arguably the most important fact is that previous liquid-based sensors rely upon changes in optical absorbance. Since the Beer-Lambert Law states that absorbance has a linear dependence on path length, colloidal liquid-based sensors are not feasible in microfluidic diagnostic systems. The colorimetric substrate presented here bypasses these shortcomings. Previous reports of disassembling colorimetric nanomaterials on a substrate have included the resuspension of nanoparticles released from nanolattices dried on hydrophobic paper or constructed on a fluid lipid bilayer membrane.19,20 However, unlike previous biosensors, we present the first self-assembled substrate that yields a structural color change directly at the site of disassembly when imaged in DF. The small optical path length of the biosensing substrate (~nm) is better suited for the length scales of on-chip diagnostic systems in comparison to suspended nanoassembled networks that require longer path lengths (~cm) for colorimetric changes to be observable. The utility of this localized disassembly mechanism may enable functionality for spatiotemporal monitoring of cellular secretions and POC capability using innovative handheld DF viewing techniques.
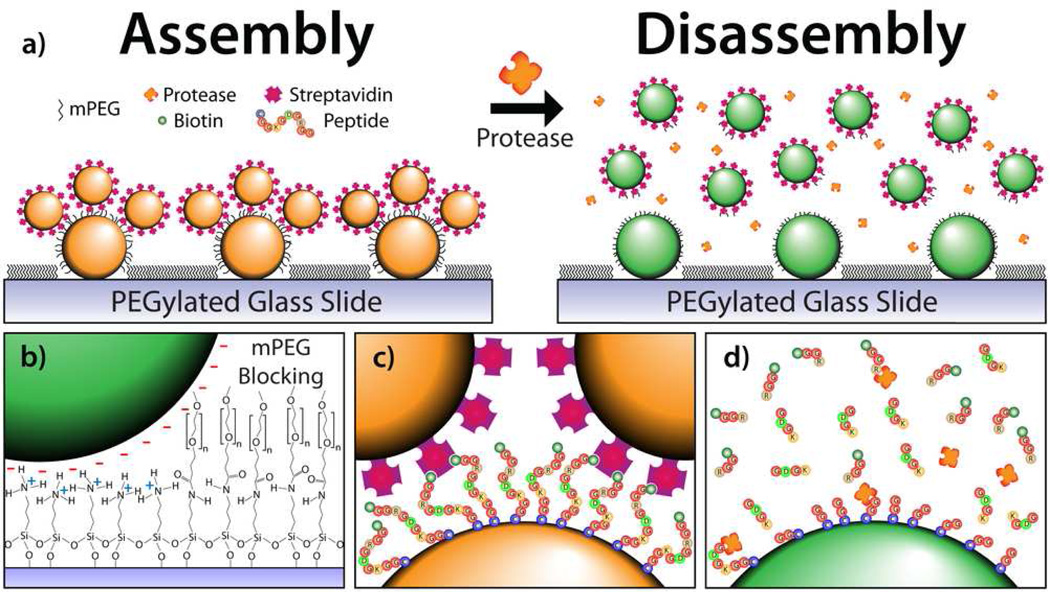
Protease diagnostics are greatly underutilized in clinical settings, thus a substrate able to detect protease activity was designed as a proof-of-concept to demonstrate the structurally induced spectral scattering shift of the biosensor. However, we stress that by altering the core-satellite tether, detection platforms for other biomolecular species can be created. Detectable levels of proteases present in peripheral fluids taken ex vivo have been shown to act as diagnostic markers that are presymptomatically indicative of disease.21–24 This article presents the use of scattered light from coupled gold nanoparticle assemblies and their subsequent disassembly via proteolysis as a colorimetric assay to detect protease activity. Cleavage of the nanoassemblies disengages the plasmon coupling between nanoparticles and shifts the observed DF scattered light from orange to green (Figure 1a). This model system is not only limited to protease activity monitoring, but is applicable to the detection of numerous biomolecules.
Figure 1.
a) Core-satellite Au nanoassemblies before and after proteolytic cleavage. b) Immobilization of the negatively charged AuNP cores is enabled by Coulombic electrostatic attraction to an APTES monolayer on glass. The glass slide is subsequently passivated with an mPEG chain to block nonspecific satellite adsorption. c) Satellite attachment is enabled by a directional cysteine/biotin-streptavidin peptide tether. d) The addition of trypsin cleaves the peptide, releasing the satellites into solution and enabling colorimetric detection of the protease.
RESULTS AND DISCUSSION
Gold nanoparticles (AuNPs) were first deposited onto APTES (3-aminopropyltriethoxysilane) functionalized glass microscope slides. The sodium citrate reduction (Turkevich) method25 produces negatively charged AuNPs and the AuNPs, which served as the core of the nanoconstruct, were electrostatically immobilized onto the positively charged surface of the APTES glass slide through Coulombic attraction (Figure 1b, left). Next, a biotinylated ten amino acid long peptide substrate, cleavable by the serine protease trypsin, was incubated on the substrate. The C-terminus of the peptide is a cysteine residue, enabling the thiol-mediated adsorption of the peptide onto the surface of the core nanoparticle. The peptide sequence is biotin-GGRGDGKGGC-OH. Streptavidin conjugated satellite nanoparticles bind to the biotinylated N-terminus of the peptide for controlled, directional self-assembly (Figure 1c). The incubation of methoxy polyethylene glycol succinimidyl valerate (mPEG-SVA, MW = 2 kDa) on the substrate before satellite deposition blocks the microscope slide such that the satellites only attach to the core nanoparticles. This essential step prevents the satellites from nonspecifically adsorbing onto the substrate and facilitates a reversible color shift. The APTES functionalized slide enables passivation as the mPEG-SVA reacts with the exposed primary amine, releasing 1-Hydroxy-2,5-pyrrolidinedione and immobilizing the PEG chain onto the slide (Figure 1b, right). It should be noted that the linking peptide chain could be replaced by a variety of other biomolecules such as an oligonucleotide or aptazyme for diverse analyte identification. All substrates were stored in deionized (DI) water to prevent capillary remodeling caused by evaporation. The substrates remained functional for more than 28 days when stored in DI water at 4°C.
Pairs of slides were created for each geometrical combination of core and satellite size and analyzed under DF. A spectrophotometer measured the far-field scattering spectra from the substrate and data taken from five random positions on each substrate were averaged to determine peak position and full width at half maximum (FWHM). AuNPs cores were deposited at high densities, ~33.1 cores µm−2 for 50 nm AuNP cores, enabling easy visualization of the scattered color yet substrates were not overly dense, which would cause unfavorable plasmonic coupling between core particles. Four variations of core diameter were investigated. 30 nm and 50 nm cores scattered green in comparison to 80 nm and 100 nm cores, which appeared chartreuse (Table S1 and Figure S1 in the Supporting Information). Additionally, the two larger core sizes have broader peakwidths (FWHM). Radiation damping has been shown to be responsible for the redshift and bandwidth broadening observed in both the near- and far-field scattering as nanoparticles increase in size.26 Core sizes with green scattering, which is the most sensitive color to the human eye, are optimal as their plasmon peak will be redshifted upon satellite attachment and plasmon coupling. The redshift for assemblies with a large core size cannot be visually discerned as the far-field scattering spectra shifts into the near IR, however such shifts are detectable with spectroscopy. Another important property for core size is the scattering cross section (σs). When a particle radius (r) is much smaller than the wavelength of light (r ≪ λ), Rayleigh theory states σs ∝ r6/λ4. Thus, core sizes of 30 nm and smaller have a small scattering cross section, resulting in dimly scattered green light which is difficult to visualize (Table S1). The optimal core size, as determined by its spectral peak position and scattering cross section, is 50 nm.
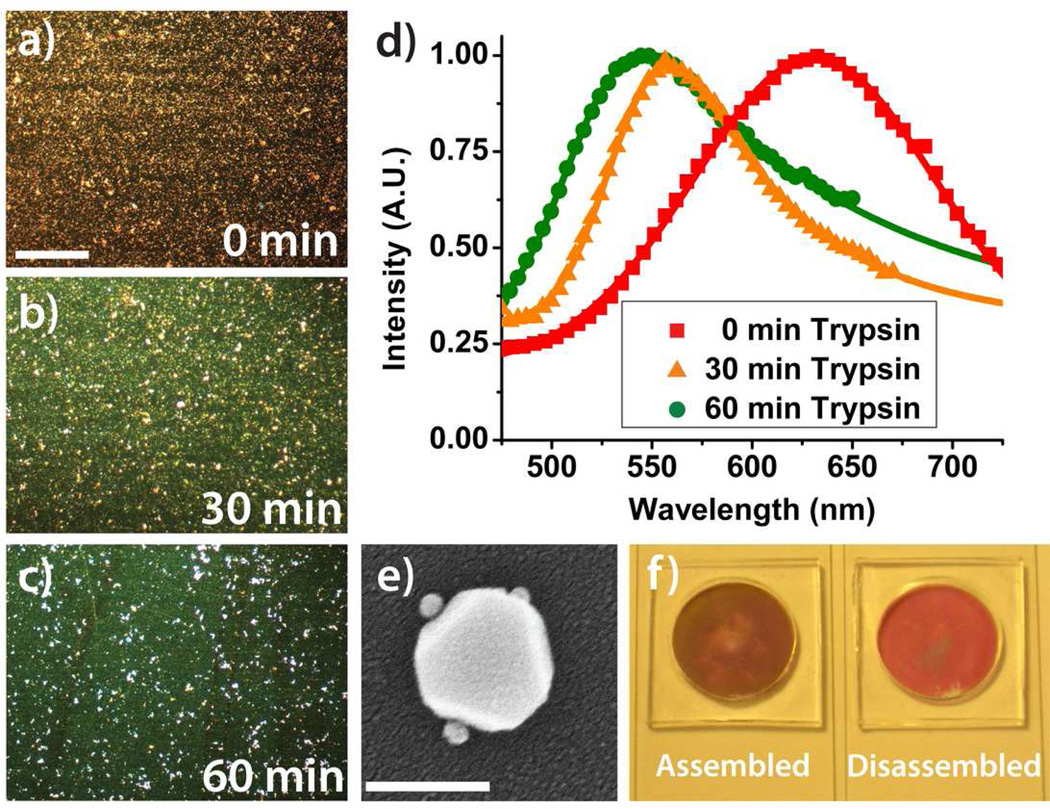
The serine protease trypsin, known to cleave the carboxyl end of lysine and arginine, was used to initiate substrate disassembly (Figure 1d). Trypsin (100 µM) was spotted directly onto the substrate at room temperature (25°C) and produced a complete color change in less than 40 minutes as seen in the images captured by a true-color charge-coupled device (CCD) camera (Figure 2). Figures 2a, 2b, and 2c are time-lapse images of a substrate at 0 min, 30 min, and 60 min after trypsin exposure. The white spots observable in Figure 2c are believed to be scattering from mPEG-SVA aggregates that accumulated during surface passivation. A representative DF spectrum depicts a colorimetric shift greater than 70 nm for a substrate with 50 nm cores and 50 nm satellites (Figure 2d). Modified Gaussian curves have been fit to the data. An SEM image of a 100 nm core with 10 nm satellites is shown in Figure 2e. Core sizes larger than 100 nm scatter light greater than 600 nm and satellite disassembly is visually indiscernible. Thus, larger sized core particles were eliminated from the study. A nanoassembled substrate with 50 nm cores and 50 nm satellites, before and after trypsin-mediated disassembly, is shown under bright field illumination in Figure 2f. The short path length of the monolayer of core-satellites does not scatter enough of the incident light for a colorimetric change to be noticeably observable by eye. A technique similar to resonance-enhanced adsorption may be able to amplify this faint color shift under bright field illumination and facilitate detection without the constraint of DF illumination.27 Polydimethylsiloxane (PDMS) droplet immobilizers, visible in Figure 2f, were used to contain the droplet and prevent capillary-induced remodeling of the nanostructures due to evaporation. All of the images presented in Figure 2 are unaltered.
Figure 2.
Dark field microscopy images representative of the observable colorimetric shift upon disassembly by proteolytic cleavage with 100 µM trypsin at a) 0 minutes, b) 30 minutes, and c) 60 minutes. The substrate is composed of 50 nm core AuNPs and 50 nm satellite AuNPs. (scale bar: 100 µm) d) The corresponding normalized spectra taken simultaneously with the images. e) SEM image of an individual core-satellite nanoassembly with a 100 nm core and 10 nm satellites. (scale bar: 100 nm) f) A 50 nm core and 50 nm satellite nanoassembled substrate before and after trypsin cleavage via bright field illumination. The short path length of the core-satellite nanoassembled substrate does not scatter enough of the incident light for a colorimetric change to be noticeably observable by eye.
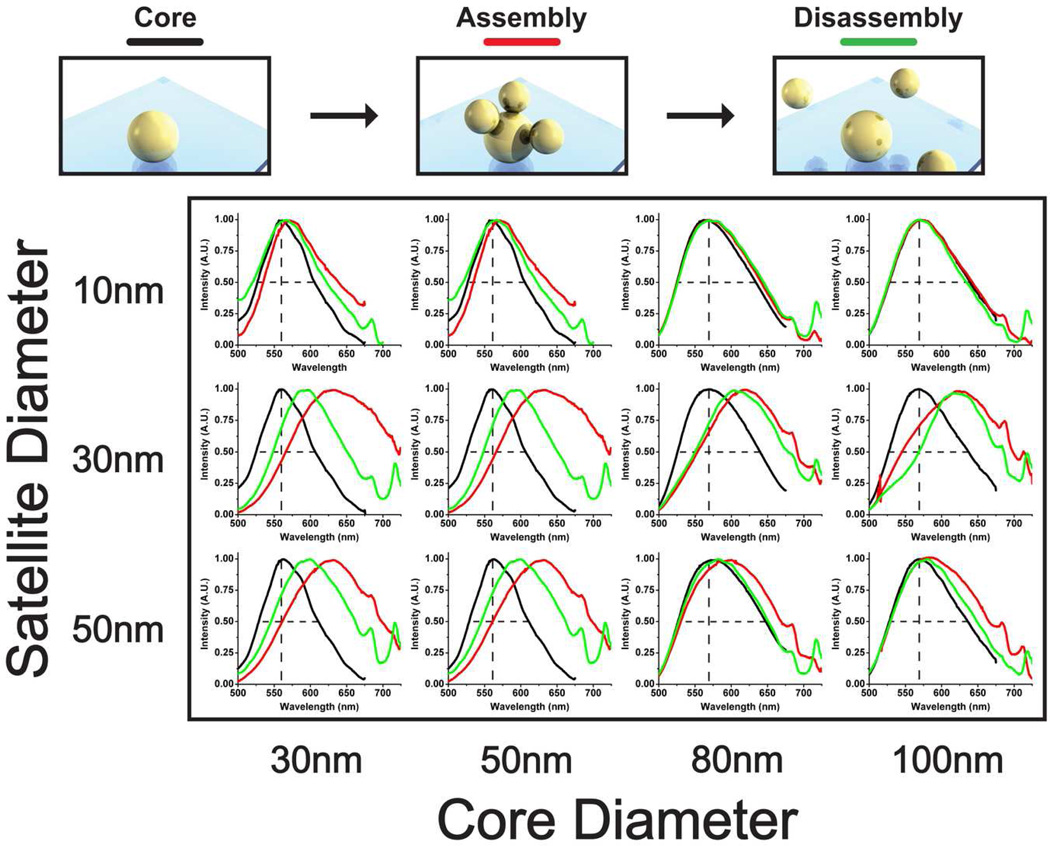
A large-scale study was performed for an array of nanostructures to characterize assembly and disassembly (Figure 3). Core sizes ranged from 30 nm to 100 nm and satellite sizes ranged from 10 nm to 50 nm. Satellites larger than 50 nm are difficult to conjugate with streptavidin as the surface-to-volume ratio is large and the colloid becomes unstable during physio-adsorption of the streptavidin onto the AuNP surface. Substrates were prepared in duplicate and optical measurements were acquired at random locations in quintuplicate for each slide (n=10). The acquisitions were collected over a large field of view such that substrate inconsistencies and variations were averaged over the acquired spectrum. Disassembly was achieved by exposing the substrates to trypsin for 60 minutes at room temperature. The ten spectral measurements were averaged and are graphed in Figure 3. The peaks observed at ~670 nm and ~740 nm in Figure 3 are attributed to source normalization artifacts due to a poor signal-to-noise ratio at the spectral extremities of our spectrophotometer (see Figure S5 for the emitted spectrum from the Xenon light source). A MATLAB script (Supporting Information) individually calculated the peak shift and FWHM for each spectral measurement and the values were averaged to obtain peak shift and FWHM statistics.
Figure 3.
Spectral characterization study for a combination of core and satellite diameters. Scatter from single AuNP cores (black), assembly redshift (red), and disassembly blue shift (green) are depicted. Spectra are averaged results (n=10). Core-satellite nanostructures with combinations of 30 nm and 50 nm nanoparticle sizes consistently produced the greatest peak shifts. Dashed black lines depict initial peak position and FWHM as a visual aid for the reader.
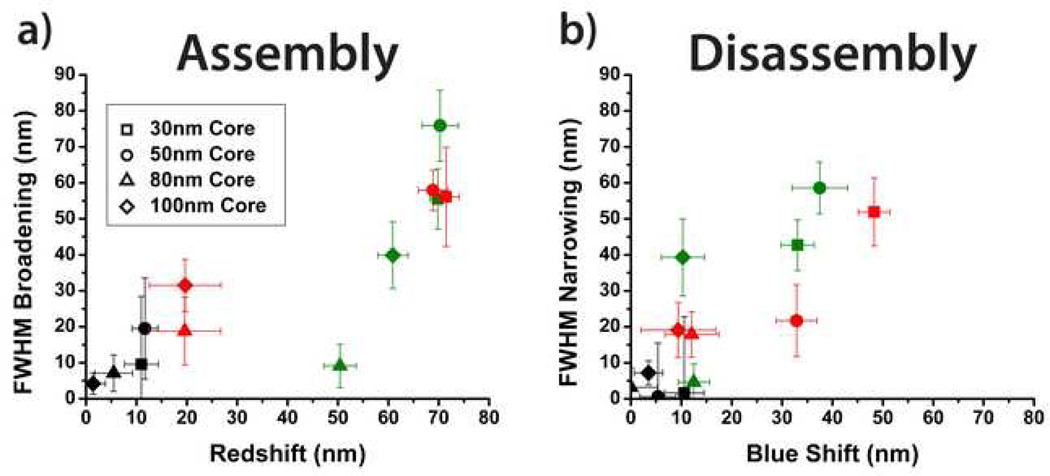
Two important optical characteristics for visual detection are the peak shift and change in FWHM upon assembly and disassembly. Peak shift upon assembly is defined as: Δλpeak = λpeak,core+sat. - λpeak,core and is depicted on the x-axis of Figure 4a. The maximum attained assembly redshifts were approximately ~70 nm for all combinations of 50 nm and 30 nm AuNPs. 30 nm satellites consistently produced the largest peak shift of all the satellite sizes. This behavior seems contrary to intuition as 50 nm satellites cause a larger structural change than 30 nm satellites. Limited streptavidin conjugation onto the larger 50 nm AuNPs may have resulted in fewer satellites capable of binding to the core particles. Next, the peak shift upon disassembly was analyzed and is depicted on the x-axis of Figure 4b. Combinations of 30 nm and 50 nm AuNPs provided the largest blue shift, with 30 nm core and 50 nm satellite assemblies causing a maximum shift of 48.3 nm. The investigation reveals that the assembly redshift is not completely reversable, which may be caused by nonspecific aggregation of the satellites to the substrate and/or irreversible core-satellite attachment. Overall, core-satellite combinations of 30 nm and 50 nm result in the largest spectral peak shifts. The findings of this analysis correlate well with previous theoretical simulations of core-satellite plasmon band redshifting due to interparticle dipole coupling and radiation damping.10,28
Figure 4.
Correlation between peak shift and FWHM broadening/narrowing upon a) assembly and b) disassembly. Data points are extracted from Figure 3 with the shape corresponding to the core size and the color corresponding to the satellite size. 10 nm, 30 nm, and 50 nm satellites are represented by black, green, and red, respectively.
The second optical characteristic analyzed was the FWHM assembly broadening and disassembly narrowing properties of the nanoassemblies. Analysis of the FWHM is necessary to maximize the signal-to-noise ratio during biosensing as large FWHM increases may cause disassembly blue shifts to be indiscernible. The change in the bandwidth upon assembly, defined as ΔFWHM = FWHMcore+sate. − FWHMcore, was a maximum, 75.9 nm, for the 50 nm core and 30 nm satellite assembly (Figure 4a). Other combinations of 30 nm and 50 nm AuNP produced ~56 nm in bandwidth broadening. During disassembly, 50 nm cores with 30 nm satellites obtained the maximum bandwidth narrowing (Figure 4b). Minimizing the bandwidth upon assembly and disassembly is essential such that only one color, a single peak, can be easily resolved by human eye for both cases. Thus optimal core-satellite combinations would lie in the lower right side of Figures 4a and 4b. However, our data reveals that shifts in peak position appear to be positively correlated to changes in bandwidth. These results coincide with recent findings that larger satellite sizes broaden the plasmon band when assembled onto core AuNPs.28 This characteristic is mainly due to retardation effects as the nanoassembly size becomes significant in comparison to the wavelength of light. Radiation damping has been shown to be a large factor in the redshifts observed in the far-field scattering as nanoparticles increase in size.26 Ultimately, larger satellite sizes should lead to greater changes in FWHM upon assembly and disassembly. Similar to the nanoassemblies’ characteristics with regard to peak shift, 30 nm satellites consistently produced the greatest fluctuations in bandwidth for both assembly and disassembly.
In order to quantify the overall biosensing capability of each nanoassembly variation, we have proposed the use of a figure of merit (FOM) for detecting plasmon peak shifts28
where 〈FWHM〉 is the average of the two FWHM values. The FOM takes into account both the peak shift and plasmon bandwidth, quantifying the nanoassemblies’ ability to provide a resolved and observable colorimetric change. The experimentally obtained FOM values are shown in Table 1 for the nanoassembly variations we have studied. The substrate with 30 nm cores and 50 nm satellites has the highest FOM value, 0.37. However, the small scattering cross section of a 30 nm core AuNP made this substrate very difficult to observe by naked eye. The second and third nanoassemblies with the next highest FOM values, 50 nm cores with 30 nm and 50 nm satellite sizes, yielded spectacular color shifts for our given experimental setup (Figure 2). These two combinations of nanoassembly are optimal.
Table 1.
Figure of Merit for Various Core-Satellite Assemblies
| Satellite Size |
10 nm | 0.10 | 0.05 | 0.01 | 0.07 |
| 30 nm | 0.26 | 0.28 | 0.10 | 0.08 | |
| 50 nm | 0.37 | 0.25 | 0.09 | 0.03 | |
| 30 nm | 50 nm | 80 nm | 100 nm | ||
| Core Size | |||||
CONCLUSION
We have proposed an advancement upon the current suspension-based modality of colloidal biomolecular sensors that is better suited for on-chip diagnostic system integration while preserving the capability for diverse species detection. We present the fabrication of nanoassemblies self-assembled on a two-dimensional glass substrate, demonstrate the colorimetric functionality of biomolecular sensing via trypsin-mediated disassembly, and systematically analyze the optical detection properties of the substrate. A FOM was determined to quantify visual resolution the of the color change and nanoassemblies with 50 nm cores and either 30 nm or 50 nm satellites were concluded to be optimal. To the best of our knowledge, this plasmonic biosensor is the first self-assembled substrate that yields a structural color change directly at the site of disassembly. This new modality may enable a new generation of robust POC diagnostic devices with enhanced sensing capabilities.
METHODS
Nanoassembled Substrate Fabrication
Droplet immobilizers were prepared by cutting 1 in. × 1 in. squares of cured, 4 mm thick PDMS (Dow Corning) and punching a 20 mm circle through the center. One droplet immobilizer was placed in the middle of a 1 in. × 3 in. 3-aminopropyltriethoxysilane (APTES) treated microscope slide (The Microscope Store). The slides were cleaned with a stream of nitrogen and immediately placed in a Petri dish. The surface adhesion of the PDMS to the APTES treated slide prevented liquid leakage and capillary remodeling due to surface dehydration. Gold colloid (800 µL, British Biocell International) at stock concentration was pipetted into the PDMS well and incubated at 4°C for 18 hrs. The wells were then washed multiple times with DI water (Milli-Q). A 50 µg/mL solution of the biotinylated peptide, biotin-GGRGDGKGGC-OH (Chi Scientific), in 1× PBS buffer (Gibco, Invitrogen) was prepared. The peptide solution was pipetted (800 µL) into the PDMS well and incubated at 4°C for 18 hrs. The wells were again washed with DI water. A 2 mM mPEG-SVA (MW = 2kDa, Laysan Bio) solution was prepared in 100 mM NaHCO3 (pH 8.25, Sigma-Aldrich) and sonicated briefly.29 The mPEG-SVA solution was then pipetted (800 µL) into the PDMS well and incubated at room temperature (25°C) for 6 hrs. The wells were again washed with DI water. Finally, streptavidin conjugated AuNPs were pipetted (800 µL) into the well and incubated at 4°C for 12 hrs. The slides were then washed with DI water and stored at 4°C before exposure to 0.25% Trypsin-EDTA 1× (Gibco, Invitrogen).
Streptavidin Conjugation onto AuNPs
A modified version of a previous protocol was used.30 A sodium borax buffer was prepared by titrating 10 mL of 200 mM H3BO3 (pH 5.4, Sigma-Aldrich) with 2 mM Na2B4O7 (pH 8.9, Sigma-Aldrich) to a final pH 7.1 (~18 mL of 2 mM Na2B4O7 was added). The pH of the buffer is essential as it must be above the isoelectric point of streptavidin (pI ≈ 5.0) such that streptavidin can displace the negatively charged citrate groups adsorbed on the surface the AuNPs, but not too high of a pH as excess salt ions from Na2B4O7 will induce flocculation. 1 mL of 10 nm, 30 nm, and 50 nm AuNPs (stock concentration) were spun down at 14,000, 10,000, and 7,000 RPM for 10, 8, and 5 min, respectively. The supernatant was discarded and the particles were resuspended in an equal volume (1 mL) of 5 µg/mL streptavidin (SouthernBiotech) in the sodium borax buffer. The suspension was placed on a rocker for 15 min to allow physio-adsorption. The streptavidin conjugated AuNPs were spun down again and the supernatant was discard. The conjugated AuNPs were resuspended in DI water. A 1% w/v NaCl (Sigma-Aldrich) test was performed to ensure streptavidin conjugation. Conjugated AuNPs can withstand salt concentrations up to 2–3% before crashing out of solution.
Dark Field Spectroscopy
The nanoassemblies were imaged in dark field with an inverted microscope (Carl Zeiss Axiovert 200) under illumination from a Xenon light source (Karl Storz). The scattered light was collected by a 20× microscope objective (Carl Zeiss, LD Achroplan) with a numerical aperture (NA = 0.40) smaller than that of the Nikon DF dry condenser lens (NA = 0.80 – 0.95). The spectra were acquired with a Princeton Instruments Acton SP2300 using WinSpec and images were taken with a true-color CCD (QImaging Micropublisher 3.3 RTV). All measurements were taken with the nanoassemblies immersed in DI water and at room temperature (25°C).
Supplementary Material
Acknowledgements
J. Waldeisen gratefully acknowledges support from an NSF Graduate Research Fellowship. The authors acknowledge financial support from the National Institutes of Health (NIH) Nanomedicine Development Center for the Optical Control of Biological Function (Grant No. 3PN2 EY01824), Center for Nanostructured Materials and Technology (CNMT), National Academies Keck Futures Initiative funding (Grant No. NA-KFI Nano09), DARPA MF3 (Award No. HR0011-06-10050), and DARPA SERS S&T Fundamental Program under LLNL subcontract (Grant No. B573237). We thank N. Choudhary for her early contributions to the project.
Footnotes
Supporting Information Available. Details concerning the scattering properties of various sized AuNPs, sample standard deviation measurements for peak and FWHM measurements, microfluidic device bonded onto core-satellite substrate, emission spectra of Xenon light source, AuNP fabrication data, predicted detection times for physiologically relevant biosamples, and MATLAB scripts for spectral data analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-Based Method for Rationally Assembling Nanoparticles into Microscopic Materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 2.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-Guided Crystallization of Colloidal Nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 3.Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Jr, Schultz PG. Organization of ‘Nanocrystal Molecules’ using DNA. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 4.Loweth CJ, Caldwell WB, Peng X, Alivisatos AP, Schultz PG. DNA-based Assembly of Gold Nanocrystals. Angew. Chem. Int. Ed. 1999;38:1808–1812. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1808::AID-ANIE1808>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 5.Yao H, Yi C, Tzang CH, Zhu J, Yang M. DNA-Directed Self-Assembly of Gold Nanoparticles into Binary and Ternary Nanostructures. Nanotechnology. 2007;18 No. 015102. [Google Scholar]
- 6.Xu X, Rosi NL, Wang Y, Huo F, Mirkin CA. Asymmetric Functionalization of Gold Nanoparticles with Oligonucleotides. J. Am. Chem. Soc. 2006;128:9286–9287. doi: 10.1021/ja061980b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maye MM, Nykypanchuk D, Cuisinier M, van der Lelie D, Gang O. Stepwise Surface Encoding for High-Throughput Assembly of Nanoclusters. Nat. Mater. 2009;8:388–391. doi: 10.1038/nmat2421. [DOI] [PubMed] [Google Scholar]
- 8.Sadasivan S, Dujardin E, Li M, Johnson CJ, Mann S. DNA-Driven Assembly of Mesoporous Silica/Gold Satellite Nanostructures. Small. 2005;1:103–106. doi: 10.1002/smll.200400039. [DOI] [PubMed] [Google Scholar]
- 9.Li B, Li CY. Immobilizing Au Nanoparticles with Polymer Single Crystals, Patterning and Asymmetric Functionalization. J. Am. Chem. Soc. 2007;129:12–13. doi: 10.1021/ja0668318. [DOI] [PubMed] [Google Scholar]
- 10.Sebba DS, Mock JJ, Smith DR, LaBean TH, Lazarides AA. Reconfigurable Core-Satellite Nanoassemblies as Molecularly-Driven Plasmonic Switches. Nano Lett. 2008;8:1803–1808. doi: 10.1021/nl080029h. [DOI] [PubMed] [Google Scholar]
- 11.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. A Molecular Ruler Based on Plasmon Coupling of Single Gold and Silver Nanoparticles. Nature Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 12.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Lu Y. Fast Colorimetric Sensing of Adenosine and Cocaine Based on a General Sensor Design Involving Aptamers and Nanoparticles. Angew. Chem. Int. Ed. 2006;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Lu Y. A Colorimetric Lead Biosensor Using DNAzyme-Directed Assembly of Gold Nanoparticles. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 15.Jun YW, Sheikholeslami S, Hostetter DR, Tajon C, Craik CS, Alivisatos AP. Continuous Imaging of Plasmon Rulers in Live Cells Reveals Early-Stage Caspase-3 Activation at the Single-Molecule Level. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17735–17740. doi: 10.1073/pnas.0907367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsch LR, Jackson JB, Lee A, Halas NJ, West JL. A Whole Blood Immunoassay Using Gold Nanoshells. Anal. Chem. 2003;75:2377–2381. doi: 10.1021/ac0262210. [DOI] [PubMed] [Google Scholar]
- 17.Myers FB, Lee LP. Innovations in Optical Microfluidic Technologies for Point-of-Care Diagnostics. Lab Chip. 2008;8:2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- 18.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with Plasmonic Nanosensors. Nature Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 19.Zhao W, Ali MM, Aguirre SD, Brook MA, Li Y. Paper-Based Bioassays Using Gold Nanoparticle Colorimetric Probes. Anal. Chem. 2008;80:8431–8437. doi: 10.1021/ac801008q. [DOI] [PubMed] [Google Scholar]
- 20.Charrier A, Candoni N, Liachenko N, Thibaudau F. 2D Aggregation and Selective Desorption of Nanoparticle Probes: A New Method to Probe DNA Mismatches and Damages. Biosens. Bioelectron. 2007;22:1881–1886. doi: 10.1016/j.bios.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Ramseier CA, Kinney JS, Herr AE, Braun T, Sugai JV, Shelburne CA, Rayburn LA, Tran HM, Singh AK, Giannobile WV. Identification of Pathogen and Host-Response Markers Correlated with Periodontal Disease. J. Periodontol. 2009;80:436–446. doi: 10.1902/jop.2009.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tchetverikov I, Lohmander LS, Verzijl N, Huizinga TWJ, TeKoppele JM, Hanemaaijer R. DeGroot, MMP Protein and Activity Levels in Synovial Fluid from Patients with Joint Injury, Inflammatory Arthritis, and Osteoarthritis. J. Ann. Rheum. Dis. 2005;64:694–698. doi: 10.1136/ard.2004.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brawley OW, Ankerst DP, Thompson IM. Screening for Prostate Cancer. CA: Cancer J. Clin. 2009;59:264–273. doi: 10.3322/caac.20026. [DOI] [PubMed] [Google Scholar]
- 24.Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK. Microfluidic Immunoassays as Rapid Saliva-Based Clinical Diagnostics. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turkevich J, Stevenson PC, Hillier J. A Study of the Nucleation and Growth Processes in the Synthesis of Colloidal Gold. Discuss. Faraday Soc. 1951;11:55–75. [Google Scholar]
- 26.Ross BM, Lee LP. Comparison of Near- and Far-Field Measures for Plasmon Resonance of Metallic Nanoparticles. Opt. Lett. 2009;34:896–898. doi: 10.1364/ol.34.000896. [DOI] [PubMed] [Google Scholar]
- 27.Maier I, Morgan MRA, Lindner W, Pittner F. Optical Resonance-Enhanced Absorption-Based Near-Field Immunochip Biosensor for Allergen Detection. Anal. Chem. 2008;80:2694–2703. doi: 10.1021/ac702107k. [DOI] [PubMed] [Google Scholar]
- 28.Ross BM, Waldeisen JR, Wang T, Lee LP. Strategies for Nanoplasmonic Core-Satellite Biomolecular Sensors: Theory-Based Design. Appl. Phys. Lett. 2009;95:193112. doi: 10.1063/1.3254756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y-P, Oh E, Oh Y-H, Moon DW, Lee TG, Kim H-S. Protein Kinase Assay on Peptide-Conjugated Gold Nanoparticles by Using Secondary-Ion Mass Spectrometric Imaging. Angew. Chem. Int. Ed. 2007;46:6816–6819. doi: 10.1002/anie.200701418. [DOI] [PubMed] [Google Scholar]
- 30.Oliver C. Immunocytochemical Methods and Protocols. In: Javois LC, editor. Methods in Molecular Biology. Vol. 115. Totowa, New Jersey: Humana Press; 1999. pp. 327–339. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.