Abstract
Although protein–RNA interactions (PRIs) are involved in various important cellular processes, compiled data on PRIs are still limited. This contrasts with protein–protein interactions, which have been intensively recorded in public databases and subjected to network level analysis. Here, we introduce PRD, an online database of PRIs, dispersed across several sources, including scientific literature. Currently, over 10,000 interactions have been stored in PRD using PSI-MI 2.5, which is a standard model for describing detailed molecular interactions, with an emphasis on gene level data. Users can browse all recorded interactions and execute flexible keyword searches against the database via a web interface. Our database is not only a reference of PRIs, but will also be a valuable resource for studying characteristics of PRI networks.
Availability
PRD can be freely accessed at http://pri.hgc.jp/
Keywords: protein-RNA interaction, Biomolecular interaction, RNA binding protein, Database
Background
Interactions between proteins and RNAs (Protein–RNA interactions; PRIs) are essential events that control a variety of cellular processes, such as RNA splicing, transport, stabilization, and translation [1]. There are some well-known databases (e.g., BioGRID [2] and IntAct [3]) comprising comprehensive collections of biomolecular interactions that include PRIs. However, the available PRI data remain limited compared with protein–protein interaction data, which have been collected and intensively analyzed at the network level. In addition, it is difficult to compile a list of PRIs by searching and browsing these databases. Therefore, the nature of networks comprising interactions between proteins and RNAs is still unclear. There are several other databases that deal with PRIs (e.g., [4, 5]); however, those databases focus on relatively limited aspects or subsets of available PRI data, such as interactions based on 3D complex structures. In this paper, we introduce PRD, an online database for consolidating various types of PRI data dispersed across several online data sources. Our database is not only a reference of PRIs, but will also be a valuable resource for exploring the characteristics of PRI networks.
Methodology
Overview of the database:
PRD is a database for storing physical interaction data between protein and RNA molecules, with particular emphasis on interactions at the gene level. Currently, PRD contains data on 10,817 interactions, which is the equivalent of 1,539 unique gene pairs. More than two-thirds of the data in PRD are from humans and yeast. A large proportion of the rest are derived from model organisms, such as Mus musculus and Escherichia coli. Although PRD primarily stores information about interactions between proteins and protein-coding RNAs, it also contains interaction data on transfer RNAs, ribosomal RNAs, microRNAs, and viral RNAs.
Interaction data model and curation:
The interaction data model in PRD closely follows the HUPO PSI-MI model [6]. Although the PSI-MI model was developed with a focus on the interactions among proteins, we adopted it as a base model to facilitate exchange between different biomolecular interaction databases and to allow subsequent analyses. Briefly, the data model used in PRD can store two types of information. The first one concerns interactors (i.e., proteins and RNA), such as gene and taxonomic names. Auxiliary information, such as binding region/motif and the participant identification method can also be stored. The second type of information concerns the interaction between two interactors, such as detection interaction methods and biological functions deduced from those experiments. Using this data model, we extracted as much information as possible from the literature and deposited it in PRD. Moreover, controlled vocabulary terms defined in PSI-MI 2.5 [7] were used in the curation process, when appropriate terms were available.
Imported data:
To increase the coverage of the database, we imported data from other public databases: BioGRID [2] and IntAct [3]. In addition, we obtained a list of complexes containing both protein and RNA chains from the Protein Data Bank of Japan (PDBj), and imported them as PRI pairs if the atoms without hydrogen were within 5Å of each other. Currently, we have confirmed 65, 267, and 10,145 interactions from BioGRID, IntAct, and PDBj, respectively.
PRD interface and access
Searching and browsing interactions:
PRD can be searched by gene names, Entrez GeneIDs, PubMed IDs, or by free keywords (Figure 1). Users can also browse all the data in the database by clicking the ‘browse’ button. PRD supports several field specifiers to limit the scope of searches. The use of logical operators (AND, OR, and NOT) to join two or more expressions within a single query is permitted.
Figure 1.
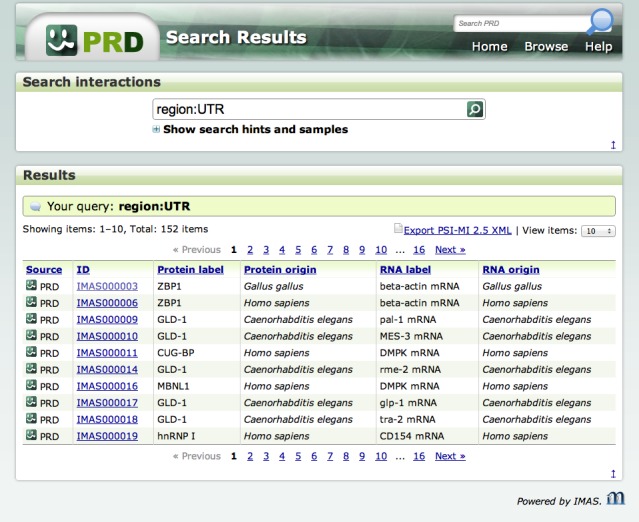
A snapshot of the PRD web interface. The Figure is a screenshot of the search results page for a certain query.
Displaying interaction information:
Each ‘Interaction information’ page corresponds to an individual interaction, and contains information about the two participants of the interaction (i.e., protein and RNA molecules), and related information. In the ‘Protein-’ and ‘RNA-’ information section on each page, gene names, taxonomy names, and corresponding accession numbers for each molecule are displayed as basic information with hyperlinks to external databases. In this section, information about binding sites, Gene Ontology terms, protein motifs/domains based on the InterPro [8], and RNA sequence motifs based on the UTRsite [4] and Rfam [9] databases are also displayed, when those have been assigned. An ‘Experiments’ section displays the detection methods used for each interaction. In addition, biological functions are displayed if they have been recorded. References describing the corresponding interactions are presented on each page with hyperlinks to PubMed.
Data export:
PRD has the ability to export PSI-MI XML 2.5 files [6], which are widely supported by various existing software (e.g., Cytoscape [10]), and allow users to save and analyze exported data on local computers.
Future developments
Many articles remain to be curated. To compensate for missed articles describing PRIs, we plan to implement a function allowing users to directly deposit interaction data via a web form.
Acknowledgments
We would like to thank Dr. Toru Tsuji for data curation. We also thank Dr. Nobuhide Doi, Dr. Hiroshi Yanagawa, and Dr. Koichi Takeda for their helpful comments on the database construction. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas “Integrative Systems Understanding of Cancer for Advanced Diagnosis, Therapy and Prevention (No. 4201)” (23134510) of The Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Citation:Fujimori et al, Bioinformation 8(15): 729-730 (2012)
References
- 1.JD Keene, et al. Endocrinology. 2010;151:1391. [Google Scholar]
- 2.C Stark, et al. Nucleic Acids Res. 2011;39:D698. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S Kerrien, et al. Nucleic Acids Res. 2012;40:D841. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.G Grillo, et al. Nucleic Acids Res. 2010;38:D75. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.BA Lewis, et al. Nucleic Acids Res. 2011;39:D277. doi: 10.1093/nar/gkq1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S Kerrien, et al. BMC Biol. 2007;5:44. [Google Scholar]
- 7. http://www.psidev.info/mif/
- 8.S Hunter, et al. Nucleic Acids Res. 2012;40:D306. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.PP Gardner, et al. Nucleic Acids Res. 2011;39:D141. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ME Smoot, et al. Bioinformatics. 2011;27:431. [Google Scholar]


