Abstract
The Runx2 factor is an essential component of the regulatory mechanisms that control transcription during skeletogenesis. Runx2/p57 expression in osteoblastic cells is controlled by the P1 promoter, which is recognized by key regulators of osteoblast differentiation including homeodomain factors and Wnt- and BMP-signaling mediators. Here, we report that the transcription factor C/EBPβ up-regulates Runx2/p57 expression by directly binding to the Runx2 P1 promoter in mesenchymal, pre-osteoblastic and osteoblastic cells. This C/EBPβ-mediated up-regulation is principally dependent on C/EBP site II that is located within the first 180 bp of the proximal P1 promoter region and is highly conserved among mouse, rat, and human Runx2 genes. Our studies reveal how the C/EBPβ factor, known to have a key role during osteogenesis, contributes to regulating the expression of Runx2, the master regulator of osteoblast differentiation.
Keywords: Runx2, Bone-specific transcription, C/EBPβ
Introduction
The Runx2 transcription factor is an essential regulatory component during bone formation in mammals as it controls the expression of several key osteoblast-related genes (Lian et al. 2004; Karsenty et al. 2008; Marie, 2008). Runx2 null mice exhibit dramatic defects in osteogenesis and hereditary mutations in the human RUNX2 gene can be linked to ossification defects such as those reported in Cleidocranial Dysplasia (Otto et al. 1997; Otto et al. 2002).
Runx2 expression in osteoblastic cells is controlled by several regulatory elements distributed within the P1 promoter (Drissi et al. 2000; Zambotti et al. 2002; Lengner et al. 2005; Gaur et al. 2005; Lee et al. 2005; Hassan et al. 2006, 2007), which are recognized by cognate transcription factors during osteogenesis. Among these factors the homeodomain proteins Dlx3, Dlx5, Msx2, CDP/Cut (Lee et al. 2005; Hassan et al. 2006), and Hoxa10 (Hassan et al. 2007), as well as transcriptional regulators like AP-1 (Zambotti et al. 2002), β-catenin/TCF (Gaur et al. 2005), Nkx3.2 (Lengner et al. 2005), NF-1 (Zambotti et al. 2002), Sp1 and Ets (Zhang et al. 2009). We have recently shown that in osteoblastic cells Runx2 expression involves a specific chromatin remodeling process within the first 400 bp of the Runx2 P1 promoter region (Cruzat et al. 2009). The changes in chromatin structure occur with elevated levels of histone H3 and H4 acetylation and are independent of SWI/SNF complex activity (Cruzat et al. 2009). However, the precise mechanism(s) involved in this remodeling process and the specific chromatin remodelers responsible remain undefined.
CCAAT/Enhancer Binding-Proteins (C/EBPs) are members of the CREB/ATF family of bzip (Basic Leucine Zipper) transcription factors (Nerlov, 2007). Most of these factors are broadly expressed in mammalian cell types, where they significantly contribute to control of tissue-specific genes (Nerlov, 2007). C/EBPβ exists as three protein isoforms generated by alternative translation initiation: p38/C/EBPβ-1 (also known as Liver Activating Protein* or LAP*), p33/C/EBPβ-2 (LAP), and p20/C/EBPβ-3 (Liver Inhibitory Protein or LIP). All three isoforms contain the bzip DNA-binding domain and retain all (C/EBPβ-LAP* and C/EBPβ-LAP) or only part (C/EBPβ-LIP) of the N-terminal transcription regulatory domain (Calkhoven et al. 2000; Xiong et al. 2001). C/EBPβ-LIP has been shown to have dominant-negative activity at C/EBPβ-LAP*- and C/EBPβ-LAP-target genes (Ramji and Foka, 2002).
Several reports support an important role for C/EBPβ during osteogenesis (Marie, 2008). C/EBPβ knock out mice exhibit delayed bone formation and decreased expression of osteoblast marker genes such as alkaline phosphatase and osteocalcin (Tominaga et al. 2008; Smink et al. 2009). Also, bone-targeted overexpression of C/EBPβ-LIP in mice results in osteopenia from reduced bone formation (Harrison et al. 2005). C/EBPβ can up-regulate osteoblast-related genes during osteogenesis by synergizing with Runx2 (Gutierrez et al. 2002; Hata et al. 2005; Shirakawa et al. 2006) and ATF4 (Tominaga et al. 2008) transcription factors as well as by recruiting SWI/SNF chromatin-remodeling activity to target promoters (Villagra et al. 2006). It has been also demonstrated that C/EBPβ can interact with key components of the Mediator complex (Mo et al. 2004; Li et al. 2008) as well as with transcriptional co-regulators like CBP/p300 (Mink et al. 1997), G9A (Pless et al. 2008), and PRMT4/CARM1 (Kowenz-Leutz et al. 2010) which contain histone-modifying activities. Therefore, this factor may be regulating target genes in osteoblastic cells through several different pathways.
Sequence analysis of the first 1 kb of the Runx2 P1 promoter predicts four high-affinity C/EBPβ–binding sites, which are conserved among mouse, rat and human genomes (see Figure 1A). Because C/EBPβ (C/EBPβ-LAP* and C/EBPβ-LAP) is expressed in osteoblastic cells and its levels increase during osteogenesis (Gutierrez et al. 2002 and data not shown), it is important to define the role of this transcription factor in the P1-driven transcription of the Runx2 gene in osteoblasts. Here, we report that C/EBPβ up-regulates the Runx2/p57 expression by directly interacting with the Runx2 P1 proximal promoter in mesenchymal, pre-osteoblastic and osteoblastic cells.
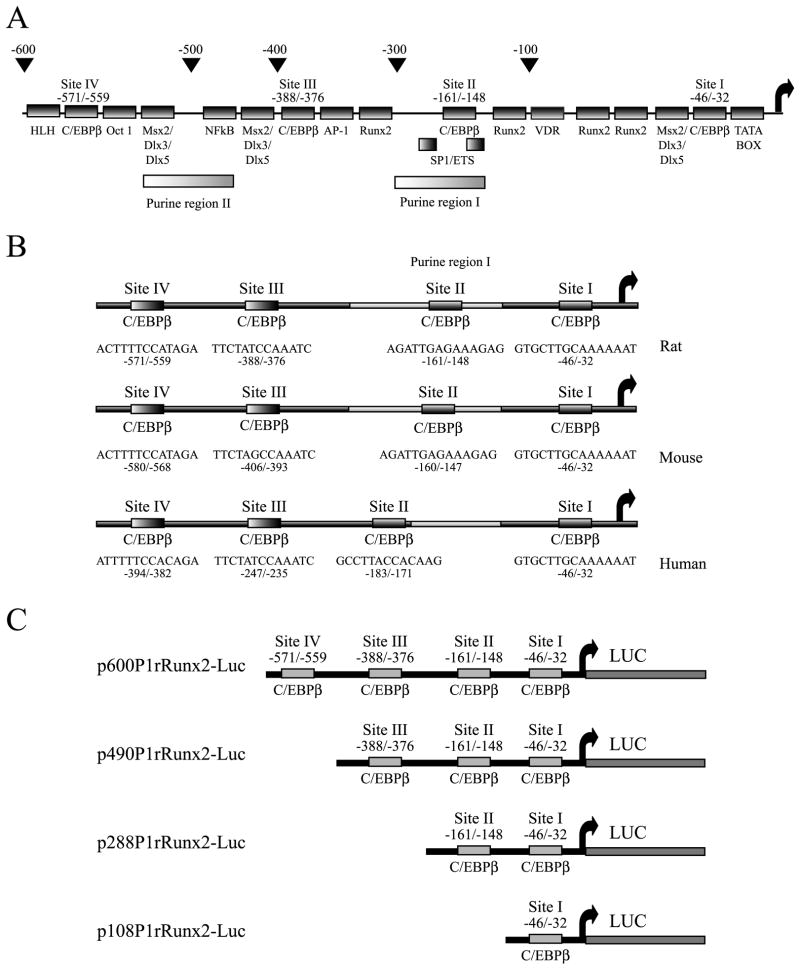
Figure 1. Regulatory sites in the Runx2 P1 promoter.
A) Schematic representation of the regulatory elements present in the P1 promoter (first 600 bp) of the Runx2 gene. The diagram shows the positions of the different binding sites for transcription factors and the putative C/EBP sites. B) Comparison of the rat, mouse and Human Runx2 P1 promoters, showing the sequence of the four putative C/EBPβ-interaction sites, which are conserved among all three species. C) Schematic representation of the constructs carrying deletions of the rat Runx2 P1 promoter controlling the expression of the luciferase reporter gene. The presence of the putative C/EBP sites in the different constructs is indicated.
Materials and Methods
Cell Culture
Rat-derived ROS17/2.8 osteoblastic cells and mouse C2C12 cells were cultured as described previously (Majeska et al. 1980; Cruzat et al. 2009). Mouse MC3T3-E1 and Runx2-null TERT-immortalized osteoprogenitor cells (Bae et al. 2007) were grown in α-minimal essential medium (MEM) supplemented with 10% fetal bovine serum.
Expression constructs
Luciferase-reporter constructs driven by the Runx2 P1 promoter (p2.8P1mRunx2-Luc, p600P1rRunx2-Luc, p490P1rRunx2-Luc, p288P1rRunx2 and p108P1rRunx2) were described previously (Drissi et al. 2000; Zhang et al. 2009). All mutant versions of the p288P1rRunx2-Luc vector (MUT1, MUT2 and MUT1+2) were generated with the Quick-Change Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. For oligonucleotides utilized see supplementary figure S3. Expression vectors pC/EBPβ-LAP*, pC/EBPβ-LAP and pC/EBPβ-LIP were kindly provided by Dr. Jose L. Gutierrez (University of Concepcion, Concepcion, Chile). The pC/EBPβ plasmid containing the mouse C/EBPβ cDNA was generated from MSV/C/EBPβ (Gutierrez et al., 2002). mC/EBPβ cDNA was cleaved and then inserted into the EcoRI/XhoI sites of pcDNA3.1/His (Invitrogen, Carlsbad, CA, USA), which contains both 6xHis and Xpress tags. The bacterial vector coding for His-C/EBPβ was generated from MSV/C/EBPβ (Gutierrez et al. 2002) by digestion with SphI/HindIII and subsequent insertion into the pQE-80L plasmid (QIAGEN Inc., Valencia, CA, USA). The bacterial expression vector coding for GST-Runx2 was described previously (Paredes et al. 2002; Sierra et al. 2003).
Transient transfections and reporter assays
C2C12 and MC3T3-E1 cells were plated in 24- and 12-well plates (21 and 15 mm diameter wells, respectively) and transiently transfected with the FuGENE 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN, USA). ROS17/2.8 cells were plated in 24-well plates and transfected with Lipofectamine 2000 reagent (Invitrogen). Mouse Runx2-null TERT-immortalized cells were plated in 24-well plates and transfected with the FuGENE 6 Transfection Reagent (Roche Diagnostics). The cells were transfected with 250 or 500 ηg of the firefly luciferase reporter plasmids and 12.5 or 25 ηg, respectively, of the Renilla luciferase (pRL-SV40) plasmid as internal control (Promega Corp. Madison, WI, USA). The total amount of DNA was maintained at a constant level (1.3μg for 12-well and 650 ηg for 24-well) by adding an appropriate amount of pBluescript (pBS) plasmid (Agilent Technologies Inc.). After 24 h, the cells were harvested using 50 μL of passive lysis buffer (Promega) per well. Cell lysates (20 μl) were evaluated for firefly luciferase activity using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer’s instructions and normalized to values of Renilla luciferase activity.
Chromatin immunoprecipitation (ChIP)
ChIP studies were performed as described earlier (Soutoglou et al. 2002; Villagra et al. 2006; Cruzat et al. 2009) with modifications. ROS17/2.8 cell cultures (100 mm diameter plates) were washed with phosphate buffered saline (PBS 1X) and incubated for 10 min with 1% formaldehyde and gentle agitation at room temperature. The cross-linking was stopped by the addition of 0.125 M glycine for 5 min. The following experimental steps were performed on ice or at 4°C. The cells were washed with 5 ml of PBS, scraped off in the same volume of PBS, and collected by centrifugation at 1,000 x g for 5 min. The cell pellet was resuspended in 1 ml of lysis buffer (50 mM Hepes, pH 7.8, 20 mM KCl, 3 mM MgCl2, 0.1% Nonidet P40, and a mixture of proteinase inhibitors), incubated for 10 min on ice and homogenized by Dounce homogenizer. The cell extract was collected by centrifugation at 1,500 x g for 5 min and resuspended in 0.3 ml of sonication buffer (50 mM Hepes, pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate acid, 0.1% SDS, and a mixture of proteinase inhibitors) for each 100 mm plate. To reduce the length of the chromatin fragments to 500 bp or smaller (confirmed by electrophoretic analysis and PCR amplification), the extract was sonicated with a Misonix sonicator (model 3000), using 15-s pulses at 30% power. After centrifugation at 16,000 x g for 10 min, the supernatant was collected, frozen in liquid nitrogen, and kept at −80 °C. An aliquot was used for A260 measurements. Cross-linked extracts (8 A260 units) were resuspended in sonication buffer to a final volume of 500 μl. The samples were precleared by incubation with 50 μl of protein A/G-agarose beads (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) for 1 h at 4 °C with agitation. After centrifugation at 1,000 x g for 5 min, the supernatant was collected and immunoprecipitated with anti-C/EBPβ polyclonal antibody (C-19, Santa Cruz Biotechnology) and anti-Runx2 (M-70, Santa Cruz Biotechnology) for 12h at 4°C. The immunocomplexes were recovered with the addition of 50 μl of protein A-agarose beads and subsequent incubation for 1 h at 4°C with agitation. The complexes were washed once with: sonication buffer, sonication buffer plus 500mM NaCl, LiCl buffer (100 mM Tris-HCl, pH 8.0, 500 mM LiCl, 0.1% Nonidet P40, and 0.1% deoxycholic acid), and with Tris-EDTA (TE) buffer pH 8.0 (2mM EDTA and 50 mM Tris-HCl, pH 8.0), washing each time for 5 min at 4°C. The protein-DNA complexes were then eluted by incubation with 100 μl of elution buffer (50 mM NaHCO3 and 1% SDS) for 15 min at 65°C. After centrifugation at 10,000 x g for 5 min, the supernatant was collected and NaCl added to a final concentration of 200 mM. This mixture was incubated with 20 μg/ml of RNase A for 12–16h at 65°C to reverse the cross-linking. The proteins were digested with 100 μg/ml of proteinase K for 2 h at 50°C and the DNA recovered by phenol/chloroform extraction and ethanol precipitation using glycogen (20 μg/ml) as a precipitation carrier. The PCR primers used to evaluate the rat Runx2 P1 promoter region were: forward -179, 5′-GGAGGGGAGAAGGAAAAAGA-3′; reverse +147, 5′-AGCACTCACTGACTCGGTTG-3′.
Western Blotting
Nuclear extracts were prepared as reported previously (Paredes et al. 2004) from ROS17/2.8, C2C12, MC3T3-E1 and Runx2-null TERT-immortalized osteoprogenitor cells, transfected with the appropriate expression vector as described in each figure legend. Proteins were detected by Western blot using specific antibodies: C/EBPβ (C-19), TFIIB (C-18) (Santa Cruz Biotechnology) and Xpress (Invitrogen).
Protein Expression and Protein-DNA interaction analyses
The recombinant protein GST-Runx2 was obtained by expression in Escherichia coli (E.coli) BL21 and purified in a glutathione-Sepharose column (GE Healthcare, Pittsburgh, PA, USA) as previously reported (Paredes et al. 2002; Sierra et al. 2003; Paredes et al. 2004). His-C/EBPβ was obtained following transformation of E.coli BL21 and subsequent purification using a Ni-NTA (QIAGEN Inc.) column, according to the manufacturer’s instructions and as described previously (Paredes et al. 2004). The purity and integrity of these proteins were evaluated by SDS/PAGE and by Western blot analysis using specific antibodies (Santa Cruz Biotechnology). The specific DNA binding activity of the bacterially expressed proteins was analyzed as described previously (Paredes et al. 2002) by electrophoretic mobility shift assay (EMSA), using oligonucleotides end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (For oligonucleotide sequences, see supplementary figure S3). Binding reactions contained 50 ηg of purified proteins, 50 fmol of each labeled oligonucleotide, and were performed in 20 μl of 10 mM Tris/HCl pH 7.4, 100 mM KCl, 1 mM EDTA, 0.05 % Nonidet P40, 0.5 mM PMSF, 1 mM DTT, 10% (v/v) glycerol, 50 μg/μl BSA, 1 μg/μl poly [d(I-C)]. Protein-DNA complexes were fractionated in 3.5% (w/v) polyacrylamide gels (acrylamide:bis-acrylamide ratio 60:1) and 0.5X TBE (1x TBE is 45mM Tris/borate/1mM EDTA), dried and visualized by autoradiography.
Results
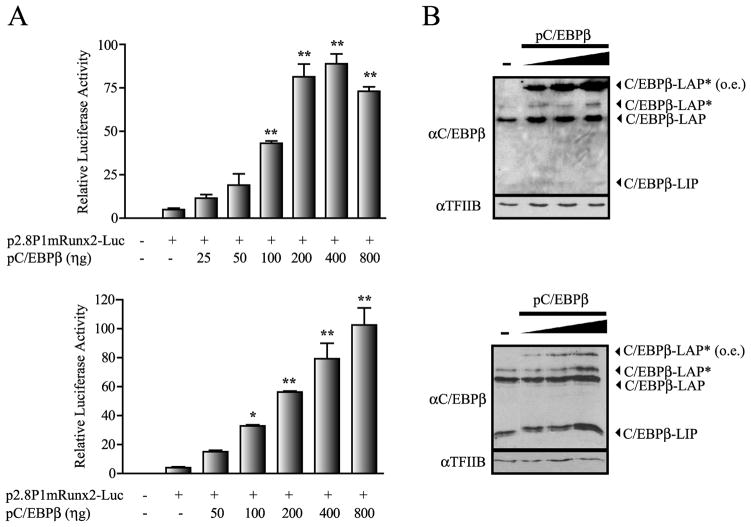
Several reports support a critical role for the transcription factor C/EBPβ during osteogenesis (Marie, 2008) by controlling the expression of various osteoblast-phenotypic genes (Tominaga et al. 2008; Smink et al. 2009; Gutierrez et al. 2002; Villagra et al. 2006). Sequence analyses of the first 1 kb of the Runx2 gene P1 promoter, reveals four high affinity C/EBP binding sites that are conserved among the mouse, rat, and human genomes (Figure 1A, B). Hence, we first addressed whether these sites contribute to transcription of this osteoblast master regulator in mesenchymal or osteoblast-lineage-committed cells. We performed transient transfection experiments using a 2.8 Kb segment of the Runx2 P1 promoter driving the expression of the Luciferase (Luc) reporter gene and found that forced expression of C/EBPβ/LAP* in both mouse mesenchymal C2C12 and pre-osteoblastic MC3T3 cells (Figure 2A, upper and lower graphs, respectively) stimulate Runx2 P1 activity in a dose-dependent manner. These results indicate that C/EBPβ/LAP* functions as a positive regulator of the Runx2 P1 promoter in both uncommitted C2C12 cells and in MC3T3 cells that are committed to the osteoblast lineage.
Figure 2. C/EBPβ up-regulates the Runx2 P1 promoter activity.
A) C2C12 and MC3T3 cells (upper and lower graphs, respectively) were transiently co-transfected during 24 h with the 2.8 kb mouse Runx2 P1 promoter construct controlling the luciferase reporter gene (500 ηg) and increasing amounts of the pC/EBPβ construct. The presence (+) or absence (−) of each construct is indicated below the bars. The results represent at least three independent experiments, each assayed in triplicate. Each bar represents the mean +/− standard error. Statistical significance was determined by the ANOVA test (*: p<0.05; **: p<0.01). B) Analysis by Western blot of the pC/EBPβ over-expression. Nuclear extracts (10μg) isolated from transiently transfected C2C12 and MC3T3 cells (upper and lower blots, respectively) were analyzed by Western blot using polyclonal antibodies against C/EBPβ. The figure shows the presence of the different endogenous C/EBPβ isoforms and the ectopically over-expressed C/EBPβ (o.e.). Re-blotting against TFIIB was used to control for equal protein loading.
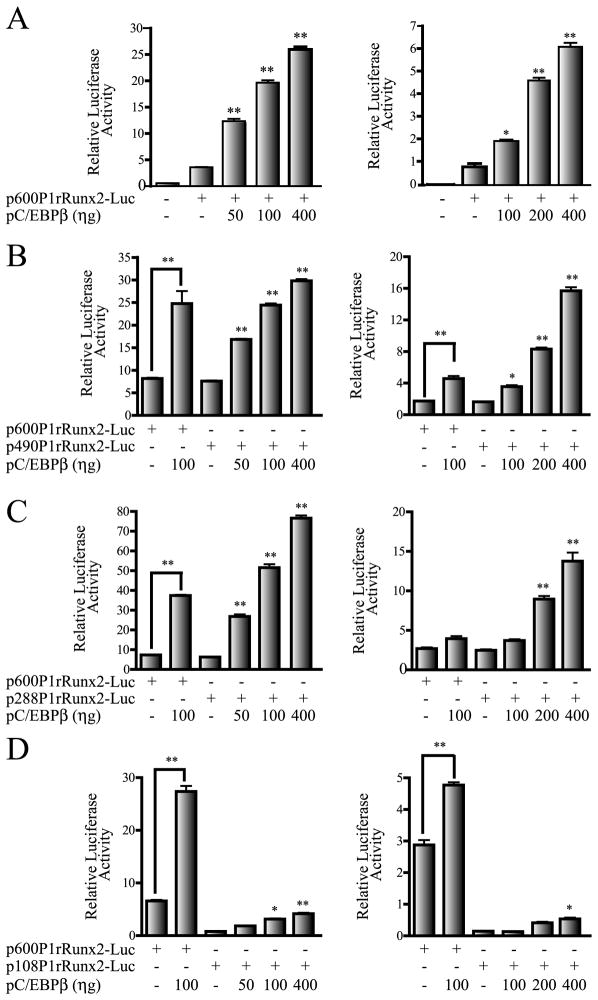
We next determined whether the four C/EBP elements within the first 600 bp of the Runx2 P1 promoter were contributing to the C/EBPβ-mediated effect. Using a series of deletion constructs of the rat Runx2 P1 promoter (see Figure 1C), we determined the requirement of the four C/EBP sites for the C/EBPβ response. As shown in Figure 1C, these constructs contain either all four C/EBP sites I, II, III and IV (p600P1rRunx2-Luc), sites I, II and III (p490P1rRunx2-Luc), sites I and II (p288P1rRunx2-Luc), or only site I (p108P1rRunx2-Luc). We found that increased expression of C/EBPβ in both C2C12 and MC3T3 cells enhances the activity of the P1 promoter deletion constructs that retain the putative C/EBP site II (Figures 3A, 3B and 3C). The shorter construct retaining only the most proximal C/EBP site I responded poorly to the increasing levels of the C/EBPβ factor in both cell lines (Figure 3D). The C/EBPβ-mediated up-regulation of the Runx2 P1 promoter was also determined in ROS17/2.8 osteoblastic cells (supplementary Figure S1A) although in this case the stimulatory effect was less marked. This result is likely due to the high concentrations of endogenous C/EBPβ proteins already present in these cells (supplementary Figure S1A, lower panel and Shin et al. 2006). Together these results indicate that the putative C/EBP site II is a key contributor to the C/EBPβ-dependent stimulation of Runx2 gene P1 promoter activity, and most prominently in non-osseous and early progenitor cells.
Figure 3. Putative C/EBP site II is a key component in the C/EBPβ-enhanced Runx2 P1 promoter activity.
Transient co-transfection experiments were performed in C2C12 and MC3T3 cells (left and right panels, respectively), using luciferase reporter constructs carrying different deletions of the rat Runx2 P1 promoter (500 ηg) and increasing amounts of the pC/EBPβ expression plasmid. A) 600 bp rat Runx2 P1 promoter construct, B) 490 bp Runx2 P1 promoter construct, C) 288 bp Runx2 P1 promoter construct, and D) 108 bp Runx2 P1 promoter construct. The presence (+) or absence (−) of each construct in the transfection mix is indicated below the bars. These results represent at least three independent experiments, each assayed in triplicate. Each bar represents the mean +/− standard error. Statistical significance was determined by the ANOVA test (*: p<0.05; **: p<0.01).
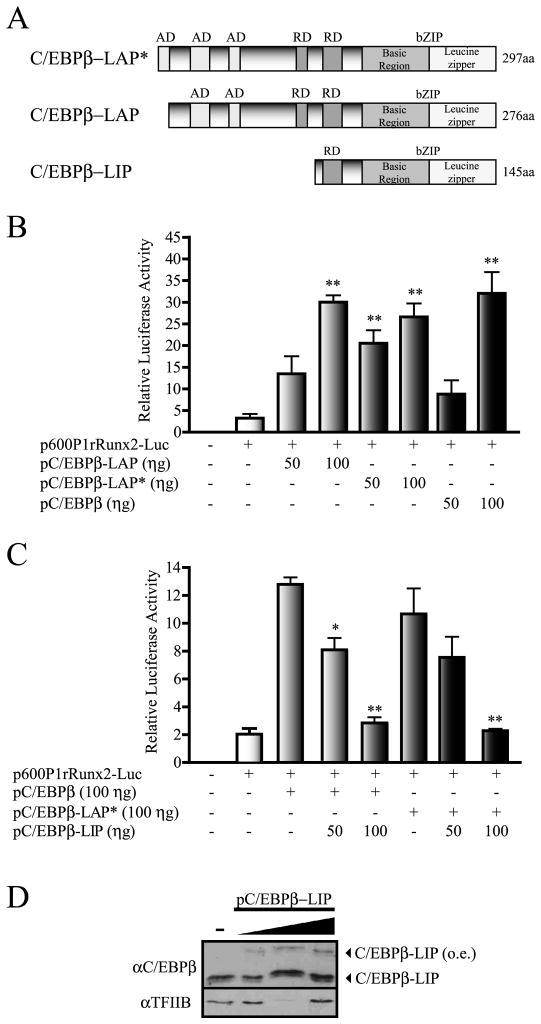
C/EBPβ exists as three protein isoforms which are generated by alternative translation initiation (Nerlov, 2007). The two larger isoforms, C/EBPβ-LAP* and C/EBPβ-LAP (Figure 4A), have been shown to function principally as activators of gene transcription, while the third shorter C/EBPβ-LIP isoform (Figure 4A) can function as a negative regulator (Nerlov, 2007). Hence, we determined whether a specific C/EBPβ protein isoform is regulating the Runx2 P1 promoter activity. As shown in Figure 4B, forced expression of either C/EBPβ-LAP* or C/EBPβ-LAP proteins (see also supplementary Figure S2) equally upregulated Runx2 P1 promoter activity in MC3T3-E1 cells in a dose-dependent manner. This result indicates that the first 21 aminoacid residues at the N-terminus of C/EBPβ-LAP* do not significantly contribute to the C/EBPβ-mediated stimulation of the Runx2 P1 promoter. Interestingly, co-expression of C/EBPβ-LIP (see Figure 4D) inhibited in a dose-dependent manner both C/EBPβ-LAP*- and C/EBPβ-LAP-enhanced Runx2 P1 promoter activity (Figure 4C). These results are in agreement with the previously described role of C/EBPβ-LIP as a negative regulator of transcription (Nerlov, 2007; Ramji and Foka, 2002) and suggest that binding of C/EBPβ-LAP* or C/EBPβ-LAP to the Runx2 promoter is required during C/EBPβ-mediated transcriptional up-regulation of Runx2. Similar results were obtained when these analyses were performed in C2C12 cells (not shown) indicating that C/EBPβ-LAP* or C/EBPβ-LAP can stimulate Runx2 P1 promoter activity in both mesenchymal and osteoblast-committed cell lineages.
Figure 4. C/EBPβ isoforms LAP* and LAP can up-regulate the Runx2 P1 promoter activity.
A) Schematic representation of the three rat C/EBPβ isoforms (C/EBPβ-LAP*, C/EBPβ-LAP, and C/EBPβ-LIP). The diagram shows the presence of activation domains (AD), repression domains (RD) and the leucine zipper basic domains (bZIP), which support both DNA-binding (basic region) and dimerization (leucine zipper). B) MC3T3 cells were transiently transfected with 250 ηg of the 600 bp rat Runx2 P1 promoter construct controlling the luciferase reporter gene and co-transfected with vectors that selectively express the C/EBPβ isoforms LAP*, LAP, or all three LAP*, LAP, and LIP. C) The effect of the selective over-expression of C/EBPβ-LIP on the C/EBPβ-LAP-enhanced Runx2 P1 promoter activity was also determined by transient transfection in MC3T3 cells. These results represent at least three independent experiments, each assayed in triplicate. Each bar represents the mean +/− standard error. Statistical significance was determined by the ANOVA test (*: p<0.05; **: p<0.01). D) Western blot analysis showing C/EBPβ-LIP over-expression in MC3T3 cells. Nuclear extracts (10 μg) were resolved by SDS-PAGE (12% acrylamide) and revealed using specific antibodies against C/EBPβ to detect the over-expressed (o.e.) xpress-tagged C/EBPβ-LIP protein. Detection of TFIIB was used to control for equal protein loading.
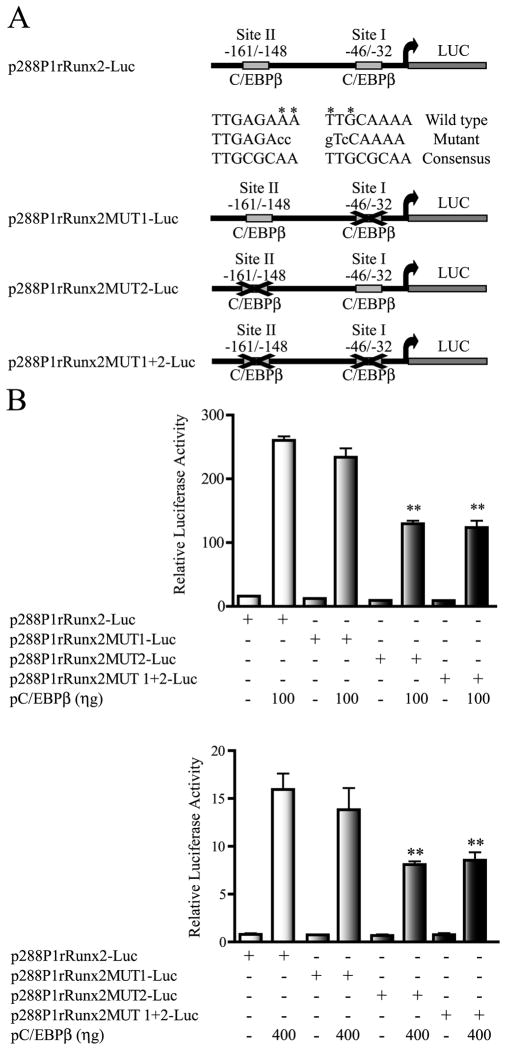
We next determined whether an intact C/EBP site II was required for C/EBPβ-mediated up-regulation of the Runx2 P1 promoter activity. For this purpose, the p288P1rRunx2-Luc reporter construct, containing the C/EBP sites I and II (see Figure 5A), was used as a template to mutate both elements, either independently or in combination, by site-directed mutagenesis (see Methods). Changes in two consensus nucleotides of the C/EBP sites (see Figure 5A), which abolish C/EBPβ-binding, were selected based on previous reports (Gutierrez et al. 2002; Chen et al. 2005) and confirmed by protein-DNA interaction analyses (see below). As shown in Figure 5B, mutation of the C/EBP site II (MUT2) significantly, although not completely, inhibits the ability of the Runx2 P1 promoter construct to respond to C/EBPβ over-expression, both in C2C12 (Figure 5B, upper graph) and in MC3T3 (Figure 5B, lower graph) cells. In contrast, mutation of C/EBP site I (MUT1) does not affect the ability of C/EBPβ factors to stimulate this Runx2 P1 promoter construct (Figure 5B, upper and lower graphs). Similarly, a double mutation of C/EBP sites I and II (MUT1+2) reduces the C/EBPβ-mediated up-regulation of the Runx2 P1 promoter to a level equivalent to that obtained when only C/EBP site II is mutated. Together, these results confirm that C/EBP site II is a principal contributor during C/EBPβ-mediated activation of the Runx2 P1 promoter, both in mesenchymal and osteoblast-committed lineages.
Figure 5. C/EBP site II is a principal component in the C/EBPβ-mediated transcriptional up-regulation of the Runx2 P1 promoter.
A) Schematic representation of the wild type and mutated versions of the 288P1rRunx2-Luc reporter constructs. The figure shows the sequences of the putative C/EBP sites I and II present in these reporter constructs. Nucleotides marked with an asterisk were replaced by the nucleotides showed in lowercase letters. B) C2C12 and MC3T3 cells (upper and lower graphs, respectively) were transiently transfected with 250 ηg of the reporter constructs containing intact putative C/EBP sites I and II (p288P1rRunx2-Luc), a mutated C/EBP site I (p288P1rRunx2MUT1-Luc), a mutated C/EBP site II (p288P1rRunx2MUT2-Luc), or both C/EBP sites I and II mutated (p288P1rRunx2MUT1+2-Luc). Cells were also co-transfected with a vector for C/EBPβ over-expression (pC/EBPβ) in the amounts indicated under each graph. These results represent at least three independent experiments, each assayed in triplicate. Each bar represents the mean +/− standard error. Statistical significance was determined by the ANOVA test (*: p<0.05; **: p<0.01).
As shown in Figure 5B, even in the absence of a fully functional C/EBP site II, the p288P1rRunx2-Luc construct exhibits enhanced promoter activity during C/EBPβ over-expression. This result may be related to an indirect effect through a different transcription factor that is induced by the forced expression of C/EBPβ in the cells. Alternatively, it may be the result of C/EBPβ functioning at this Runx2 promoter region through a different regulatory element. Previous reports indicate that C/EBPβ can form functional complexes with the transcription factor Runx2 to up-regulate the expression of bone-related genes (Gutierrez et al. 2002; Villagra et al. 2006; Tominaga et al. 2008; Shin et al. 2006). Moreover, these Runx2-C/EBPβ-containing complexes can still bind to target promoters in the presence of an intact consensus element for only one of these two factors (Gutierrez et al. 2002).
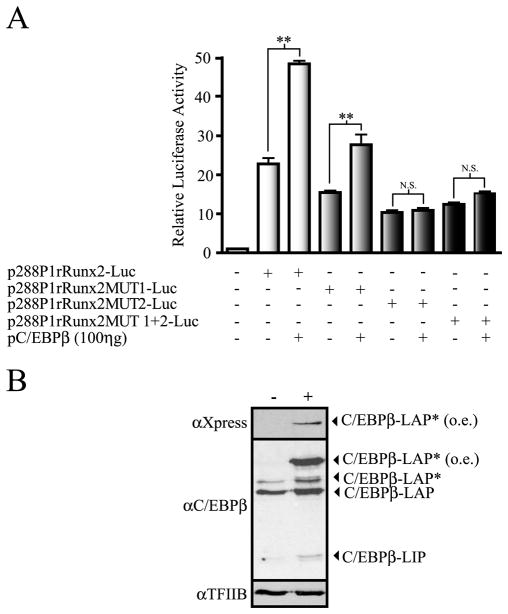
The p288P1rRunx2-Luc promoter construct contains 3 conserved Runx binding sites (−119/−114; −85/80; and −74/−69, see supplementary Figure S3), which have been shown to be recognized by Runx2 (Drissi et al. 2000 and see Figure 7). We determined whether the presence of the transcription factor Runx2 in the cells is important during C/EBPβ-mediated up-regulation of Runx2 P1 promoter activity. The plasmid coding for C/EBPβ-LAP* and the p288P1rRunx2-Luc construct (wild-type and with the C/EBP sites mutated) were co-transfected into calvarial-derived osteoprogenitor cells from Runx2 null mice (Bae et al. 2007). Interestingly, we observed that the overall basic activity of these Runx2 P1 promoter constructs is higher in these Runx2-null cells than in both C2C12 and MC3T3 cells. This result suggests that the lack of Runx2 generates a cellular background that is more suitable for Runx2 gene transcription (see Discussion). As shown in Figure 6A, mutation of C/EBP site II (MUT2) results in 50 percent reduction of the p288P1rRunx2-Luc construct basal activity, thus confirming the relevant role of C/EBP site II in Runx2 P1 promoter activity. In contrast, mutation of C/EBP site I (MUT1) produces only a minor effect on the Runx2 P1 promoter activity (~ 25 percent reduction) indicating that in the absence of Runx2, this site contributes partially to maintenance of the overall promoter activity. Importantly, we determined that over-expression of C/EBPβ-LAP* stimulates the wild-type basal activity of the p288P1rRunx2-Luc construct as well as the activity of the P1 promoter sequence carrying a mutated C/EBP site I (Figure 6). However, high concentrations of C/EBPβ are unable to enhance the activity of the reporter constructs with mutations in the C/EBP site II (MUT2 and MUT1+2) (Figure 6 and data not shown). Therefore, our results demonstrate that in Runx2-null cells Runx2 P1 promoter activity can also be stimulated by C/EBPβ. In addition, we conclude that in the absence of an intact C/EBP site II, the C/EBPβ factor is unable to up-regulate Runx2 P1 promoter activity when the transcription factor Runx2 is not present.
Figure 7. C/EBPβ and Runx2 factors interact independently and simultaneously with the Runx2 P1 promoter in vitro and in intact osteoblastic cells.
The ability of C/EBPβ and Runx2 recombinant proteins to interact specifically with their cognate binding sequences in the P1 promoter was evaluated by EMSA as described in materials and methods. The combinations used in each binding reaction are indicated at the top of the gels. The positions of the specific protein-DNA complexes are also marked. A) Binding of purified His-C/EBPβ protein to the radio-labeled oligonucleotides (Probe) containing the C/EBP site I (left panel) or C/EBP site II (right panel). Competition assays were performed by adding an excess (100 fold) of the cold-specific oligonucleotide (competitor). B) Binding of His-C/EBPβ protein to a radio-labeled consensus C/EBP element (consensus probe) and competition assays using an excess of cold-oligonucleotides (competitor) containing either the wild-type sequence for C/EBP site I, C/EBP site II, or their mutated versions (MUT 1 and MUT2, respectively). C) Binding of purified His-C/EBPβ and GST-Runx2 proteins to radio-labeled Runx2 P1 promoter fragments containing the C/EBP site I (left panel) or C/EBP site II (right panel). D) Binding of endogenous C/EBPβ and Runx2 factors to the proximal Runx2 gene P1 promoter sequences in ROS17/2.8 cells, assessed by chromatin immunoprecipitation (ChIP). The enrichment of the Runx2 P1 promoter sequences in the precipitated DNA was determined by PCR amplification using specific primers and normalized to input material. IgG: non-specific immunoglobulin G.
Figure 6. C/EBPβ requires the putative C/EBP site II to up-regulate the Runx2 P1 promoter activity in Runx2-null mice osteoprogenitor cells.
A) TERT-immortalized mouse Runx2-null osteoprogenitor cells were co-transfected with 250 ηg of the wild type or mutant versions of the p288P1rRunx2-Luc reporter construct and increasing amounts of the C/EBPβ expression vector (pC/EBPβ). Each bar represents the mean +/− standard error. Statistical significance was determined by the ANOVA test (*: p<0.05; **: p<0.01; N.S. not statistically significant). B) Western blot analysis of the mouse Runx2-null cells transfected with the pC/EBPβ vector. Nuclear extracts (10 μg) were fractionated by SDS-PAGE (12% acrylamide) and revealed by Western blot using specific antibodies against C/EBPβ or the Xpress-tag to detect the over-expressed Xpress-tagged C/EBPβ/LAP* (o.e.). Detection of TFIIB was used to control for equal protein loading.
To show that C/EBP sites I and II in the Runx2 P1 promoter region are bona fide binding sites which can be recognized in vitro by C/EBPβ, we performed electromobility shift assays (EMSA) using recombinant C/EBPβ-LAP (His-tagged C/EBPβ-LAP) and radio-labeled oligonucleotide probes including either C/EBP site I (probe site I) or C/EBP site II (probe site II). As shown in Figures 7A and 7B, both probes can be bound in vitro by C/EBPβ-LAP. This interaction is specific as no retarded band is detected when mutant C/EBP site I or mutant C/EBP site II probes are utilized or when an excess (100 fold) of unlabeled specific oligonucleotide is included in the binding reaction. Similarly, formation of the specific protein-DNA complexes is inhibited when an excess (100 fold) of a unlabeled C/EBP consensus oligonucleotide (Osada et al., 1996; Ramji and Foka, 2002) is included, but not when unlabled oligonucleotides carrying mutated C/EBP sites I or II are used as competitors.
We next evaluated in vitro whether the C/EBPβ-LAP and Runx2 factors can recognize simultaneously, their cognate sites located within the most proximal 200 bp of the Runx2 P1 promoter region. For this purpose, we performed EMSA using recombinant his-tagged C/EBPβ-LAP and GST-tagged Runx2/p57 produced in bacteria (Paredes et al. 2004) and PCR-generated radio-labeled DNA probes spanning the Runx2 P1 promoter sequences −179/−98 (82 bp probe) and −111/+15 (126 bp probe) (see details in supplementary Figure S3). As shown in Figure 7C, C/EBPβ-LAP and Runx2 factors bind both independently and simultaneously to the Runx2 P1 promoter segments including the C/EBP sites I and II and the Runx elements. These results demonstrate that in vitro, these C/EBP sites are bona fide C/EBPβ-binding elements, although site II shows more relevant functional regulatory activity. Our results also confirm previous reports indicating that Runx2 binds in vitro to its own P1 promoter (Drissi et al. 2000).
It was important to demonstrate that these two transcription factors interact with the Runx2 gene P1 promoter in the in vivo cellular environment of osteoblastic cells expressing Runx2. Hence, we performed chromatin immunoprecipitation (ChIP) analyses and found that both Runx2 and C/EBPβ bind to the proximal Runx2 P1 promoter region in ROS17/2.8 osteosarcoma cells (Figure 7D), that actively transcribe the Runx2 gene (Paredes et al. 2004; Sierra et al. 2003; Cruzat et al. 2009). Taken together, these results indicate that C/EBPβ can up-regulate transcription of the Runx2/p57 gene by directly binding to the proximal region of the P1 promoter.
Discussion
The C/EBPβ transcription factor has been shown to be a key regulator of early and late osteogenesis (Marie, 2008). Here, we find that C/EBPβ up-regulates Runx2/p57 expression by direct association with bona-fide C/EBP sites at the Runx2 gene P1 promoter sequence in mesenchymal, osteoprogenitor, pre-osteoblastic, and osteoblastic cells. The C/EBPβ-mediated enhancement of Runx2 P1 promoter activity is principally, although not exclusively (see below), dependent on C/EBP site II, located within the proximal 180 bp promoter region immediately up-stream of the transcriptional start site. This C/EBP site II is highly conserved among the mouse, rat, and human Runx2 P1 promoter sequences, likely indicating that is also a conserved mechanism to regulate Runx2 transcription during bone cell formation.
In a recent report it was proposed that C/EBPβ functions through the most distal C/EBP site IV of the Runx2 P1 promoter to down-regulate Runx2 transcription in uncommitted mesenchymal cells, expressing low levels of C/EBPβ factor (Wiper-Bergeron et al. 2007). Although our results appear to contradict these findings, there are a number of reasons that may explain this discrepancy. In their study Wiper-Bergeron and colleagues did not consistently observe the inhibitory effect of C/EBPβ on the Runx2 P1 promoter activity. This was the case when mesenchymal cell lines committed to the osteoblastic lineage and expressing C/EBPβ were analyzed (Wiper-Bergeron et al. 2007). Several groups, including ours, have shown that osteoblastic cells co-express the Runx2/p57 and C/EBPβ genes at high levels (Gutierrez et al. 2002; Shen and Christakos, 2005; Dhawan et al. 2005; Villagra et al. 2006), thus providing support for the idea that both factors function cooperatively during osteoblast differentiation to promote the expression of other key bone-related genes. Additionally, in their report these investigators did not determine whether direct binding of C/EBPβ to the Runx2 P1 promoter was occurring under their experimental conditions. This opens the possibility of an indirect repressive mechanism activated by the forced expression of C/EBPβ in the cells analyzed in their study. Also, these investigators neither evaluated the contribution of C/EBP sites II and III nor assessed the binding of C/EBPβ to these bona-fide C/EBP sites. Despite these issues, it is still possible that in the study of Wiper-Bergeron et al. (2007) the arrival at different conclusions about the role of C/EBPβ in Runx2 gene transcription may be related to the dissimilar cell backgrounds utilized in their over-expression experiments. Further studies that clarify this point will be required in the future.
TERT-immortalized calvarial cells from Runx2-null mice represent an osteoprogenitor cell model (Bae et al. 2007). We find that these cells not only express high levels of C/EBPβ, but also exhibit high Runx2 P1 promoter activity when transiently transfected, further indicating that C/EBPβ can function as an activator and not a repressor, of Runx2/p57 gene transcription. Interestingly, our results show that the absence of a functional C/EBP site II reduces proximal P1 promoter activity and abrogates the enhancement in this promoter activity that follows C/EBPβ over-expression. This site represents a bone-fide C/EBP binding element as shown in our EMSA experiments using both short oligonucleotides carrying wild-type and mutated versions of this sequence and longer segments of the P1 proximal promoter region spanning C/EBP site II. In contrast, although mutation of C/EBP site I (also shown to be recognized in vitro by C/EBPβ) slightly reduces Runx2 P1 promoter activity, it does not prevent C/EBPβ-mediated up-regulation. Therefore, our results support a model in which C/EBPβ-enhanced Runx2 transcription occurs principally through C/EBP site II. The fact that a Runx2-null cellular environment results in an apparently higher Runx2 P1 promoter activity may indicate that the transcriptional mechanisms operating through C/EBPβ (and perhaps through other transcriptional regulators too) are over-stimulated in these cells, as part of a compensatory response seeking to elevate Runx2 expression. In agreement with this suggestion, it has been reported that this Runx2-null osteoprogenitor cell model responds to forced expression of Runx2 by rapidly becoming engaged in osteoblast differentiation and expressing key bone phenotypic markers (Bae et al. 2007).
Our results show that when cells expressing low (C2C12 and MC3T3) or high (ROS17/2.8 cells) levels of Runx2 protein, are transfected with Runx2 P1 promoter constructs carrying mutated C/EBP sites (e. g. sites I and II in the 288 bp Runx2 P1 promoter construct), these reporter plasmids still retain the ability to at least partially respond to C/EBPβ over-expression. Nevertheless, transfection of these same reporter constructs in the osteoprogenitor cell line lacking Runx2 (TERT-immortalized Runx2-null cells), does not result in C/EBPβ-mediated enhancement. These results indicate that C/EBPβ-mediated transcriptional up-regulation may also partially operate through Runx2 factors bound to the Runx2 P1 promoter. Alternatively, these findings may suggest that C/EBPβ-mediated activation of the P1 promoter includes an indirect, yet undetermined component, which is absent in cells lacking Runx2. In support of the first possibility and in agreement with previous reports (Drissi et al. 2001), we demonstrate here that the Runx2 protein binds to the proximal P1 promoter region both in vitro and in intact osteoblastic cells expressing Runx2. Importantly, both C/EBPβ and Runx2 factors can simultaneously bind to this P1 promoter region in vitro. In addition, we and others have previously shown that C/EBPβ can up-regulate osteoblast-related genes during osteogenesis by synergizing with Runx2 (Gutierrez et al. 2002; Hata et al. 2005; Shirakawa et al. 2006; Villagra et al. 2006). Moreover, these Runx2-C/EBPβ-containing complexes can still bind target promoters in the presence of an intact consensus element for only one of these two factors (Gutierrez et al. 2002). A potential cooperative mechanism between C/EBPβ and Runx2 to up-regulate Runx2 gene expression in osteoblasts is currently being investigated.
Supplementary Material
A) ROS17/2.8 osteoblastic cells were transiently transfected with 250 ηg of the p600P1rRunx2-Luc reporter construct alone or in combination with increasing amounts of the pC/EBPβ (C/EBPβ/LAP*) expression plasmid. C/EBPβ protein over-expression was confirmed by Western blot (bottom panel) using antibodies against X-press to detect the over-expressed Xpress-tagged C/EBPβ/LAP* (o.e.) or antibodies against C/EBPβ. To control for equal protein loading the membranes were re-blotted against TFIIB. B) The contribution of the putative C/EBP sites I and II to the Runx2 P1 promoter activity in osteblastic cells, was assessed by transient transfection in ROS17/2.8 cells. Cells were transfected with the p288P1rRunx2-Luc reporter plasmids (250 ηg each), including intact or mutated versions of the C/EBP sites I and II, either independently or combined, as indicated under each graph. The results were expressed as percentage (%) of control activity, considering as the control the relative luciferase activity of the wild-type p288P1rRunx2-Luc construct (100%). The reporter activity and statistical significance were determined as described in figure legend 3.
C2C12 cells were transiently transfected with the p600P1rRunx2-Luc reporter construct (250 ηg), alone or in combination with the C/EBPβ-expression plasmid (pC/EBPβ) or the constructs that selectively express the different C/EBPβ isoforms LAP* (pC/EBPβ-LAP*) or LAP (pC/EBPβ-LAP). The reporter activity and statistical significance were determined as described in figure legend 3.
A) Schematic representation of the Runx2 P1 promoter indicating the C/EBP and Runx binding sites identified in this region and the positions of the primers used to amplify the promoter segments for EMSA and ChIP. B) Sequences of the different primers and oligonucleotides used for EMSA and ChIP experiments.
Acknowledgments
This work was supported by: Contract grant sponsor: FONDECYT; Contract Grant Number: 1095075 (To MM). Contract grant sponsor: FONDAP; Contract grant number: 15090007 (to MM). Contract grant sponsor: NIH; Contract grant number: PO1 AR48816 (To GSS). BH was supported by a Doctoral Scholarship and a Doctoral Thesis grant from CONICYT.
References
- Bae J-S, Gutierrez S, Narla R, Pratap J, Devados R, Wijnen AJ, Stein JL, Stein GS, Lian JB, Javed A. Reconstitution of Runx2/Cbfa1-Null Cells Identifies a Requirement for BMP2 Signaling Through a Runx2 Functional Domain During Osteoblast Differentiation. J Biol Chem. 2007;100:434–449. doi: 10.1002/jcb.21039. [DOI] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhao M, Rao R, Inoue H, Hao C-M. C/EBPβ and Its Binding Element Are Required for NFκB-induced COX2 Expression Following Hypertonic Stress. J Biol Chem. 2005;280:16354–16359. doi: 10.1074/jbc.M411134200. [DOI] [PubMed] [Google Scholar]
- Cruzat F, Henriquez B, Villagra A, Hepp M, Lian JB, van Wijnen AJ, Stein JL, Imbalzano AN, Stein GS, Montecino M. SWI/SNF-Independent Nuclease Hypersensitivity and an Increased Level of Histone Acetylation at the P1 Promoter Accompany Active Transcription of the Bone Master Gene Runx2. Biochemistry. 2009;48:7287–7295. doi: 10.1021/bi9004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan P, Peng X, Sutton ALM, MacDonald PN, Croniger CM, Trautwein C, Centrella M, McCarthy TL, Christakos S. Functional Cooperation between CCAAT/Enhancer-Binding Proteins and the Vitamin D Receptor in Regulation of 25-Hydroxyvitamin D3 24-Hydroxylase. Mol Cell Biol. 2005;25:472–487. doi: 10.1128/MCB.25.1.472-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi H, Luc Q, Shakoori R, Chuva de Sousa Lopes S, Choi J-Y, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PVN, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT Signaling Promotes Osteogenesis by Directly Stimulating Runx2 Gene Expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- Gutierrez S, Javed A, Tennant D, van Rees M, Montecino M, Stein GS, Stein JL, Lian JB. CCAAT/Enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- Harrison JR, Huang Y-J, Wilson AK, Kelly PL, Adams DJ, Gronowicz GA, Clark SH. Col1a1 Promoter-targeted Expression of p20 CCAAT Enhancer-binding Protein β (C/EBPβ), a Truncated C/EBPβ Isoform, Causes Osteopenia in Transgenic Mice. J Biol Chem. 2005;280:8117–8124. doi: 10.1074/jbc.M410076200. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. BMP2 Commitment to the Osteogenic Lineage Involves Activation of Runx2 by DLX3 and a Homeodomain Transcriptional Network. J Biol Chem. 2006;281:40515–40526. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- Hata K, Nishimura R, Ueda M, Ikeda F, Matsubara T, Ichida F, Hisada F, Nokubi T, Yamaguchi A, Yoneda T. A CCAAT/Enhancer Binding Protein β Isoform, Liver-Enriched Inhibitory Protein, Regulates Commitment of Osteoblasts and Adipocytes. Mol Cell Biol. 2005;25:1971–1979. doi: 10.1128/MCB.25.5.1971-1979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Bae J-S, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Structural Coupling of Smad and Runx2 for Execution of the BMP2 Osteogenic Signal. J Biol Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G. Transcriptional Control of Skeletogenesis. Annu Rev Genom Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Pless O, Dittmar G, Knoblich M, Leutz A. Crosstalk between C/EBPβ phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 2010;29:1105–1115. doi: 10.1038/emboj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-H, Kim Y-J, Yoon W-J, Kim J-I, Kim B-G, Hwang Y-S, Wozney JM, Chi X-Z, Bae S-C, Choi K-Y, Cho J-Y, Choi J-Y, Ryoo H-M. Dlx5 specifically regulates Runx2-II expression by binding to homeodomain response elements in the Runx2 distal promoter. J Biol Chem. 2005;280:35579–35587. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Hassan MQ, Serra RW, Lepper C, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Nkx3.2-mediated repression of Runx2 promotes chondrogenic differentiation. J Biol Chem. 2005;280:15872–15879. doi: 10.1074/jbc.M411144200. [DOI] [PubMed] [Google Scholar]
- Li H, Gade P, Nallar SC, Raha A, Roy SK, Karra S, Reddy JK, Reddy SP, Kalvakolanu DV. The Med1 Subunit of Transcriptional Mediator Plays a Central Role in Regulating CCAA/Enhancer-binding Protein-β-driven Transcription in Response to Interferon-γ. J Biol Chem. 2008;283:13077–13086. doi: 10.1074/jbc.M800604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen A, Stein J, Stein G. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- Majeska RJ, Rodan SB, Rodan GA. Parathyroid hormone-responsive clonal cell lines from rat osteosarcoma. Endocrinology. 1980;107:1494–1503. doi: 10.1210/endo-107-5-1494. [DOI] [PubMed] [Google Scholar]
- Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- Mink S, Haenig B, Klempnauer K-H. Interaction and Functional Collaboration of p300 and C/EBPβ. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras Induces Mediator Complex Exchange on C/EBPβ. Mol Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA Binding Specificity of the CCAAT/Enhancer-binding Protein Transcription Factor Family. Biochemistry. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- Otto F, Kanegane H, Mundlos S. Mutations in the RUNX2 Gene in Patients With Cleidocranial Dysplasia. Hum Mutat. 2002;19:209–216. doi: 10.1002/humu.10043. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, Beddington RSP, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa, a candidate gene for Cleidocranial Dysplasia Syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Paredes R, Arriagada G, Cruzat F, Villagra A, Olate J, Zaidi K, van Wijnen A, Lian JB, Stein GS, Stein JL, Montecino M. Bone-specific transcription factor Runx2 interacts with the 1a,25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells. Mol Cell Biol. 2004;24:8847–8861. doi: 10.1128/MCB.24.20.8847-8861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes R, Gutierrez J, Gutierrez S, Allison L, Puchi M, Imschenetzky M, van Wijnen A, Lian J, Stein G, Stein J, Montecino M. Interaction of the 1alpha,25-dihydroxyvitamin D3 receptor at the distal promoter region of the bone-specific osteocalcin gene requires nucleosomal remodelling. Biochem J. 2002;363:667–676. doi: 10.1042/0264-6021:3630667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless O, Kowenz-Leutz E, Knoblich M, Lausen J, Beyermann M, Walsh MJ, Leutz A. G9a-mediated Lysine Methylation Alters the Function of CCAAT/Enhancer-binding Protein-β. J Biol Chem. 2008;283:26357–26363. doi: 10.1074/jbc.M802132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramji D, Foka P. CCAAT/enhancer-binding proteins:structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Christakos S. The Vitamin D Receptor, Runx2, and the Notch Signaling Pathway Cooperate in the Transcriptional Regulation of Osteopontin. J Biol Chem. 2005;280:40589–40598. doi: 10.1074/jbc.M504166200. [DOI] [PubMed] [Google Scholar]
- Shin CS, Jeon MJ, Yang J-Y, Her S-J, Kim D, Kim SW, Kim SY. CCAAT/enhancer-binding protein δ activates the Runx2-mediated transcription of mouse osteocalcin II promoter. J Mol Endocrinol. 2006;36:531–546. doi: 10.1677/jme.1.01944. [DOI] [PubMed] [Google Scholar]
- Shirakawa K, Maeda S, Gotoh T, Hayashi M, Shinomiya K, Ehata S, Nishimura R, Mori M, Onozaki K, Hayashi H, Uematsu S, Akira S, Ogata S, Miyazono K, Imamura T. CCAAT/Enhancer-Binding Protein Homologous Protein (CHOP) Regulates Osteoblast Differentiation. Mol Cell Biol. 2006;26:6105–6116. doi: 10.1128/MCB.02429-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A, Arriagada G, Olate J, Imschenetzky M, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol Cell Biol. 2003;23:3339–3351. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smink JJ, Begay V, Schoenmaker T, Sterneck E, de Vries TJ, Leutz A. Transcription factor C/EBPβ isoform ratio regulates osteoclastogenesis through MafB. EMBO J. 2009;28:1769–1781. doi: 10.1038/emboj.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis L. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. CCAAT/Enhancer-binding Protein β Promotes Osteoblast Differentiation by Enhancing Runx2 Activity with ATF4. Mol Biol Cell. 2008;19:5373–5386. doi: 10.1091/mbc.E08-03-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villagra A, Cruzat F, Carvallo L, Paredes R, Olate J, Wijnen AV, Stein GS, Lian JB, Stein JL, Imbalzano AN, Montecino M. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein β-dependent recruitment of SWI/SNF activity. J Biol Chem. 2006;281:22695–22706. doi: 10.1074/jbc.M511640200. [DOI] [PubMed] [Google Scholar]
- Wiper-Bergeron N, St-louis C, Lee JM. CCAAT/Enhancer Binding Protein β Abrogates Retinoic Acid-Induced Osteoblast Differentiation via Repression of Runx2 Transcription. Mol Endocrinol. 2007;21:2124–2135. doi: 10.1210/me.2006-0452. [DOI] [PubMed] [Google Scholar]
- Xiong W, Hsieh C-C, Kurtz AJ, Rabek JP, Papaconstantinou J. Regulation of CCAAT/enhancer-binding protein-β isoform synthesis by alternative translational initiation at multiple AUG start sites. Nucleic Acids Res. 2001;29:3087–3098. doi: 10.1093/nar/29.14.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambotti A, Makhluf H, Shen J, Ducy P. Characterization of an osteoblast-specific enhancer element in the CBFA1 gene. J Biol Chem. 2002;277:41497–41506. doi: 10.1074/jbc.M204271200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hassan M, Xie R-L, Hawse JR, Spelsberg TC, Montecino M, Stein JL, Lian JB, van Wijnen A, Stein GS. Co-stimulation of the Bone-related Runx2 P1 Promoter in Mesenchymal Cells by SP1 and ETS Transcription Factors at Polymorphic Purine-rich DNA Sequences (Y-repeats) J Biol Chem. 2009;284:3125–3135. doi: 10.1074/jbc.M807466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) ROS17/2.8 osteoblastic cells were transiently transfected with 250 ηg of the p600P1rRunx2-Luc reporter construct alone or in combination with increasing amounts of the pC/EBPβ (C/EBPβ/LAP*) expression plasmid. C/EBPβ protein over-expression was confirmed by Western blot (bottom panel) using antibodies against X-press to detect the over-expressed Xpress-tagged C/EBPβ/LAP* (o.e.) or antibodies against C/EBPβ. To control for equal protein loading the membranes were re-blotted against TFIIB. B) The contribution of the putative C/EBP sites I and II to the Runx2 P1 promoter activity in osteblastic cells, was assessed by transient transfection in ROS17/2.8 cells. Cells were transfected with the p288P1rRunx2-Luc reporter plasmids (250 ηg each), including intact or mutated versions of the C/EBP sites I and II, either independently or combined, as indicated under each graph. The results were expressed as percentage (%) of control activity, considering as the control the relative luciferase activity of the wild-type p288P1rRunx2-Luc construct (100%). The reporter activity and statistical significance were determined as described in figure legend 3.
C2C12 cells were transiently transfected with the p600P1rRunx2-Luc reporter construct (250 ηg), alone or in combination with the C/EBPβ-expression plasmid (pC/EBPβ) or the constructs that selectively express the different C/EBPβ isoforms LAP* (pC/EBPβ-LAP*) or LAP (pC/EBPβ-LAP). The reporter activity and statistical significance were determined as described in figure legend 3.
A) Schematic representation of the Runx2 P1 promoter indicating the C/EBP and Runx binding sites identified in this region and the positions of the primers used to amplify the promoter segments for EMSA and ChIP. B) Sequences of the different primers and oligonucleotides used for EMSA and ChIP experiments.