Abstract
Antibody heavy-chain recombination that results in the incorporation of multiple diversity (D) genes, although uncommon, contributes substantially to the diversity of the human antibody repertoire. Such recombination allows the generation of heavy chain complementarity determining region 3 (HCDR3) regions of extreme length and enables junctional regions that, because of the nucleotide bias of N-addition regions, are difficult to produce through normal V(D)J recombination. Although this non-classical recombination process has been observed infrequently, comprehensive analysis of the frequency and genetic characteristics of such events in the human peripheral blood antibody repertoire has not been possible because of the rarity of such recombinants and the limitations of traditional sequencing technologies. Here, through the use of high-throughput sequencing of the normal human peripheral blood antibody repertoire, we analysed the frequency and genetic characteristics of V(DD)J recombinants. We found that these recombinations were present in approximately 1 in 800 circulating B cells, and that the frequency was severely reduced in memory cell subsets. We also found that V(DD)J recombination can occur across the spectrum of diversity genes, indicating that virtually all recombination signal sequences that flank diversity genes are amenable to V(DD)J recombination. Finally, we observed a repertoire bias in the diversity gene repertoire at the upstream (5′) position, and discovered that this bias was primarily attributable to the order of diversity genes in the genomic locus.
Keywords: antibody responses, B-cell receptor, immunogenetics, immunoglobulins
Introduction
Since the discovery that recombination activating gene (RAG) -mediated recombination of variable (V), diversity (D) and joining (J) gene segments generates virtually unlimited sequence diversity in the antibody repertoire,1–5 much progress has been made in determining the genetic and mechanistic elements that participate in the antibody recombination process. It is generally understood that recombination signal sequences (RSS), which are composed of conserved AT-rich heptamer and nonamer sequences separated by spacers of either 12 or 23 nucleotides, are recognized and bound by RAG1 and RAG2 proteins at the initiation of the recombination process.6,7 RAG binding is highly dependent on the heptamer and nonamer sequences, and alterations to either sequence result in decreased RAG binding.8–10 It has long been understood that spacer length is critical to recombination, and there is evidence of sequence conservation within the spacer region.11–13 Recombination typically occurs only between RSS elements of different spacer lengths, in a model commonly referred to as the 12/23 rule of recombination.14–17 After binding to one 12-bp RSS and one 23-bp RSS, the RAG complex induces single-strand DNA nicks between the coding sequence and the heptamer of each RSS, resulting in hairpin formation on each of the coding ends and a blunt double-stranded break on each signal end.18–21 The hairpins are opened, nucleotides may be added to or removed from the coding ends, and the double-strand breaks at the coding ends are joined into a single coding strand.22–27 In antibody heavy-chain genes, D gene segments are flanked by 12-bp RSSs on either side, whereas V and J gene segments are flanked by 23-bp RSSs.28,29 Recombination therefore proceeds in a stepwise fashion, with D-JH recombination preceding VH-D recombination, resulting in a complete heavy-chain variable region.30,31
The human diversity gene locus consists of 27 different genes, which are arranged into four tandemly oriented groups,32,33 and are located approximately 8 kb upstream of Cμ, the nearest constant region gene. The unique positioning of diversity genes in regularly spaced 9-kb clusters suggests that the locus may have been created by a series of duplications.32,34 Of the 27 diversity genes, 25 are considered functional and two are thought to encode pseudogenes.33
D-D fusions, which result in VH(DHDH)JH recombinations, are thought to be prohibited by the 12/23 rule, but such non-12/23 recombination events have been demonstrated to occur in both in vitro and in vivo systems.35–40 Even in model systems designed to induce non-12/23 recombinations, these recombination events are much less efficient than those that adhere to the 12/23 rule.6,41,42 V(DD)J recombinations have been shown to be more frequent in self-reactive and polyreactive antibody populations than in the normal repertoire.43 This is possibly a result of the long complementarity determining region 3 (CDR3) loops created by D-D fusions, which have been associated with autoreactive properties.44–46
Recent work has suggested, however, that many previously identified D-D fusions are likely to be artefacts of the random N-addition process.39 Over-estimation of the frequency of D-D fusions in the peripheral blood repertoire is typically the result of reliance on very short regions of homology between the recombinant sequence and germline diversity genes when identifying putative D-D fusions. These short regions of homology are probably the result of random matches to the germline genes caused by non-templated N-addition rather than D-D fusion. The most stringent analyses of D-D fusions in the human repertoire, identifying sequences that are highly likely to be true D-D fusions and not coincidental N-addition matches to germline diversity genes, were performed by Sanz35,37 and by Raaphorst et al.,35,37 but a thorough genetic analysis of the V(DD)J repertoire has not been possible because of the rarity of D-D fusions.
In this report, we present an extensive genetic analysis of the human peripheral blood antibody repertoire using high throughput antibody gene analysis and, for the first time, describe the frequency and detailed genetic characteristics of V(DD)J recombination events in the naive and memory subsets of the circulating human B-cell repertoire.
Materials and methods
Sample preparation and sorting
Peripheral blood was obtained from healthy adult donors following informed consent, under a protocol approved by the Vanderbilt Institutional Review Board. Mononuclear cells were isolated by density gradient centrifugation with Histopaque 1077 (Sigma, St. Louis, MO) and B cells were enriched by paramagnetic separation (Miltenyi Biotec, Cambridge, MA). Cells from particular B-cell subsets were sorted as separate populations on a high-speed sorting cytometer (FACSAria III; Becton Dickinson, Franklin Lakes, NJ) using the following phenotypic markers, naive B cells: CD19+ CD27− IgM+ IgG− CD14− CD3−, IgM memory B cells: CD19+ CD27+ IgM+ IgG− CD14− CD3− and IgG memory B cells: CD19+ CD27+ IgM− IgG+ CD14− CD3−. Total RNA was isolated from each sorted cell subset using a commercial RNA extraction kit (RNeasy; Qiagen, Valencia, CA).
cDNA synthesis and PCR amplification of antibody genes
Reverse transcription (RT-) PCR primers were originally described by the BIOMED-2 consortium47 and were slightly modified to suit amplification for large-scale parallel pyrosequencing. One hundred nanograms of each total RNA sample was used in duplicate 50-μl RT-PCRs using the OneStep RT-PCR system (Qiagen) and gene-specific RT-PCR primers (see Supplementary material, Table S1). Thermal cycling was performed as described elsewhere.47 Five microlitres of each pooled RT-PCR was used for each 454-Adapter PCR, performed in quadruplicate. Thermal cycling was carried out as before, but for 10 cycles. Sequences for all primers are found in the supplementary information (Table S1).
Amplicon nucleotide sequence analysis
The amplicon libraries were quantified using a Qubit fluorometer (Invitrogen, Carlsbad, CA). The size and quality of the DNA libraries were evaluated on an Agilent Bioanalyzer 2100 using the DNA 7500 labchip (Agilent, Palo Alto, CA). The samples were then diluted to a working concentration of 1 × 106 molecules per microlitre.
Quality control of the amplicon libraries and emulsion-based clonal amplification and sequencing on the 454 Genome Sequencer FLX Titanium system were performed by the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign, according to the manufacturer's instructions (454 Life Sciences, Branford, CT).
Antibody sequence analysis
The ImMunoGeneTics (IMGT) High/V-Quest webserver (http://www.imgt.org) was used for germline gene assignments and initial analysis. As the 454 sequencing platform typically results in a higher frequency of insertion and deletion mutations than other sequencing platforms, High/V-QUEST analysis of the antibody sequences was performed using the option to identify and remove insertions or deletions. When performing further analysis, any insertions or deletions that were not codon-length (i.e. that resulted in frameshifts) were not considered. Antibody sequences returned from IMGT were considered to be high-quality sequences if they met the following requirements: sequence length of at least 300 nt; identified V and J genes; an intact, in-frame recombination; and absence of stop codons or ambiguous nucleotide calls within the reading frame.
Stringent filtering for putative V(DD)J recombinations
The antibody sequence region identified by IMGT as a putative diversity gene is designated here as the ‘match region’. The length of the match region, minus any mismatches between the match region and the germline diversity gene, is designated the ‘match score’. Our filtering process required the match regions to contain a maximum of one nucleotide difference from the germline diversity gene segment. The match score, which represents the number of identically matched nucleotides between the match region and the germline diversity gene segment, was required to be at least 60% of the overall length of the germline diversity gene segment, except in the case of the short IGHD7-27 gene segment, for which we required a match score of 72% of the germline diversity gene length.
Results
Stringent filtering procedure for identification of putative V(DD)J recombinations
We separately isolated naive, IgM memory and IgG memory B cells from four healthy individuals using flow cytometric sorting and subjected the transcribed antibody genes from those cells to high throughput sequencing. We selected the transcribed antibody genes (mRNA) for amplification to reduce the number of non-expressed, non-functional sequences. After selecting only high-quality, non-redundant antibody sequences, we obtained a total of 294 232 naive cell sequences, 161 313 IgM memory cell sequences and 94 841 IgG memory cell sequences (Table S3).
To limit the number of falsely identified V(DD)J recombinations (that is, recombinations with N-addition regions that contain stretches of similarity to diversity genes), we developed a stringent filtering procedure. All sequences were analysed with the IMGT High/V-QUEST webserver, with the number of accepted diversity genes set to 2. The antibody region identified by IMGT as a putative diversity gene is designated here as the ‘match region’ (Fig. 1). Our filtering process required the match regions to contain a maximum of one nucleotide difference from the germline diversity gene segment. The length of the match region, minus any mismatches between the match region and the germline diversity gene, is designated the ‘match score’. The match score, which represents the number of identically matched nucleotides between the match region and the germline diversity gene segment, was required to be at least 60% of the overall length of the germline diversity gene segment, except in the case of the short IGHD7-27 gene segment, for which we required a match score of 72% of the germline diversity gene length (eight of 11 nucleotides). Minimum acceptable match scores and false discovery rate calculations for each diversity gene are shown in the supplementary material, Table S2. Absolute counts of V(DD)J recombinants are shown by donor and subset in Table S4.
Figure 1.
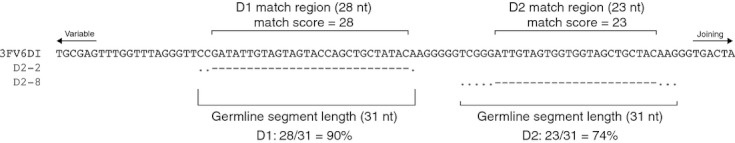
Stringent filtering of V(DD)J recombinants. A sample V(DD)J recombinant (3FV6DI) is shown, along with the germline diversity gene assignments for both the 5′ D (D2-2) and 3′ D (D2-8) positions. Dashes indicate conservation between germline and 3FV6DI and dots indicate mismatches. The match region is indicated above the 3FV6DI sequence, along with the match score (the match region length minus any mismatches within the match region). The germline segment length is shown below the sequence alignment. Below the germline segment length, the scoring calculation is shown. Sequences that contained scores of > 60% for both diversity gene positions were considered V(DD)J rearrangements.
This stringent filtering process results in sequences that contain long stretches of sequence identity with germline diversity genes (Table 1).
Table 1.
Alignment of representative V(DD)J recombinants with associated germline diversity genes. Dashes indicate identity of the antibody sequence with the germline gene segment; dots indicate nucleotides in the gene segment not present in the mature antibody
 |
Analysis of N-addition regions indicates that identified V(DD)J recombinations probably result from D-D fusion
Although it is highly unlikely that random insertion of nucleotides at the VD or DJ junction would result in long sequence stretches that identically match germline diversity gene segments, we examined the N-addition lengths of all recombination sites in the total peripheral blood repertoire to determine if the N-addition regions flanking both diversity genes matched that of the normal V(D)J repertoire. We determined the mean N-addition length for all VD and DJ recombination sites in the total repertoire and the VD (also referred to as N1), DD (N2) and DJ (N3) recombination sites in the putative V(DD)J repertoire (Fig. 2a). We found that there was no statistically distinguishable difference between the mean length of the recombination sites in the total repertoire and in the putative V(DD)J repertoire.
Figure 2.
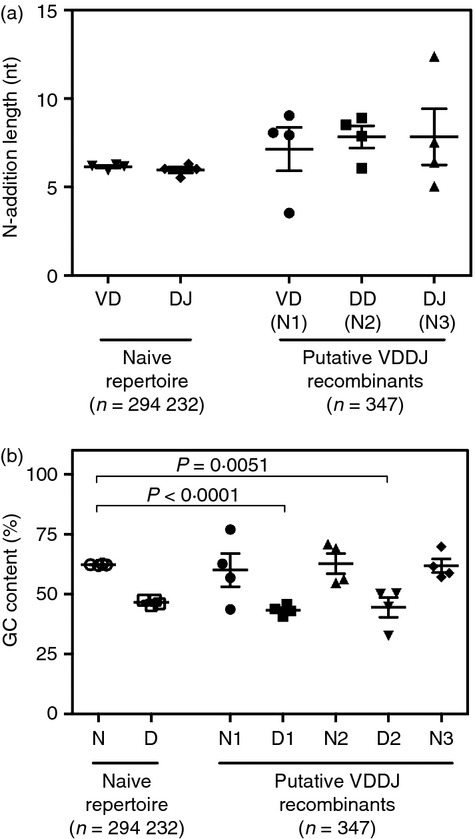
Putative V(DD)J recombinants contain normal N-addition lengths and diversity genes, with low GC content. (a) N-addition length for each recombination site in the total naive repertoire or in the V(DD)J repertoire. The mean N-addition length for each of four healthy individuals ± SEM is shown for each recombination site. (b) GC content (as a percentage of the region sequence) for each N-addition region or diversity gene segment in the V(DD)J repertoire or the total naive repertoire. Combined N-addition at the VD and DJ junctions (N) or diversity gene region (D) are shown for the total repertoire. Diversity genes at the 5′ D position (D1) and 3′ D position (D2) and N-addition sites at the VD junction (N1), DD junction (N2) and DJ junction (N3) are shown for putative V(DD)J recombinants.
We next compared the GC content of all N-addition (N) and D gene regions in the total naive repertoire to the GC content of both assigned diversity genes (D1 and D2) and the N1, N2 and N3 N-addition sites in the V(DD)J repertoire (which correspond to the VD, DD and DJ recombination sites, respectively) (Fig. 2c). There was a highly significant decrease in the GC content of both D1 (P < 0·0001) and D2 (P = 0·0051) regions when compared with the N-addition regions in the total naive repertoire. In contrast, neither of the assigned D gene segments in the V(DD)J repertoire differed in the GC content of their assigned D genes from the total naive repertoire, Hence, the D1 and D2 regions better resembled the GC content of D gene segments than N-addition regions.
Frequency of putative V(DD)J recombinants is greatly reduced in peripheral blood memory B-cell subsets
The sequences obtained from each of the three cell subsets were examined for the presence of junctions containing multiple diversity gene segments using a high stringency filtering procedure. The frequency of V(DD)J recombination in each of the three sorted B-cell subsets is shown in Fig. 3(a). The mean V(DD)J recombinant frequency in the naive population (0·12%) was more than 10-fold higher than in the IgM memory population (0·01%, P = 0·0095). Interestingly, the IgG memory population did not contain a single predicted V(DD)J recombination event that passed our filtering procedure.
Figure 3.
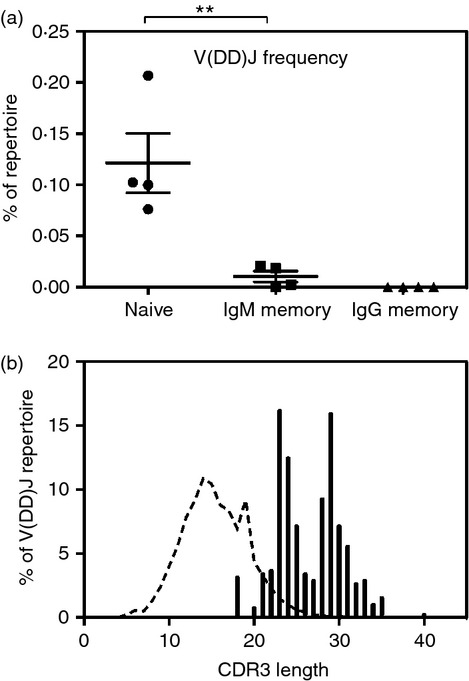
Frequency and diversity gene use of putative V(DD)J recombinants. (a) V(DD)J recombinant frequency (as a percentage of the subset repertoire) of naive, IgM memory or IgG memory B-cell subsets isolated from the peripheral blood of four healthy individuals. The V(DD)J frequency for each donor ± SEM for each subset is shown. Pairwise comparisons of V(DD)J recombinant frequency between different subsets were determined using a one-way analysis of variance with Bonferroni's correction. (b) Histogram of CDR3 length distribution of V(DD)J recombinations (filled bars). The length distribution for the entire repertoire (dashed line) is also shown for comparison. **P < 0·01.
It was possible that our filtering procedure, which allowed only a single mutation in the match region, preferentially rejected prediction of V(DD)J recombinants from the somatically mutated memory populations while retaining V(DD)J recombinants in the mutation-free naive population. We performed the analyses a second time using a less stringent filtering protocol, in an effort to correctly predict V(DD)J recombination even in the somatically mutated memory subset. This ‘loose’ filter, which allowed mismatches within the match region and reduced the match length to 55% of the germline gene, revealed a higher V(DD)J recombinant frequency in all subsets compared with the more stringent filter, but still showed a significant reduction in V(DD)J recombination frequency in the IgM memory (0·08%, P = 0·0077) or IgG memory (0·04%, P = 0·0048) subsets when compared with the naive subset (0·23%). Memory subsets have a reduced frequency of long heavy chain complementarity determining region 3 (HCDR3) loops48 and the V(DD)J population is dominated by long HCDR3-containing antibodies (Fig. 3b), with the average HCDR3 length of 26·5 amino acids.
Germline diversity gene usage in putative V(DD)J recombinants
The D gene usage in V(DD)J recombinants was compared to D gene usage in the total naive cell repertoire to identify if there was a preferential use of particular D genes in V(DD)J recombinants (Fig. 4). Interestingly, D gene use at the 3′ D position in V(DD)J recombinants was very similar to D gene usage in the total naive repertoire, showing only an increase in D7 gene family usage (difference between means of 6·93 ± 2·58, P = 0·036) and an approximately equivalent decrease in D2 gene family usage (difference between means of −10·02 ± 2·40, P = 0·0058). D gene use at the 5′ D position in V(DD)J recombinants showed a significant increase in the D2 gene family (P = 0·0022) and a significant decrease in the D1, D3 and D4 gene families (P < 0·0001, P = 0·0132 and P = 0·0414, respectively). The 5′ D position also showed a strong trend toward increased D5 gene family use (P = 0·06) and a notable absence of D7 family members.
Figure 4.
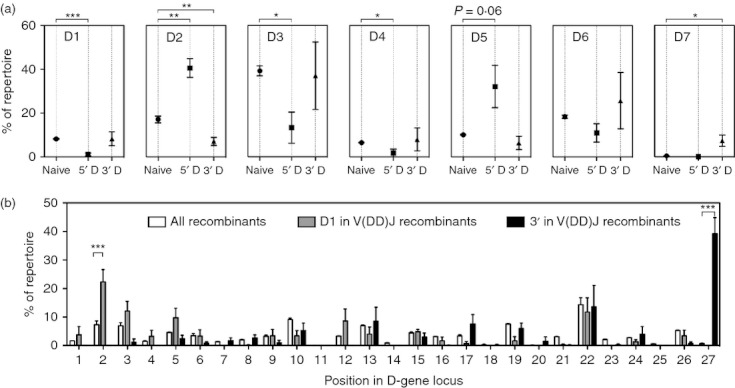
Diversity gene use in putative V(DD)J recombinants differs from that in the total naive repertoire. (a) Diversity gene family use for the total naive repertoire (Naive) or for the 5′ D and 3′ D positions in V(DD)J recombinants. (b) Diversity gene use in the total repertoire, at the D1 position in V(DD)J recombinants, and at the D2 position of V(DD)J recombinants. Mean ± SEM for each donor is shown. Pairwise comparisons were determined using a two-way analysis of variance with Bonferroni's correction. *P < 0·05; **P < 0·01; ***P < 0·001.
D gene order in V(DD)J recombinants matches the order of those D genes in the genome
We analysed 5′ D and 3′ D pairings in the V(DD)J repertoire and discovered that every V(DD)J recombinant contained D-D pairings in an orientation that matched the orientation of the genomic locus (Fig. 5a). We also found that V(DD)J recombination occurred across the spectrum of D genes, using every D gene with the exceptions of D4-11, D4-14 and D6-25. D4-11 (< 0·001%), D4-14 (0·895%) and D6-25 (0·60%) were the least frequently observed D genes in the total repertoire (Fig. 5c), so the lack of these D genes in the V(DD)J repertoire was probably a result of their rarity. As expected, the 5′ D position contained a high proportion of D gene segments located at the 5′ end of the genomic locus (Fig. 5b), with the frequency of D gene presence in the 5′ D position decreasing exponentially with distance from the 5′ end of the genomic locus. A similar preference for upstream D genes was not seen in when analysing the frequency of D gene use in the entire repertoire (Fig. 5c). Only three of the 10 furthest downstream (3′) D genes were ever found in the 5′ D position of a V(DD)J recombinant. There was also a weak trend toward increased usage of downstream D genes in the 3′ D position of V(DD)J recombinants (P = 0·2089).
Figure 5.
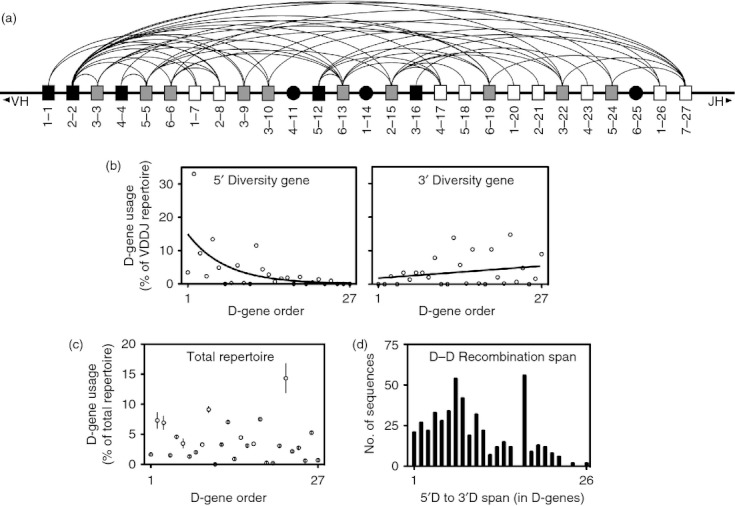
Genomic orientation of diversity genes matches the orientation in putative V(DD)J recombinants and explains diversity gene use bias at 5′ D and 3′ D positions. (a) All functional diversity genes are represented in the genomic orientation, with arcs connecting diversity genes found paired in a V(DD)J recombinant. Black boxes indicate diversity genes found only in the 5′ D position, white boxes indicate diversity genes found only in the 3′ D position, and grey boxes indicate diversity genes found in both positions. D4-11, D1-14 and D6-25, the only diversity genes not found in any V(DD)J recombinants, are represented by black circles. The orientation of diversity genes in V(DD)J recombinants matches the genomic orientation in every instance, so the leftmost member of any linked pair of diversity genes shown in this diagram was always in the 5′ D position of the V(DD)J recombination. (b) The frequency of each diversity gene in either the 5′ D or 3′ D positions is shown with the diversity genes ordered and labelled by position from the 5′ end of the genomic locus. (c) The frequency of each diversity gene in the total repertoire (including all B-cell subsets). Shown are the mean frequency ± SEM for each donor. (d) The frequency of each recombination span, defined as the distance between the 5′ D gene and the paired 3′ D gene in V(DD)J recombinants (measured in diversity gene segments and including non-functional genes). Recombination between adjacent diversity genes results in a recombination span of 1.
We next determined whether or not there was a preference for D-D recombination events between D genes located close to each other in the genomic locus. For each V(DD)J recombination, we determined a recombination span, calculated by subtracting the position number of the 5′ D gene from the position of the 3′ D gene. Recombination between adjacent D genes resulted in a recombination span of 1, whereas recombination between the first and last (27th) diversity genes resulted in a recombination span of 26. We observed a strong trend toward decreased use of the most distant pairings (Fig. 5d; P = 0·0568). Notably, although there was a global trend toward decreased pairings of distant diversity genes, the most frequently observed recombination span was 17, of which there were several D-D combinations accounting for over 10% of all V(DD)J recombinants.
Skewed germline gene usage in 5′ D and 3′ D positions was probably the result of diversity gene orientation in the genomic locus
Understanding that the frequency of diversity gene use in the 5′ D position of V(DD)J recombinants depended on location in the genomic locus (Fig. 5b), we investigated whether or not orientation of the genomic locus was likely to be the cause of the skewed diversity gene usage seen in 5′ D and 3′ D positions of V(DD)J recombinants, or whether other genetic or mechanistic factors were the dominant force behind the skewed diversity gene repertoire.
At the 5′ D position, the complete lack of D7 family use was readily explained by the location of the only D7 family member, D7-27, at the 3′ end of the genomic locus; in-order V(DD)J rearrangements with D7-27 in the 5′ D position are not possible. The increase in D2 family use at the 5′ D position was also probably attributable to genomic orientation. The most commonly used D2 gene member in the naive repertoire, D2-2, accounted for over half of D2 family use in the naive repertoire and is located one position from the 5′ end of the genomic locus. In addition, three of the four D2 family members, accounting for over 80% of D2 family use in the naive repertoire, were found in the 15 (of 27) most upstream positions of the genomic locus. Much like the increase in D2 family usage, the decrease in D1 family was probably a result of the extreme downstream position of the most commonly used D1 family member, D1-26, which accounted for over 50% of D1 family usage in the naive repertoire. Finally, the trend toward increased D5 family frequency was possibly the result of the positioning of the two most common D5 gene members, used in over 75% of D5 family use in the naive repertoire, in the 5′ half of the genomic locus.
The increase in D7 family usage in 3′ D positions was likely to be attributable to the fact that D7-27, the only D7 family member, is positioned at the furthest 3′ position of the genomic locus, allowing for in-order V(DD)J recombination with every other diversity gene. Alternatively, the decreased use of D2 family use in 3′ D positions was possibly explained by the fact that the most commonly used D2 family member, D2-2, is positioned such that only one possible V(DD)J recombination exists with D2-2 in the 3′ D position.
Discussion
Previous estimates of V(DD)J frequency have varied widely, partly because of differing criteria for identification of putative V(DD)J recombinants.35–40 Several methods used previously involved low-stringency filters that identified putative D genes using as few as four homologous nucleotides. When using such a short homology region to identify D-D fusion events, it is likely that many of the identified D-D fusions were false attributions. The presence of false positives in the identified V(DD)J repertoire could dilute or counteract genetic trends in the true V(DD)J population, so limiting false-positive results was of critical importance. Our stringent approach, designed to minimize false positives, probably under-estimated, to some degree, the actual frequency of circulating B cells that express antibodies encoded by DNAs derived from V(DD)J recombination. Hence, the V(DD)J frequency estimate produced in this study describes the minimum expected frequency of such events in the human peripheral blood repertoire.
We identified putative V(DD)J recombinants in the naive B-cell subset at a frequency of approximately one in 800 circulating cells. We also observed that the frequency of V(DD)J recombinants is markedly lower in memory subsets than in the naive subset. It is not clear why the memory subsets contain such a reduced fraction of V(DD)J recombinations, however, previous data showing reduced frequency of long HCDR3s in memory subsets48 suggest that this observation may be a more general trend among B cells encoding antibodies with long HCDR3s and not a phenomenon that is specific to V(DD)J recombinants. Alternatively, it is also possible that our stringent filtering protocol, when applied to the somatically mutated memory subsets, excluded many V(DD)J recombinations as the result of germline mismatches caused by somatic hypermutation.
To further show that the sequences in putative V(DD)J recombinants identified in this study were generated by true D-D fusion and not by alignment of coincidental N-addition-mediated sequences, we closely analysed the N-addition length, GC-content and orientation of the 5′ and 3′ D gene segments in the identified V(DD)J recombinants and determined that it was highly unlikely that N-addition mimicry was the primary cause of these putative V(DD)J recombinants. If N-addition mimicry were to produce such sequences frequently, this process would require prolonged stretches of N-addition by TdT that are not GC-rich, which contradicts the extensively studied characteristics of the enzyme. Further, this mechanism would require that TdT must, in sequence regions both preceding and following the diversity gene mimic region, revert to production of characteristic GC-rich regions. Finally, TdT would somehow have to position the uncharacteristic stretches of non-GC-rich N-addition within the larger N-addition region such that the flanking GC-rich sequences match the length of typical GC-rich N-addition regions. The frequent occurrence of such a process seems highly unlikely.
Interestingly, we did not see any clonally related sequences in our sample of V(DD)J recombinants. To be considered clonally related, we required use of the same V, D1, D2 and J genes, and the same HCDR3 length. The absence of clonally related sequences in our sample was likely to be the result of the extremely low frequency of V(DD)J recombinants in either of the memory subsets, which results in very few clonal families available for identification. The vast majority of our V(DD)J sequence pool consisted of naive sequences, which further reduced the odds of finding clonally related sequences.
Analysis of diversity gene use in V(DD)J recombinants revealed two surprising observations. First, every diversity gene, with the sole exception of D4-11, was found in at least one V(DD)J recombination. While differences in the RSSs that flank diversity genes may affect the frequency of D-D fusion between certain diversity genes, our data indicate that the vast majority of RSSs that flank diversity genes are amenable to D-D fusion. Second, diversity gene use at the downstream (3′ D) position was very similar to diversity gene use in normal V(D)J recombinations, whereas diversity gene use at the upstream (5′ D) position was significantly different from normal V(D)J recombinations. Also, the 5′ D bias appears to be explained primarily by position of the germline gene segments in the genomic locus. Taken together, these observations suggest that 3′ D to JH recombination occurs normally in V(DD)J recombinants and that 5′ D gene usage is skewed primarily by the requirement that diversity genes in the 5′ D position must be positioned upstream of the 3′ D gene segment in the genomic locus.
Disclosure
The authors have no conflicts to disclose concerning the work in this paper.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Flow cytometric gating of naive, IgM memory and IgG memory subsets.
Table S1. Primers used in RT-PCR and 454-Adapter PCR.
Table S2. Minimum accepted match score and false match probability for each diversity gene.
Table S3. Flow cytometric sorting and 454 sequencing results.
Table S4. Absolute counts of V(DD)J recombinants by donor and subset.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Brack C, Hirama M, Lenhard-Schuller R, Tonegawa S. A complete immunoglobulin gene is created by somatic recombination. Cell. 1978;15:1–14. doi: 10.1016/0092-8674(78)90078-8. [DOI] [PubMed] [Google Scholar]
- 2.Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci USA. 1982;79:4118–22. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 4.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–48. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 5.Oettinger M, Schatz D, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–23. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 6.Hesse J, Lieber M, Mizuuchi K, Gellert M. V(D)J recombination – a functional definition of the joining signals. Genes Dev. 1989;3:1053–61. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 7.Alt FW, Oltz EM, Young F, Gorman J, Taccioli G, Chen J. VDJ recombination. Immunol Today. 1992;13:306–14. doi: 10.1016/0167-5699(92)90043-7. [DOI] [PubMed] [Google Scholar]
- 8.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–62. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 9.Cuomo CA, Mundy CL, Oettinger MA. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol Cell Biol. 1996;16:5683–90. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadel B, Tang A, Lugo G, Love V, Escuro G, Feeney AJ. Decreased frequency of rearrangement due to the synergistic effect of nucleotide changes in the heptamer and nonamer of the recombination signal sequence of the V kappa gene A2b, which is associated with increased susceptibility of Navajos to Haemophilus influenzae type b disease. J Immunol. 1998;161:6068–73. [PubMed] [Google Scholar]
- 11.Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–96. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee AI, Fugmann SD, Cowell LG, Ptaszek LM, Kelsoe G, Schatz DG. A functional analysis of the spacer of V(D)J recombination signal sequences. PLoS Biol. 2003;1:E1. doi: 10.1371/journal.pbio.0000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montalbano A, Ogwaro KM, Tang A, Matthews AG, Larijani M, Oettinger MA, Feeney AJ. V(D)J recombination frequencies can be profoundly affected by changes in the spacer sequence. J Immunol. 2003;171:5296–304. doi: 10.4049/jimmunol.171.10.5296. [DOI] [PubMed] [Google Scholar]
- 14.van Gent DC, Ramsden DA, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–13. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 15.Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–206. [PMC free article] [PubMed] [Google Scholar]
- 16.Steen S, Gomelsky L, Roth D. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–53. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 17.Schatz DG. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- 18.Roth DB, Menetski JP, Nakajima PB, Bosma MJ, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–91. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 19.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–32. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 20.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–95. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 21.Sadofsky MJ. The RAG proteins in V(D)J recombination: more than just a nuclease. Nucleic Acids Res. 2001;29:1399–409. doi: 10.1093/nar/29.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 23.Mahajan KN, Gangi-Peterson L, Sorscher DH, Wang J, Gathy KN, Mahajan NP, Reeves WH, Mitchell BS. Association of terminal deoxynucleotidyl transferase with Ku. Proc Natl Acad Sci USA. 1999;96:13926–31. doi: 10.1073/pnas.96.24.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shockett PE, Schatz DG. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:4159–66. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 26.Mansilla-Soto J, Cortes P. VDJ recombination: artemis and its in vivo role in hairpin opening. J Exp Med. 2003;197:543–7. doi: 10.1084/jem.20022210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth DB. Restraining the V(D)J recombinase. Nat Rev Immunol. 2003;3:656–66. doi: 10.1038/nri1152. [DOI] [PubMed] [Google Scholar]
- 28.Early P, Huang H, Davis M, Calame K, Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980;19:981–92. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 29.Kurosawa Y, Tonegawa S. Organization, structure, and assembly of immunoglobulin heavy chain diversity DNA segments. J Exp Med. 1982;155:201–18. doi: 10.1084/jem.155.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alt F, Blackwell T, Yancopoulos G. Development of the primary antibody repertoire. Science. 1987;238:1079–87. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- 31.Schatz DG, Oettinger MA, Schlissel MS. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–83. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 32.Siebenlist UU, Ravetch JVJ, Korsmeyer SS, Waldmann TT, Leder PP. Human immunoglobulin D segments encoded in tandem multigenic families. Nature. 1981;294:631–5. doi: 10.1038/294631a0. [DOI] [PubMed] [Google Scholar]
- 33.Corbett SJ, Tomlinson IM, Sonnhammer EL, Buck D, Winter G. Sequence of the human immunoglobulin diversity (D) segment locus: a systematic analysis provides no evidence for the use of DIR segments, inverted D segments, “minor” D segments or D-D recombination. J Mol Biol. 1997;270:587–97. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]
- 34.Ichihara YY, Matsuoka HH, Kurosawa YY. Organization of human immunoglobulin heavy chain diversity gene loci. EMBO J. 1988;7:4141–50. doi: 10.1002/j.1460-2075.1988.tb03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanz I. Multiple mechanisms participate in the generation of diversity of human H chain CDR3 regions. J Immunol. 1991;147:1720–9. [PubMed] [Google Scholar]
- 36.Kiyoi H, Naoe T, Horibe K, Ohno R. Characterization of the immunoglobulin heavy chain complementarity determining region (CDR)-III sequences from human B cell precursor acute lymphoblastic leukemia cells. J Clin Invest. 1992;89:739–46. doi: 10.1172/JCI115650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raaphorst FM, Raman CS, Tami J, Fischbach M, Sanz I. Human Ig heavy chain CDR3 regions in adult bone marrow pre-B cells display an adult phenotype of diversity: evidence for structural selection of DH amino acid sequences. Int Immunol. 1997;9:1503–15. doi: 10.1093/intimm/9.10.1503. [DOI] [PubMed] [Google Scholar]
- 38.Koralov SB, Novobrantseva TI, Hochedlinger K, Jaenisch R, Rajewsky K. Direct in vivo VH to JH rearrangement violating the 12/23 rule. J Exp Med. 2005;201:341–8. doi: 10.1084/jem.20041577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson LC, Moffatt-Blue CS, McDonald RZ, et al. Paucity of V-D-D-J rearrangements and VH replacement events in lupus prone and nonautoimmune TdT−/− and TdT+/+ mice. J Immunol. 2006;177:1120–8. doi: 10.4049/jimmunol.177.2.1120. [DOI] [PubMed] [Google Scholar]
- 40.Koralov SB, Novobrantseva TI, Königsmann J, Ehlich A, Rajewsky K. Antibody repertoires generated by VH replacement and direct VH to JH joining. Immunity. 2006;25:43–53. doi: 10.1016/j.immuni.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 41.Akira S, Okazaki K, Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987;238:1134–8. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- 42.Akamatsu Y, Tsurushita N, Nagawa F, Matsuoka M, Okazaki K, Imai M, Sakano H. Essential residues in V(D)J recombination signals. J Immunol. 1994;153:4520–9. [PubMed] [Google Scholar]
- 43.Shlomchik M, Mascelli M, Shan H, Radic MZ, Pisetsky D, Marshak-Rothstein A, Weigert M. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171:265–92. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ichiyoshi Y, Casali P. Analysis of the structural correlates for antibody polyreactivity by multiple reassortments of chimeric human immunoglobulin heavy and light chain V segments. J Exp Med. 1994;180:885–95. doi: 10.1084/jem.180.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ditzel HJ, Itoh K, Burton DR. Determinants of polyreactivity in a large panel of recombinant human antibodies from HIV-1 infection. J Immunol. 1996;157:739–49. [PubMed] [Google Scholar]
- 46.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–7. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 47.van Dongen JJ, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 48.Tian C, Luskin GK, Dischert KM, Higginbotham JN, Shepherd BE, Crowe JE., Jr Evidence for preferential Ig gene usage and differential TdT and exonuclease activities in human naïve and memory B cells. Mol Immunol. 2007;44:2173–83. doi: 10.1016/j.molimm.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


