Abstract
Almost half of breast Ductal Carcinoma in situ are likely to remain non threatening in situ lesions with no invasion to the surrounding stroma and no metastases. The majority of focal disruptions in myoepithelial (ME) cell layers indicative of invasion onset were found to be overlying epithelial cell clusters with no or substantially reduced estrogen receptor α (ERα) expression. Here we report the down-regulation of Tyrosine kinase-2 (TYK2) and up-regulation of Strumpellin expression, among other proteins in ERα(−) cells located at disrupted ME layers compared to adjacent ERα(+) cells overlying an intact myoepithelial layer. ERα(+) and ERα(−) cells were microdissected from the same in vivo human breast cancer tissues, proteins were extracted and separated utilizing Differential in-Gel Electrophoresis (DIGE) followed by trypsin digestion, MALDI-TOF analysis, and protein identification. Proteins expressed by ERα(−) cell clusters were found to express higher levels of strumpellin that binds to Valosin-containing protein (VCP) to slow-down wound closure and promote growth; and lower levels of TYK2, a jak protein necessary for lineage specific differentiation. TYK2 levels were further analyzed by immunohistochemistry (IHC) in a cohort composed of 70 patients with broad clinical characteristics. TYK2 levels were minimal in TxN1M0 breast cancers which is the stage where the initial regional lymph node metastasis is observed. Our data highlight the role of TYK2 downregulation in breast cancer cell de-differentitation and initiation of regional metastasis. In addition, the aggressiveness of the ERα(−) cell clusters compared to ERα(+) ones present in the same duct of the same patient was confirmed.
Keywords: DCIS, Estrogen receptor α, Proteomics, Tissue microdissection, TYK2
Introduction
Human ductal carcinoma in situ (DCIS) is one of the earliest identifiable breast cancer pathologies. DCIS tumor cells remain confined to the duct and surrounded by a layer of myoepithelial (ME) cells and the basement membrane [1]. Only half of DCIS cases may eventually progress to invasive cancer. Many are likely to remain non-threatening lesions, complicating the determination of the best treatment regimen for DCIS patients. Normal breast ducts are surrounded by ME cell layer and basement membrane that form a physical barrier between the stroma and the luminal epithelium. The ME cells bound to each others by tight intercellular junctions and adhesion molecules, form a continuous layer that encircles the entire duct system. The basement membrane forms a continuous lining surrounding and attaching to ME cells via hemidesmosomes and focal adhesion complexes. Therefore, the epithelium is dependent upon the stroma for its survival needs. Communication between the epithelium and the stroma is regulated by the gate-keeping function of the ME layer and the basement membrane forcing ductal tumor cells to first break their way into the stroma through the ME cell layer, and the basement membrane followed by invasion and metastases [2].
Immunohistochemical analysis and genetic profiles in adjacent epithelial cells were carried out to analyze their correlation with the structural integrity of the myoepithelial (ME) cell layer. Focal disruptions were found to be bound by epithelial cell clusters with no or substantially reduced ERα expression, while adjacent cells within the same duct expressed high levels of ERα and were surrounded by an intact ME cell layer [3]. This implies that disruption of this ME layer is a pre-requisite for invasion. Immunohistochemical assessment of KI-67 showed that cell clusters overlying focally disrupted ME layers showed a substantially higher proliferation rate when compared to adjacent cells within the same duct [4; 5]. The roles played by estrogen receptors, in general, and ERα, in particular, during breast cancer progression and treatment have been previously reviewed [6]. In addition, we recently compared the protein profiles of whole ERα(+) breast cancer tissues to whole ERα(−) ones and identified 22 differentially-expressed proteins [7]. Unlike whole breast tissues, which are usually contaminated with ME cell layer and stromal cells, microdissected samples yield more homogeneous ERα(+) and ERα(−) cells. These microdissected samples can now give a better assessment of the differential protein profiles present in ERα(+) and ERα(−) breast cancer cells.
Using standard proteomic analysis, we identified differentially-expressed proteins in 3 pairs of microdissected ERα(+) and ERα(−) breast cancer cells using DIGE. Among the identified candidates, TYK2, a protein that belongs to the JAK family of non-receptor tyrosine kinases was found to be consistently down-regulated in ERα(−) compared to adjacent ERα(+) cancer cells. TYK2 was picked for further validation due to its paramount role in the transduction of cytokine-mediated signals, namely type I interferon (IFN) signaling using the JAK-STAT pathway [8; 9]. Validation of our proteomics data utilizing tissue microarrays (TMA) composed of a cohort of 70 patients (140 tissues) with different breast pathologies revealed that TYK2 is mostly down-regulated during metastasis to the regional lymph node. This finding supports the hypothesis that primary immunodeficiency through TYK2 down-regulation is required for initial nodal metastasis in breast cancer. The role that TYK2 down-regulation plays in ERα(−) cells neighboring disrupted myoepithelial layers was addressed and the effect of TYK2 downregulation in ERα(+) mammary epithelial cells (MCF-10A) was also analyzed.
Materials and Methods
Research involving human subjects was performed in accordance with the principles outlined in the declaration of Helsinki (1964). Freshly-frozen breast tissues samples and corresponding slide-mounted, serial-sectioned paraffin-embedded tissues were provided by the tissue bank at Georgetown University Medical Center, Washington, DC 20007. The tissue microarray (TMA) was purchased from Biomax.us (Cat # BR1503). Clinical characteristics are in Table 1. Specification sheet is in Table S1 (supplemental material).
Table 1.
Clinical characteristics of the breast cancer samples
| Characteristic | Status | N | % |
|---|---|---|---|
| Age | < 50 | 36 | 51.4 |
| > 50 | 34 | 48.6 | |
| Node Status | Negative | 54 | 77.1 |
| Positive | 16 | 22.9 | |
| Tumor size | < 2cm | 12 | 17.1 |
| > 2cm | 58 | 82.9 | |
| Stage | 1 | 6 | 8.6 |
| 2 | 44 | 62.9 | |
| 3 | 9 | 12.9 | |
| 4 | 0 | 0.0 | |
| ER Status | + | 17 | 24.3 |
| − | 44 | 62.9 | |
| PR Status | + | 14 | 20.0 |
| − | 46 | 65.7 | |
| Her2 Status | + | 41 | 58.6 |
| − | 11 | 15.7 | |
| triple | + | 6 | 8.6 |
| − | 9 | 12.9 | |
| TYK2 Levels | + | 32 | 45.7 |
| − | 38 | 54.3 |
Immunohistochemical Staining (IHC) and Tissue Microdissection
IHC was performed on the TMA and on the Breast DCIS sections as described previously [7]. Rabbit anti-smooth muscle actin (Epitomics Inc., Burlingame, CA) and Mouse anti-ERα (NCL-ER-6F11, Vector Labs, Burlingame, CA) were used at 1:1000 dilution. Rabbit anti-TYK2 (Abcam, Cambridge, MA) was used at a 1:1500 dilution. Unstained adjacent serial sections were used to microdissect clusters overlying myoepithelial disruptions, composed primarily of ERα (–) cells by laser-capture microdissection (LCM). Clusters of ERα (+) cells from the same duct, but bound by an intact ME layer, were similarly obtained. Sections were 5 to 20 µm thick. One out of 3 sections was immunostained for ERα to differentiate between ERα(+)/ERα(−) clusters , and epithelial/stromal components and to allow for a precise localization by extrapolation of the ERα(+) and ERα(−) status on the unstained tissues that will be microdissected. This allowed for the isolation of ~300K cells resulting in the extraction of ~210 µg of proteins from all three patients, suitable for differential proteome profiling procedures.
Protein Extraction and Quantitation
Microdissected tissues were homogenized in a tissue lysis buffer composed of 10 mM Tris, 6M UREA, 4% CHAPS (pH 7.5). The ratio of the volume of tissue lysis buffer added to the weight of the tissue was 1/1 (v/v) and quantitated as described previously [10].
Protein-Dye Labeling, DIGE, and Image Analysis
These experiments were optimized and performed as described previously [11] with the following alterations: 15µg of each of the 3 pairs of biological replicates of ERα(+) and ERα(−) cell extracts were labeled with 200 pmoles of either Cy3 or Cy5. Cy3 and Cy5 were swapped among ERα(+) and ERα(−) samples. An internal standard sample composed of equal amounts of ERα(+) and ERα(−) samples was labeled with Cy2 dye using the same procedure. “Labeled proteins were pooled into 3 different fractions. Each fraction was diluted with rehydration buffer (50 mM Tris-HCl pH 8.8, 6 M Urea, 4% CHAPS (w/v), 1% DTT (w/v), 1% Bio-lyte 3/10 Ampholyte) to a final volume of 450 µL and run on 3 separate gels (3 analytical gels). The remaining protein samples (120 ug) were combined and loaded on the “pick gel”. The pick gel is the gel that will be used for spot picking and protein identification after matching the spots which exhibited differential intensities on the analytical gels with those on the “pick gel”.
Sample Preparation for MALDI-TOF/TOF analysis and Protein Identification
Protein digestion and peptide recovery procedures leading to the deposition of the peptide-matrix mixture onto the MALDI target plate were performed as described previously [11]. MS analysis was performed using a 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems, Foster city, CA). The instrument was calibrated using Applied Biosystems calibration standards. MALDI-TOF spectra were acquired by accumulating 1000 laser shots in reflector mode for positive ion detection between 800 and 4000 m/z as described in [11]. MS Mass lists were picked by the GPS Explorer v3.5 software and submitted to the MASCOT v.2.0.00 search engine utilizing the same settings as in [12].
Western Blotting
Western Blotting was performed as described previously [13].
IHC Grading and Statistical Analysis
IHC evaluation was performed by a pathologist according to the semi-quantitative IHC scoring system that assesses intensity and distribution of TYK2 immunostaining. TYK2 staining covering less than 10% of tumor cells was given a score of 0; if it covered 10–30% of tumor cells the assigned score was 1; if 30–60% of tumor cells are stained, the score was 2; a score of 3 was given if more than 60% of the tumor cells were TYK2 positive. Intensity was assigned a score of 0 for negative; 1 for weak; 2 for moderate; and 3 for intense. The final score is the product of the distribution score (0, 1, 2, 3) by the intensity score (0, 1, 2, 3) resulting in score values between 0 and 9. TYK2 intensity mean and standard deviation were calculated for each breast condition. For the comparison of 3 or more means, a one-way analysis of variance (ANOVA) was performed on the means of TYK2 IHC scores in different pathologies. For the comparison of 2 means, a two-tailed unpaired t-test was performed. P values were generated for both tests. Values of P < 0.05 corresponding to a confidence interval > 95% indicated that differential TYK2 levels between analyzed breast pathologies are statistically-significant.
MCF-10A Cell Culture, TYK2 Knock-down, and Protein Extraction
MCF-10A cells (ATCC: CRL10317) were cultured, passaged, and maintained according to ATCC recommendation. TYK2 knock-down was performed using 200 picomoles of ON-TARGETplus SMARTpool TYK2 siRNA (Dharmacon, cat. # L003182) or scrambled siRNA per million cells. TYK2 knock-down and protein extraction were performed as described previously [14].
Wounding Assay
Migration of TYK2 knocked-down MCF-10A ERα (+) breast epithelial cells versus control were assessed by measuring the movement of cells into a scrape, "wound assay". The assay was terminated when the wound in MCF10A cells treated with TYK2 siRNA was completely closed, which occurred at 20 h. The assay was performed three times and standard deviations were computed.
Boyden Chamber Assay
Matrigel invasion assay (chemoinvasion) of TYK2 knocked-down MCF-10A cells was assessed using BD Fluoroblok inserts (8µm pore size, 24 well format, BD Biosciences) and EGF as chemoattractant. Using sterile forceps, cell wall inserts were added to each well (24 well plate). 30µl of 1:5 diluted Matrigel was added to the center of each well. Coated inserts were incubated for 30 min to allow the Matrigel to solidify. 2×105 cells per sample were plated in 200 µl DMEM/F12 onto the upper chamber. Immediately 400µl / well of complete medium were added in addition to EGF chemo-attractant to the lower chamber. After 24 hours, cells that passed through the pores of the membranes and attached themselves to the bottom side were stained with Calcein AM (Invitrogen) and fixed using 10% formalin/0.1 % triton. Pictures were taken using a confocal microscope (Zeiss LSM 510 - 10x). The assay was perfomed 3 times and t-test P values were generated. Invading cells were counted using the “Counting cell” application module of Metamorph software.
Collagen Gel Assay
A pre-set layer of bovine collagen gel (3.0 mg/ml) was prepared in the culture dish. Cells were seeded onto mats of Collagen I. A 1:1 ratio of collagen gel and TYK2 knocked-down MCF-10A or control cells were added on top of the pre-set layer at time t = 0 min and incubated at 37°C. Complete media was added at time t = 30 min. Pictures were taken at t = 0; 5; and 24 hrs using an inverted microscope (Olympus IX 71 - 20X) to examine the morphology of the cells.
Results
IHC and Microdissection of ERα(+) and ERα(−) cell clusters
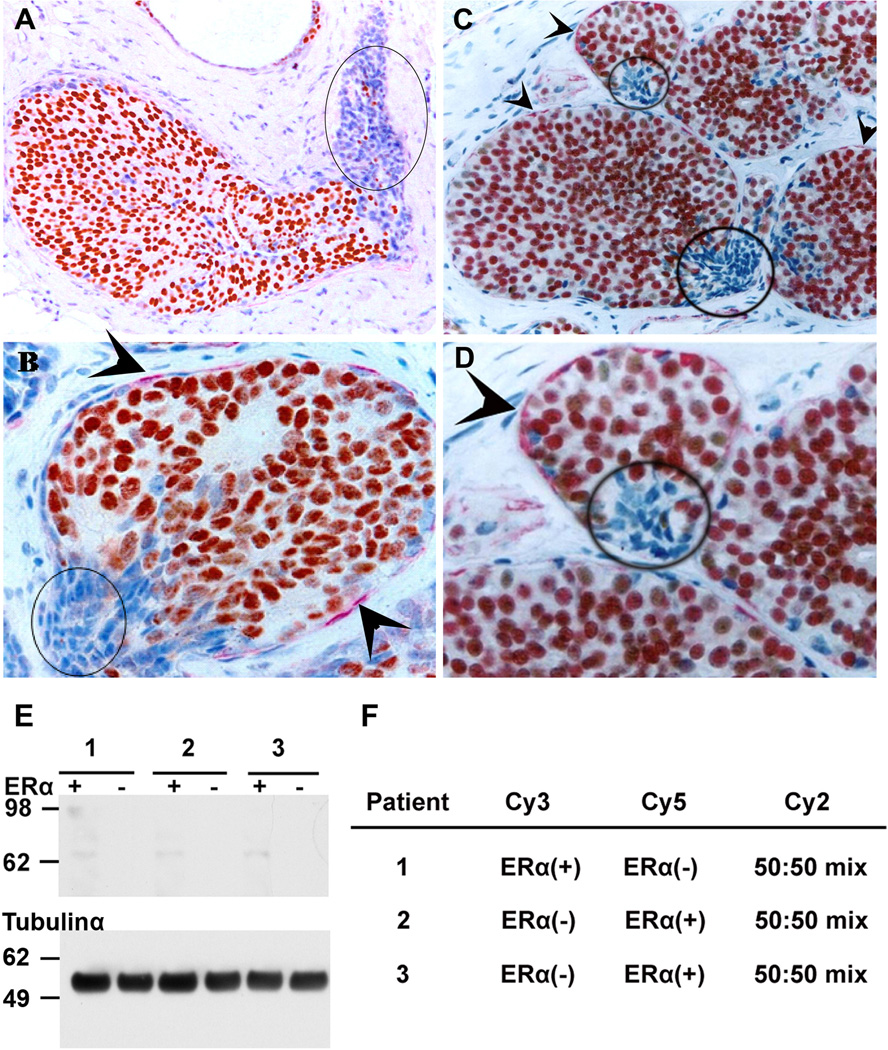
Less than 2% of the analyzed breast samples showed the described clusters of cells with ERα(−) cells bound by a focally-disrupted ME layer as described in [4]. Breast tissues from three patients matching the above conditions were isolated after immunostaining for ERα. IHC revealed the presence of ERα(−) cells overlying a focally-disrupted ME layer and ERα(+) cells overlying an intact ME layer (Fig. 1A–D). Microdissection of the epithelial ERα(+) and ERα(−) breast cancer tissues yielded homogeneous samples minimizing the presence of contaminating myoepithelial and stromal cells. ERα(+) and ERα(−) cells were microdissected from up to 200 ducts/patient. After cutting thick tissue sections, up to 1000 cells were obtained from each duct, the majority of which are ERα(+) cells. A total of three pairs of samples were generated. Although not combined initially, the final yield of total proteins extracted from these cells amounted to ~210 µg. Western blotting against ERα was performed as a quality control to assess the presence of ERα in the samples after loading 1 µg of total proteins from each sample. Immunoblotting for tubulin-α was performed as control. ERα was only detected in ERα(+) and not in ERα(−) microdissected cells (Fig. 1E).
Fig. 1.
Ducts are immunostained for ERα (brown), SMA (red), and counterstained with hematoxylin (blue) (A, B, C) IHC tissues from 3 patients showing ERα(−) cell clusters overlying focally-disrupted ME layer (circled); (D) higher magnification of C showing the ME layer (arrowhead) and the ME disruption adjacent to ERα(−) cells; (E) ERα Western blotting on microdissected tissues; (F) Dye labeling of the 3 pairs of protein samples which were extracted from the microdissected tissues.
TYK2 Down-regulation in ERα(−) Microdisected Cells
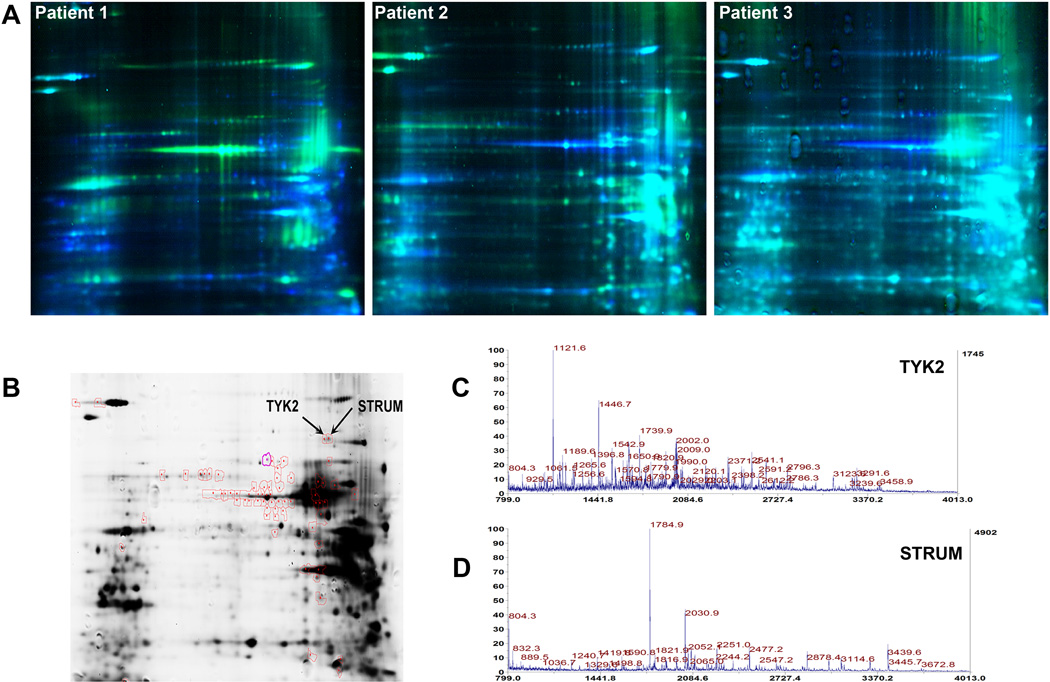
Fifteen µg of proteins from each of the 6 samples were labeled with Cy3 or Cy5 fluorophores. Dyes were swapped across the samples as listed in (Fig. 1F). Each pair of samples was separated on the same gel. Protein spots were detected for each sample using distinct excitation wavelengths for each dye and detecting the corresponding emission. The remaining protein samples (~120 µg) were combined and loaded on the pick gel. Differential spot intensities on the analytical gels (Fig. 2A) were analyzed and quantitated by the DeCyder2D and Biological Variation Analysis (BVA) tool respectively (GE Healthcare, Buckinghamshire, UK). t-test was applied on 2 populations: population 1 consists of n = 3 microdissected ERα(+) and population 2 consists of n = 3 microdissected ERα(−) breast cancer cells. Spots that were detected on the analytical gels across the 3 pairs of samples with a confidence interval of 95% were picked for excision from the Pick gel, digestion and MS-based identification.
Fig. 2.
(A) DIGE images of the 3 biological replicates; (B) Sypro Ruby Pick gel showing red circled spots that picked for mass spectrometry analysis. (C) Protein mass fingerprints (PMF) of TYK2 and (D) STRUM.
A total of 111 spots were differentially-expressed across the 3 paired samples (t < 0.05) and 63 of these spots had a match on the pick gel The pick gel was loaded with 120 µg of the remaining mixture of proteins extracted from both ERα(+) and ERα(−) cells and stained with Sypro Ruby (Fig. 2B). Excised gel fragments were digested with trypsin and analyzed utilizing a 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Foster city, CA). MS and MS/MS peak lists were picked by GPS Explorer v3.5 (Applied Biosystems, Foster city, CA) and submitted to Paragon v3.0 algorithm (Applied Biosystems, Foster city, CA) for scanning against a combined Uniprot + SwissProt database composed of more than 255,000 human protein entries. Proteins were identified based on their molecular mass, isoelectric point, and trypsin-generated peptide fingerprints. Out of the 63 spots, 10 were identified (Table 2). Differentially-expressed proteins that were identified included highly abundant proteins (e.g. tubulins, HSPs, NUMA) in addition to other proteins that are repeatedly identified as being differentially-expressed (e.g. peroxiredoxins, enolases, piruvate kinases). These proteins were not considered in the study. The remaining list of differentially-expressed proteins was composed of Tyrosine kinase 2 (TYK2) with 16 matching peptide fingerprints (Fig. 2C; Supplemental Table S2) and Strumpellin (STRUM) with 15 matching peptide fingerprints (Fig. 2D; Supplemental Table S3). Isoelectric points and molecular weights of identified proteins were confirmed on the analytical and pick gels. Strumpellin levels were 3.05 times higher in ERα(−) when compared to the ERα(+) expression across the 3 paired samples with a t = 0.041. Non-receptor tyrosine kinase 2 levels were 6.02 times lower in ERα(−) cells when compared with ERα(+) across the 3 paired samples with a t = 0.0054.
Table 2.
Differentially-expressed proteins between ERα(+) and ERα(−) cells
| Protein Name | Peptide Count |
Protein Score C.I. % | T-test | Av. Ratio ER− /ER+ |
|---|---|---|---|---|
|
P08107|HSP71_HUMAN Heat shock 70 kDa protein 1 OS=Homo sapiens GN=HSPA1A PE=1 SV=5 |
19 | 100 | 0.0035 | −2.91 |
|
Q86VS8|HOOK3_HUMAN Protein Hook homolog 3 OS=Homo sapiens GN=HOOK3 PE=1 SV=2 |
13 | 93.69239578 | 0.0051 | 4.53 |
|
P29597|TYK2_HUMAN Non-receptor tyrosine- protein kinase TYK2 OS=Homo sapiens GN=TYK2 PE=1 SV=3 |
15 | 94.37834182 | 0.0054 | −6.03 |
|
P14618|KPYM_HUMAN Pyruvate kinase isozymes M1/M2 OS=Homo sapiens GN=PKM2 PE=1 SV=4 |
13 | 99.87981121 | 0.019 | −5.56 |
|
Q14980|NUMA1_HUMAN Nuclear mitotic apparatus protein 1 OS=Homo sapiens GN=NUMA1 PE=1 SV=2 |
22 | 96.3703539 | 0.02 | −2.85 |
|
Q4AC94|C2CD3_HUMAN C2 domain-containing protein 3 OS=Homo sapiens GN=C2CD3 PE=1 SV=3 |
23 | 99.73705548 | 0.035 | 2.28 |
|
P18669|PGAM1_HUMAN Phosphoglycerate mutase 1 OS=Homo sapiens GN=PGAM1 PE=1 SV=2 |
10 | 99.9865146 | 0.041 | −3.81 |
|
Q12768|STRUM_HUMAN Strumpellin OS=Homo sapiens GN=KIAA0196 PE=1 SV=1 |
15 | 96.3703539 | 0.041 | 3.05 |
|
P29992|GNA11_HUMAN Guanine nucleotide- binding protein subunit alpha-11 OS=Homo sapiens GN=GNA11 PE=1 SV=2 |
10 | 96.11076481 | 0.043 | −4.42 |
|
P07437|TBB5_HUMAN Tubulin beta chain OS=Homo sapiens GN=TUBB PE=1 SV=2 |
12 | 99.93692396 | 0.044 | −4.61 |
TYK2 Expression Levels Relative to Hormone Receptors and Node Status
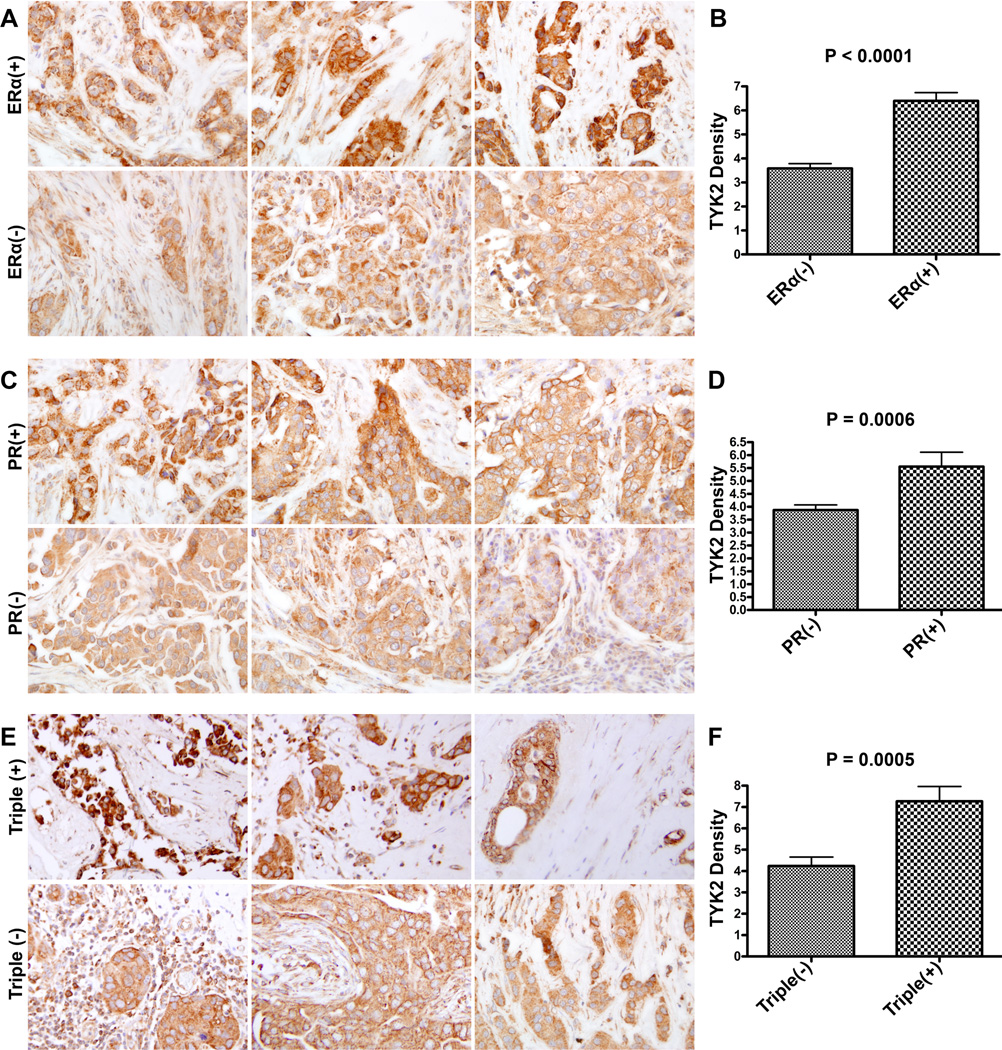
While using a large cohort of ERα(+) and ERα(−) microdissected tissues to yield a statistically-significant validation of our proteomics data is the mainstay for confirming protein identities, such experiment is limited by the low number of patients with a focal ME disruption adjacent to an ERα(−) cell cluster. We instead opted to analyze TYK2 differential-expression levels in breast cancer tissues as it relates to hormone receptors and initial invasion since these are the characteristics of the microdissected cells where TYK2 levels were originally found to be downregulated. A TMA containing 140 cores from 70 patients was stained against TYK2. The clinical characteristics of the samples are in Table S1. IHC staining revealed a significant decrease in TYK2 expression levels in ERα(−) breast cancer tissues compared to ERα(+) ones regardless of other factors (t-test; P < 0.0001) (Fig. 3A, B). Further analysis revealed a significant decrease in TYK2 levels in PR(−) compared to PR(+) breast tissues (t-test; P = 0.0006) (Fig. 3C, D) also regardless of the status of other receptors. The same observation was also noticed in triple(−) tissues versus triple(+) ones (t-test; P =0.0005) (Fig. 3E, F). Her2(−) tissues displayed lower TYK2 levels when compared to Her2(+) but the difference was not statistically significant (data not shown). Cells in all the receptor negative breast cancers showed less differentiation than their positive counterparts.
Fig. 3.
Representative set of breast tissues immunostained against TYK2 and association between TYK2 expression levels and (A, B) ERα(+) and ERα(−); (C, D) PR(+) and PR(−); (E, F) Triple (+) and Triple(−) breast cancers. P values were generated following the application of an unpaired t-test.
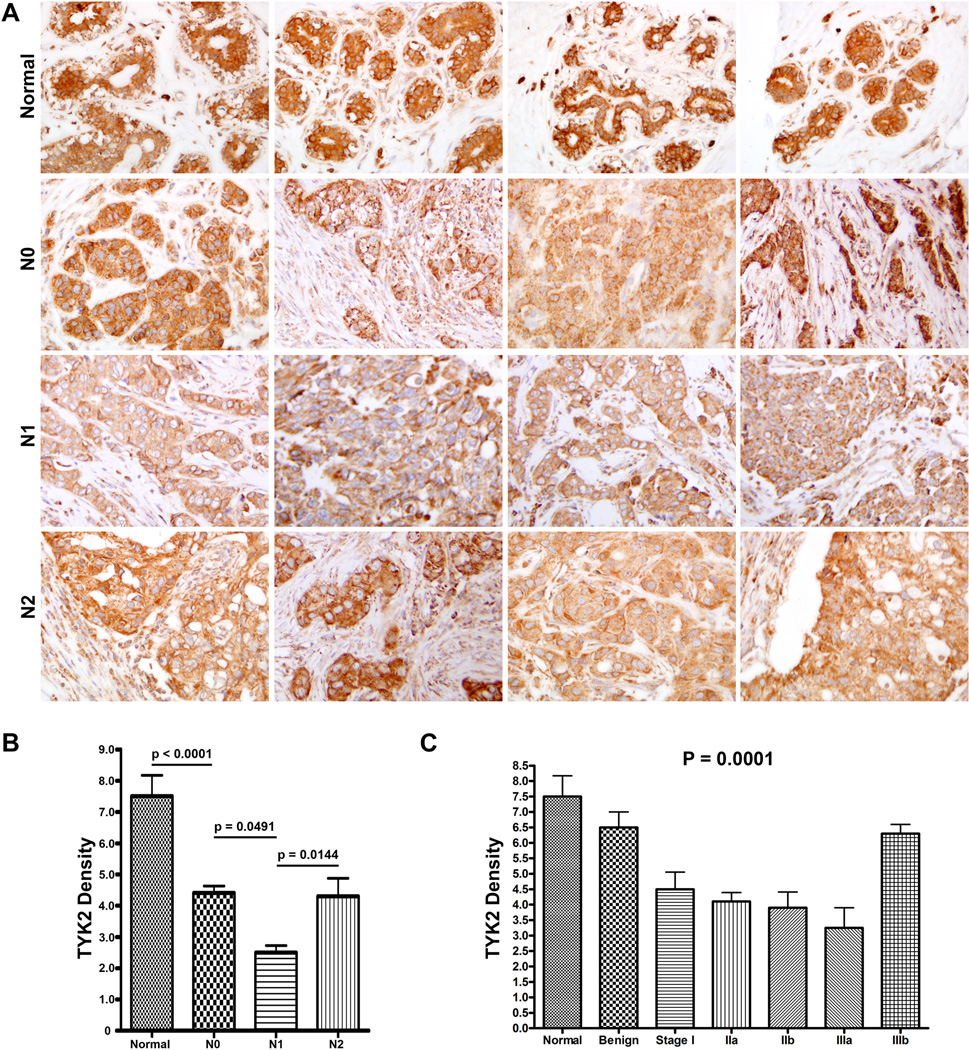
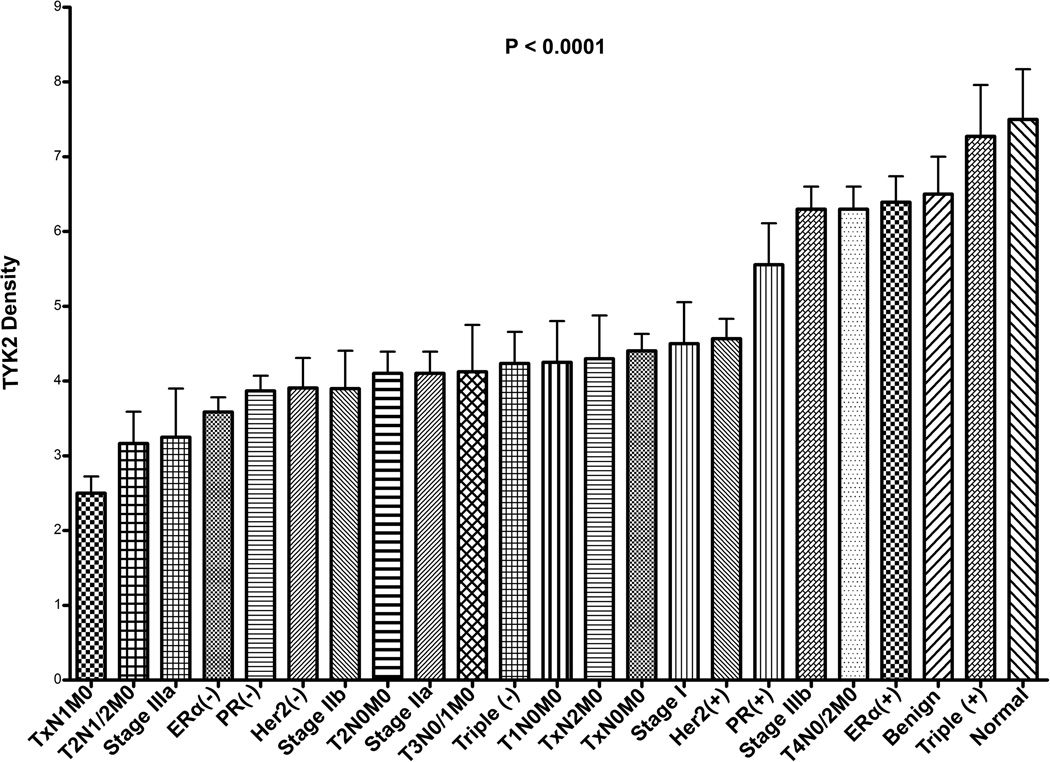
A representative set of 4 tissues of each of the following groups is shown in (Fig. 4A): normal, N0, N1, and N2. A significant association between TYK2 levels and node status was observed between the populations: Normal and N0 (t-test P < 0.0001); N0 and N1 (t-test P = 0.0491); N1 and N2 (t-test P = 0.0144) (one-way ANOVA; P = 0.0018). TYK2 levels decreased between normal and N0; N0 and N1; and then increased between N1 and N2 (Fig. 4B). The same trend was observed when TYK2 levels were analyzed relative to breast cancer stage and was found to be statistically significant. TYK2 levels decreased between normal and benign; benign and Stage I; Stage I and Stage IIa; Stage IIa and Stage IIb reaching a minimum expression level for Stage IIIa and then increasing between Stage IIIa and IIIb (one-way ANOVA; P = 0.0001) (Fig. 4C). A general diagram in figure 5 revealed that TYK2 levels were lowest in N1, T2N1/2M0, Stage IIIa, ERα(−) breast cancers and highest in normal, benign, and triple(+) breast cancers (one-way ANOVA; P < 0.0001).
Fig. 4.
(A) Representative set of breast tissues immunostained against TYK2 and association between TYK2 expression levels and (B) node status regardless of tumor size, P values were generated following unpaired t-test; and (C) breast cancer stage P values were generated following one-way ANOVA.
Fig. 5.
Summary of TYK2 expression levels in normal, benign, ERα, PR, Her2, triple-receptor status, TNM groups, breast cancer stage, and node status regardless of tumor size. P value (< 0.001) was generated for all sub-groups following one-way ANOVA. Unpaired t-tests between specific sub-groups were generated and described in the previous figures.
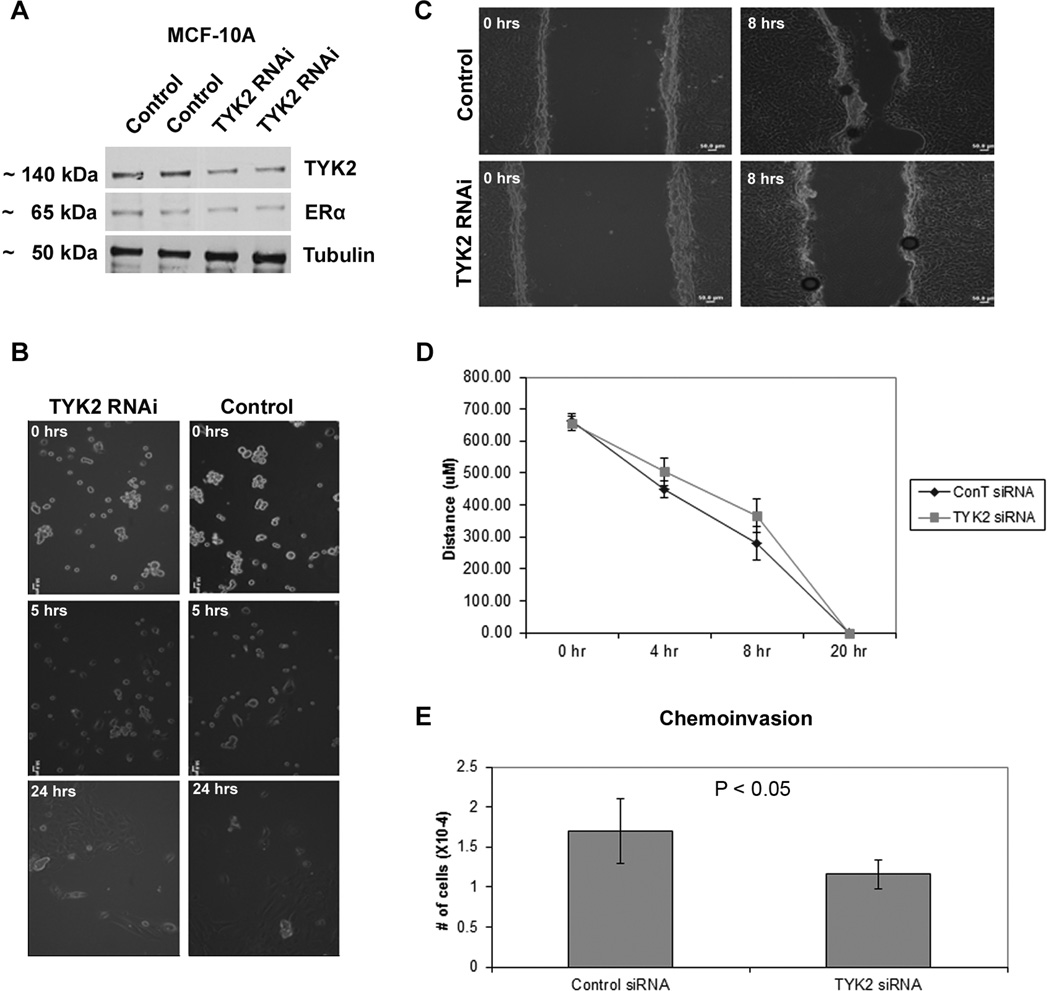
Knockdown of TYK2 Induces Decreased Migration and Cell Invasion in Erα(+) MCF-10A Human Breast Epithelial Cells
TYK2 knock-down in MCF-10A cells neither affected the levels of ERα expression (Fig. 6A) nor the cell morphology (Fig.6B). Cell morphology pictures were taken on collagen I gels, because it’s a setting that mimics the stromal microenvironment of the epithelial cells. Some of the initially seeded cells died and most of the cells showed an aggregated morphology in both TYK2 knock-down MCF-10A and control cells. Changes in migration and invasion due to TYK2 knock-down were analyzed using a scratch/wound assay and Boyden chamber assay, respectively. The rate of gap closure of the monolayer decreased by 11.93% at 4 hrs and by 31.08% at 8 hrs in TYK2 knock-down MCF-10A when compared to control cells (Fig. 6C, D). In the Boyden chamber Matrigel invasion (chemoinvasion) assay, knockdown of TYK2 caused a 31.76% decrease in chemoinvasion of mammary epithelial cells (p < 0.05) (Fig. 6E). Therefore, TYK2 loss results in decreased cell migration and chemoinvasion in ERα(+) normal human mammary epithelial cells in vitro.
Fig. 6.
Effect of TYK2 knockdown on migration, invasion, and morphology of ERα(+) breast epithelial cells (MCF10A). (A) Western blotting against TYK2, ERα, and tubulin (loading control); (B) Collagen gel assay reveals no significant morphology change between control epithelial cells and cells lacking TYK2. (C,D) Wound assay of mammary epithelial cells lacking TYK2 and control cells. The former cells exhibit a significant decrease in migration as compared to normal counterparts. (E) Chemoinvasion assay of TYK2 knocked-down shows around 50% decrease in invasion relative to the control assay.
Discussion
The heterogeneous nature of the breast physiology is hindering the research progress that should lead to breast cancer prevention and treatment. Breast tissues are composed of epithelial, myoepithelial, and stromal cells. Microdissection of the epithelial ERα(+) and ERα(−) breast cancer tissues from human in vivo breast cancer tissues was coupled with DIGE analysis which allows for the separation and the differential protein analysis of scarce samples due to the highly sensitive detection afforded by fluorophores. TYK2 and STRUM, among other proteins, were identified as down and up-regulated respectively in ERα(−) microdissected cell clusters that are adjacent to a focally-disrupted ME layer. Strumpellin per se was recently identified as a VCP-interacting protein and as a ubiquitously-expressed protein located in the cytosolic and endoplasmic reticulum fractions [15]. Overexpression of strumpellin has been associated with significantly reduced wound closure in wound healing assays [15]. Therefore, its upregulation in the ERα(−) bound by a disrupted myoepithelial layer may play a dual role in promoting tumor growth and invasion. A major discrepancy in the result is the absence of ERα from the list of differentially- expressed proteins. ERα was supposed to be one of the most downregulated proteins in ERα(−) samples since this sample is composed solely of ERα(−) extracts as shown in the Western blotting results (Fig. 1E). The isoelectric point (pI) of ERα is 8.2 making it prone to precipitation since it is close to the pH of the separation buffer (pH7.5). Furthermore, nuclear proteins have been described as moderately soluble and all successful attempts at increasing their solubility required high concentrations of thiourea and urea in the lysis buffer in addition to the use of high loads of proteins [16], which cannot be afforded in the current study. Furthermore, 2DE technology allows for the visualization of only 2–10% of the sample proteome. Granted some proteins have a higher chance of being detected on 2DE due to their high abundance and solubility in aqueous solvents, most proteins have a 2–10% detection chance.
TYK2 was picked for further analysis in breast cancers due to its important role in the transduction of cytokine-mediated signals, namely type I interferon (IFN) signaling using the JAK-STAT pathway [8; 9]. TYK2 is one of four mammalian jaks that belongs to the family of Janus Kinases (Jak) associated with cytokine receptors [17]. TYK2 was initially thought to be essential for type I IFN signaling in human fibroblast cells [18]. Lately, a study by Minegishi et al [19] showed that TYK2 deficiency in patients causes severe defects in IL-6 and IL-10 signaling resulting in hyper IgE syndrome (HIES) which is a primary immunodeficiency characterized by skin abscesses, pneumonia, and high levels of IgE [20; 21]. Although high levels of IgE are most commonly associated with allergic and parasitic diseases, a study showed that resistant breast tumor recurrence rate was significantly higher in patients with elevated IgE levels [22]. Therefore, TYK2 downregulation in ERα(−) cells resulting in HIES is a signal of bad prognosis, and therapy-resistant breast cancer. This is also in agreement with previous studies that showed that ERα(−) cell clusters in DCIS with microinvasion may represent a clone from a genetically-altered cancer stem cell, explaining why many of the differentiation markers are not expressed or are expressed at a much lower level than those of adjacent ERα(+) cells. These results suggest that ERα(−) cell clusters may arise from dedifferentiated proliferating ERα(+) breast cancer cells rendering them pathologically more aggressive and resistant to treatment [23]. Furthermore, TYK2 down-regulation in vitro results in the de-activation of downstream target STAT3 and therefore a reduced response to growth arrest conditions [24].
The above-described pathology is only present in 2% of breast cancer samples making the recruitment of a large cohort to validate the proteomic finding on breast cancers practically impossible. To compensate, we used tissue microaarays (TMA) containing 140 tissues from 70 patients stained against TYK2. These tissues are characterized by different breast cancer stages labeled according to TNM (Tumor- Nodes- Metastasis) system. In concordance with the proteomics data, TYK2 was found to be significantly down-regulated in ERα(−) breast cancers regardless of the node status, and to be minimally expressed in TxN1M0 cancers regardless of the tumor size. At cancer stages that preceded TxN1M0 (TxN0M0) and followed it (TxN2M0), breast tissues expressed significantly higher TYK2 levels. TxN1M0 is the stage of the initial metastasis of cancer cells to regional lymph nodes. The JAK-STAT pathway is known to be involved in the process of differentiation and proliferation [25]. Moreover, TYK2 was particularly found to be necessary for lineage-specific differentiation [26], and its levels were found to be elevated in invasive and metastatic cancers [27; 28]. Therefore, we believe that the drop in TYK2 levels is required for the de-differentiated ERα(−) beast cancer cells to initiate metastasis to regional lymph nodes. We performed morphology, migration, and cell invasion assays in MCF-10A ERα(+) breast cells following TYK2 knockdown. No changes in the morphology or ERα expression levels were observed as a consequence of TYK2 down-regulation. Contrary to the role it plays in the ERα(−) de-differentiated cells, TYK2 knock-down in MCF-10A ERα(+) mammary epithelial cells caused a statistically significant decrease in migration and chemo-invasion pointing that the role of TYK2 is not independent and that other members of the jak/STAT pathway are involved in the initiation of invasion.
The JAK-STAT pathway was considered a favorable prognostic marker for lymph node negative breast cancers as it is attributed to a more differentiated phenotype within the primary lesion [29]. Furthermore, the JAK-STAT pathway was found to promote homotypic adhesion, thus suppressing invasiveness [30]. Our data are in full agreement with in vitro studies that compared TYK2 levels in different breast cancer cell lines and found them to be higher in ERα(+) cell lines (T47D, MCF7, ZR-75-1) ) when compared to ERα(−) lines (MDA-MB-231, BT549, HS578T, MDA-MB-435S), and immortalized normal lines (MCF10A, MCF12A, and Hs578Bst) [31]. Interestingly, among all breast cancer cell lines analyzed in the latter study, TYK2 levels were the lowest in BT549, a cell line derived from an invasive ductal tumor that had metastasized to 3 of 7 regional lymph nodes [31].
In summary, proteins expressed by ERα(−) cell clusters overlying a focally disrupted myoepithelial layer were compared to those expressed by ERα(+) cell clusters overlying an intact myoepithelial layer. ERα(−) cell clusters showed higher levels of strumpellin that binds to VCP to slow-down wound closure and promote growth; and lower levels of TYK2 a jak protein whose levels are the lowest during the initiation of regional lymph node metastasis. Collectively, these results can explain, in part, the aggressiveness of the ERα(−) cells compared to ERα(+) counterparts present in the same duct of the same patient, and the importance of TYK2 downregulation at the initiation of nodal metastasis in de-differentiated ERα(−) breast cancer cells.
Supplementary Material
Acknowledgment
This work was supported in part by a grant from Charles and Mary Latham Fund to ZJ Sahab, grant BCTR0504465 from the Susan G. Komen Breast Cancer Foundation, grants from the Florida Breast Cancer Coalition Research Foundation, the Elsa U. Pardee Foundation, and the Florida State University to Q.-X. Sang, and a grant BCTR0706983 from the Susan G. Komen Breast Cancer Foundation, grant 2006CB910505 from the Ministry of Chinese Science and Technology to Y.-G. Man, and grants R01CA129813 and P01 CA130821 to SW Byers. The authors wish to acknowledge the support of the following Lombardi Comprehensive Cancer Center Core Facilities (NIH P30 CA51008): Tissue culture, Histopathology, Microscopy, and Proteomics shared resources.
Footnotes
Conflict of Interest
All authors declare no conflict of interest.
REFERENCES
- 1.Skinner KA, Silverstein MJ. The management of ductal carcinoma in situ of the breast. Endocrine-Related Cancer. 2001;8(1):33–45. doi: 10.1677/erc.0.0080033. [DOI] [PubMed] [Google Scholar]
- 2.Man YG, Sang QXA. The significance of focal myoepithelial cell layer disruptions in human breast tumor invasion: a paradigm shift from the "protease-centered" hypothesis. Experimental Cell Research. 2004;301(2):103–118. doi: 10.1016/j.yexcr.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 3.Man YG, Tai L, Barner R, et al. Cell clusters overlying focally disrupted mammary myoepithelial cell layers and adjacent cells within the same duct display different immunohistochemical and genetic features: implications for tumor progression and invasion. Breast Cancer Research. 2003;5(6):R231–R241. doi: 10.1186/bcr653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Man YG, Shekitka KM, Bratthauer GL, et al. Immunohistochemical and genetic alterations in mammary epithelial cells immediately overlying focally disrupted myoepithelial cell layers. Breast Cancer Research and Treatment. 2002;76:S143–S143. [Google Scholar]
- 5.Yousefi M, Mattu R, Gao CL, et al. Mammary ducts with and without focal myoepithelial cell layer disruptions show a different frequency of white blood cell infiltration and growth pattern: Implications for tumor progression and invasion. Applied Immunohistochemistry & Molecular Morphology. 2005;13(1):30–37. doi: 10.1097/00129039-200503000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Sahab ZJ, Man YG, Byers SW, et al. Putative Biomarkers and Targets of Estrogen Receptor Negative Human Breast Cancer. International Journal of Molecular Sciences. 2011;12(7):4504–4521. doi: 10.3390/ijms12074504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahab ZJ, Man YG, Semaan SM, et al. Alteration in protein expression in estrogen receptor alpha-negative human breast cancer tissues indicates a malignant and metastatic phenotype. Clinical & Experimental Metastasis. 2010;27(7):493–503. doi: 10.1007/s10585-010-9338-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoiber D, Kovacic B, Schuster C, et al. TYK2 is a key regulator of the surveillance of B lymphoid tumors. Journal of Clinical Investigation. 2004;114(11):1650–1658. doi: 10.1172/JCI22315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilks AF. 2 Putative Protein-Tyrosine Kinases Identified by Application of the Polymerase Chain-Reaction. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(5):1603–1607. doi: 10.1073/pnas.86.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahab ZJ, Suh Y, Sang QXA. Isoelectric point-based prefractionation of proteins from crude biological samples prior to two-dimensional gel electrophoresis. Journal of Proteome Research. 2005;4(6):2266–2272. doi: 10.1021/pr0501822. [DOI] [PubMed] [Google Scholar]
- 11.Sahab ZJ, Hall MD, Zhang L, et al. Tumor Suppressor RARRES1 Regulates DLG2, PP2A, VCP, EB1, and Ankrd26. J. of Cancer. 2010;1(1):14–22. doi: 10.7150/jca.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahab ZJ, Iczkowski KA, Sang QXA. Anion exchange fractionation of serum proteins versus albumin elimination. Analytical Biochemistry. 2007;368(1):24–32. doi: 10.1016/j.ab.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Khamis ZI, Sahab ZJ, Byers SW, et al. Novel Stromal Biomarkers in Human Breast Cancer Tissues Provide Evidence for the More Malignant Phenotype of Estrogen Receptor-Negative Tumors. Journal of Biomedicine and Biotechnology. 2011;2011 doi: 10.1155/2011/723650. Article ID 723650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahab ZJ, Hall MD, Sung YM, et al. Tumor suppressor RARRES1 interacts with cytoplasmic carboxypeptidase AGBL2 to regulate the a-tubulin tyrosination cycle. Cancer Research. 2011;71(4):1219–1228. doi: 10.1158/0008-5472.CAN-10-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemen CS, Tangavelou K, Strucksberg KH, et al. Strumpellin is a novel valosincontaining protein binding partner linking hereditary spastic paraplegia to protein aggregation diseases. Brain. 2010;133:2920–2941. doi: 10.1093/brain/awq222. [DOI] [PubMed] [Google Scholar]
- 16.Rabilloud T, Adessi C, Giraudel A, et al. Improvement of the solubilization of proteins in two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1997;18(3–4):307–316. doi: 10.1002/elps.1150180303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ihle JN. Cytokine Receptor Signaling. Nature. 1995;377(6550):591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 18.Firmbachkraft I, Byers M, Shows T, et al. Tyk2, Prototype of a Novel Class of Nonreceptor Tyrosine Kinase Genes. Oncogene. 1990;5(9):1329–1336. [PubMed] [Google Scholar]
- 19.Minegishi Y, Saito M, Morio T, et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25(5):745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Grimbacher B, Holland SM, Gallin JI, et al. Hyper-IgE syndrome with recurrent infections - An autosomal dominant multisystem disorder. New England Journal of Medicine. 1999;340(9):692–702. doi: 10.1056/NEJM199903043400904. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe MAE, Oda JMM, Amarante MK, et al. Regulatory T cells and breast cancer: implications for immunopathogenesis. Cancer and Metastasis Reviews. 2010;29(4):569–579. doi: 10.1007/s10555-010-9247-y. [DOI] [PubMed] [Google Scholar]
- 22.Ownby D, Ownby H, Roi L, et al. The Prognostic Value of Serum Ige Levels in Primary Breast-Cancer. Proceedings of the American Association for Cancer Research. 1982;23(MAR):150–150. doi: 10.1007/BF01806934. [DOI] [PubMed] [Google Scholar]
- 23.Zhang XC, Hashemi SS, Yousefi M, et al. Atypical E-cadherin expression in cell clusters overlying focally disrupted mammary myoepithelial cell layers: Implications for tumor cell motility and invasion. Pathology Research and Practice. 2009;205(6):375–385. doi: 10.1016/j.prp.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Sivko GS, Sanford DC, Dearth LD, et al. CCAAT/enhancer binding protein delta (C/EBP delta) regulation and expression in human mammary epithelial cells: II. Analysis of activating signal transduction pathways, transcriptional, post-transcriptional, and posttranslational control. Journal of Cellular Biochemistry. 2004;93(4):844–856. doi: 10.1002/jcb.20224. [DOI] [PubMed] [Google Scholar]
- 25.Tao L, Liu JY, Li Z, et al. Role of the JAK-STAT pathway in proliferation and differentiation of human hypertrophic scar fibroblasts induced by connective tissue growth factor. Molecular Medicine Reports. 2010;3(6):941–945. doi: 10.3892/mmr.2010.349. [DOI] [PubMed] [Google Scholar]
- 26.Chung BM, Kang HC, Han SY, et al. Jak2 and Tyk2 are necessary for lineagespecific differentiation, but not for the maintenance of self-renewal of mouse embryonic stem cells. Biochemical and Biophysical Research Communications. 2006;351(3):682–688. doi: 10.1016/j.bbrc.2006.10.081. [DOI] [PubMed] [Google Scholar]
- 27.Ide H, Nakagawa T, Terado Y, et al. Tyk2 expression and its signaling enhances the invasiveness of prostate cancer cells. Biochemical and Biophysical Research Communications. 2008;369(2):292–296. doi: 10.1016/j.bbrc.2007.08.160. [DOI] [PubMed] [Google Scholar]
- 28.Schuster C, Muller M, Freissinuth M, et al. Tyk2 expression and its signaling enhances the invasiveness of prostate cancer cells. Biochemical and Biophysical Research Communications. 2008;366(4):869–870. doi: 10.1016/j.bbrc.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 29.Nevalainen MT, Xie JW, Torhorst J, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. Journal of Clinical Oncology. 2004;22(11):2053–2060. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 30.Sultan AS, Xie JW, LeBaron MJ, et al. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24(5):746–760. doi: 10.1038/sj.onc.1208203. [DOI] [PubMed] [Google Scholar]
- 31.Song XC, Fu GY, Yang XF, et al. Protein expression profiling of breast cancer cells by dissociable antibody microarray (DAMA) staining. Molecular & Cellular Proteomics. 2008;7(1):163–169. doi: 10.1074/mcp.M700115-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.