Abstract
Wolbachia surface protein (WSP) is an eight beta-barrel transmembrane structure which participates in host immune response, cell proliferation, pathogenicity and controlled cell death program. The protein has four extracellular loops containing hyper variable regions separated by conserved regions. The WSP structure is homologous to Neisseria surface protein (Nsp A) which has about 34% similarity including antigenic variation and hydrophilicity. Recombination has a large impact on diversity of this protein including positive selection which is major constraint on protein evolution. The molecular mechanism through which Wolbachia induces various reproductive anomalies is unclear; a key feature observed for such anomalies might be because of Wolbachia undergoing extensive recombination. In Wolbachia, increased recombination is observed in ankyrin proteins, surface proteins and in some hypothetical proteins. Genetic divergence is extensive in the WSP gene, WSP is known to be a chimeric protein involved in host-symbiont interactions. Here we predicted the structural and functional variations in WSP sequences of Wolbachia present in D. melanogaster, A. tabida and in B. malayi.
Keywords: Wolbachia, OMP, WSP, Drosophila melanogaster, Asobera tabida, Brugaria malayi, Drug target, 3D protein structure
Background
Wolbachia is a type of gram negative bacterium commonly found in arthropods and nematodes. It is transmitted in host cytoplasm and is known to cause reproductive alterations such as cytoplasmic incompatibility, male killing, parthenogenesis, and feminization [1]. The mechanism that causes these reproductive alterations is unknown and it is suspected that Wolbachia proteins would play a decisive role in causing reproductive manipulations by interacting with the other host proteins [2]. The outer membrane of gram negative bacteria contains porins, which help to transfer molecules, and lipopolysaccarides [3]. Analysis of Wolbachia genomes shows a pattern of paralogous genes which have similar function, this copy is free to mutate and acquire new functions. Outer membrane protein (OMP) performs various functions including import and export of nutrients, and export of virulence factors or protein through bacterial type IV secretion; OMP also has a role in positive selection and mediates host bacterial interactions [4]. OMPs are highly variable and are among the fastest evolving microbial proteins [5, 6]. OMPs show a characteristic transmembrane β-barrel structure, formed by an even number of antiparallel sheets, connected to loops of variable length (including hyper variable and conserved regions) at the extracellular side and to short turns containing both N and C termini at the periplasmic side [7]. Keeping this in view here we investigate one such outer membrane protein of Wolbachia which is known as Wolbachia surface protein with molecular weight of 24 KD and know to be mosaic in nature. [8–10]. The reduced constraints in the overall protein structure is expected based on WSP's large amino acid variation and extensive differences in length of the exposed loops. WSP must be particularly versatile, in order to accommodate such variability, which appears to be maintained by a constant synonymous variants per protein type. Nevertheless some nucleotide haplotypes are particularly common, suggesting recent spread (via lateral gene transfer or strain transfer) rather than maintenance by selection [11]. From this we had concluded that recombination have played a role in larger extent in maintaining the diversity of Wolbachia strains causing different phenotypes. In order to provide necessary information about the variation of this protein isolated from different sources. Here we investigate the molecular evolution, the prediction of protein structure and the biophysical properties of WSP in D. melanogaster, A. tabida and B. malayi, which help in identification of drug target.
Methodology
Sequence selection:
The protein sequences of Drosophila melanogaster (NP_966785.1), Asobara tabida (AAW78819.1) and Bulgaria malayi (AAW70873.1) were selected from the SWISSPROT database, for analysis of structural variations in WSP of a Wolbachia endosymboint.
Evolutionary analysis of protein sequences and structures:
The evolutionary changes present in protein sequences on D. menalogaster, A. tabida and B. malayi was predicted using MEGA software (http://www.megasoftware.net/) and structural variations were identified using Phyre online server (http://www.sbg.bio.ic.ac.uk/~phyre/) to find the fold recognition.
Protein structure prediction:
The biophysical properties of primary protein sequences were predicted using Protparam,. Three dimensional protein structures were predicted using SOPMA and Swiss Model. The three dimensional structure can be used for potential drug targets.
Homology Modeling:
The three-dimensional structure of WSP protein was modeled using MODELLER software and structural variations present in D. menalogaster, A. tabida and B. malayi were predicted.
Structure validation and active site prediction:
The modeled WSP structure was used for structure validation using SAVS (Structure Validation and Analysis Server). The PROCHECK checks the stereo chemical quality of a protein structure, producing a number of PostScript plots analyzing its residue-by-residue geometry. The active site prediction was identified by using CastP; these results show the surface topography of active site amino acids.
Result
Sequence similarity search:
The protein sequence of WSP was submitted into Protein BLAST and picked out its homologs with the help of PDB. The template crystal structure of Neisserial Surface Protein A (NspA) [PDB ID: 1P4T] from Neisseria meningitides showing 34% identity with the given target sequence (Figure 1).
Figure 1.
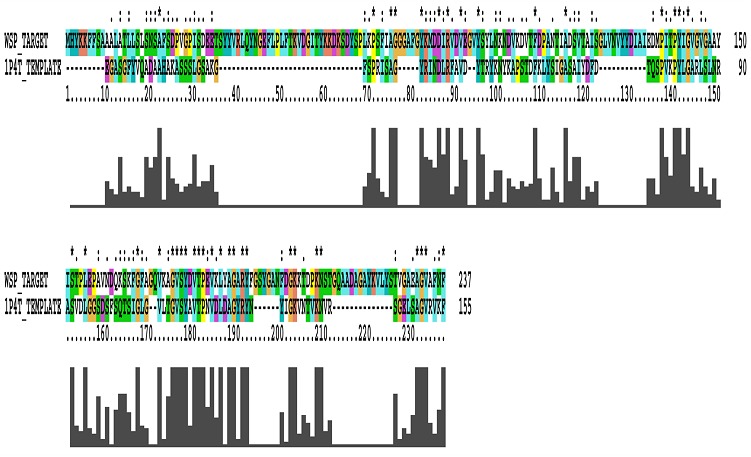
An alignment between WSP (Target) and Nspa (Template).
Genetic analysis of protein sequences:
The genetic variations of WSP sequence was analyzed using MEGA software. The multiple sequence alignment and statistical data calculations in D. melanogaster, A. tabida and B. malayi have evolutionarily diverged. The results indicate that the sequence alignment and distribution amino acids arrangement in WSP is highly variable. The multiple alignments of WSP results show 0.021, 1.271 and 1.271 distances. The total mean distance is 0.0855.
Protein structural prediction:
The primary structure prediction of WSP sequences in D. melanogaster, A. tabida and B. malayi shows instability index of 23.72, 21.99 and 30.79 respectively. The complexity of all protein structures is stable. The grand average of hydropathicity of all three organism proteins also shows -0.122, -0.227 and -0.386 respectively. The variability of amino acids in all these organisms is varying depending on molecular weight, hydropathycity and aliphatic index, indicating small evolutionary changes present in protein sequences. The secondary structural variations were predicted using SOPMA. The structural alignment of α-helices, β-sheets, and random coils for these three organisms' are shown in Table 1(see supplementary material).
Three dimensional protein structure predictions:
The three dimensional structure of WSP in D.melanogaster, A.tabida and B.malayi shows 8-stranded beta barrel of approximately 80.747 Å. The pair-wise sequence alignment is shown in (Figure 1).The three dimensional structures of protein sequences in WSP in D.melanogaster, A.tabida and B.malayi are shown in (Figure 2). The three dimensional modelled structure WSP is 8-stranded beta barrel ~80 long with extended loops on C-terminals exposed towards extracellular surface. The Nterminus of the 3D-structure is towards periplasmic space.
Figure 2.
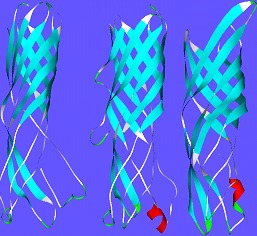
Three dimensional structure of WSP in D.melanogaster, B.malayi and A.tabida. The WSP in D.melanogaster structure, the red gating helix is absent unlike in A. tabida & B.malayi, the B.malayi has higher internal radius than others.
Homology Modelling:
The sequence identity of WSP in D.melanogaster, A.tabida and B.malayi is identical to outer membrane protein NspA from Neisseria meningitides (PDB ID: 1P4TA), which is a promising vaccine candidate because of its highly conserved character among meningococcal strains and because it induces bactericidal antibodies. NspA is a homolog of the Opa proteins, which mediate adhesion to host cells. In model protein structure the minimum and maximum barrel lengths are 18.7Å and 26.4 Å respectively. The maximum barrel length is similar to the width of porins too (2POR). This thickness is not sufficient to transverse the bacterial lipid membrane, which is approximately 80Å thick. However, in experiments with OMP W of E.coli, of similar barrel length, it has shown to work as a channel to transport small compounds across the planar lipid bilayers [12].
The template structure on WSP in D.melanogaster and A.tabida shows one salt bridge Ile74-ser195, two Tyrosines 82 and 192 blocks the channel by making a barrier, protected by salt bridge on each side. B.malayi is Glu148-val217 having salt bridge and tyrosine blocks 157, 158, 159 and end with tyr212. The coordination of proline 110, 137, 140, 154, 157, 182 residues in the cavity is charged, blocking the way unlike as in porin, and B. malayi is present the porins on 169 and 190 positions. The overall modelled structure of WSP in D. melanogaster, A. tabida shows 8-stranded beta-barrel that is approximately 80 Å long with extended loops exposed towards the extracellular surface. The N terminus of the modelled structure is towards periplasmic space. The 32 residues on the N-terminus are missing. The peptide groups are on amino acid positions 105- 163 and 70-128. In the WSP of B.malayi the peptide groups are on positions 147-161 and 162-171. For D.melangaster, A.tabida and B.malayi electrostatic surface interaction is present at -2758.73, - 2328.22 and -5086.79 respectively. The proteolytic cleavage helps for signaling peptide cleavage from protein transport in to the cytoplasm region. The active sites identified through of WSP in D. melanogaster, A. tabida and B. malayi is as shown in the Table 2 (see supplementary material).
Discussion
It seems to be a necessary application of Biophysical methods, especially structural ones (X-ray crystallography and NMR) to characterize molecular process of proteins involved in interaction with the host. There are numerous examples of studies of entire complexes between proteins and their ligands that provide direct or reliable evidence related to structure and function of the protein. Further the Insilco predictions of protein complexes will help in revealing the interactions with the host which greatly enhances the hypothesis and experimental investigations. The mechanisms of Wolbachia-host interaction are important in understanding of the bacterial life and their pathogenicity as well as generation of new drug targets. Wolbachia are a diverse group of intracellular bacteria that show impressive adaptations within invertebrate cells and in manipulating the biology of their hosts.
The WSP plays a crucial role in causing various reproductive manipulations by interacting with the host proteins. The structure of WSP contains porins and acts an ion channel which helps in the transfer of molecules. The Insilco prediction of WSP structure had similarity to NspA which shows about 34% in D. melanogaster, A. tabida and B. malayi. The genetic analysis and molecular evolution of WSP in B. malayi is less diverged than D. menalogaster & A. tabida, the evolutionary difference shows 0.021, 1.271 and 1.271 distances respectively. The total mean distance is 0.0855. This result indicates that WSP is less diverged in all these species. The structural prediction indicates that the WSP is a stable protein with various instability indexes as shown in the results. The distribution of amino acids in WSP complex protein Table 3 (see supplementary material) shows random distribution of alpha helix, extended strands, beta turns and more distribution of random coils from all these organisms.
The three dimensional structure of WSP were predicted using Swiss Model. The WSP complex structure shows 8-stranded beta barrel weight of ~80.747 Å, length 18.7Å and width of 26.4 Å which is similar to the width of porins too, it is present in cytoplasmic or periplasmic membrane of arthropods and nematodes. The C-terminal contains phenylalanine which binds to bacterial membrane and helps for apoptosis in B. malayi and A. tabida, but absent in the case of D. menalogaster. The gene encoding WSP shows homology with genes encoding outer membrane protein of the closely related to NspA. The predicted complex structure were observed in PROCHECK 3D-validation shows 84.15% of steriochemical rotation of torsion angles and no amino acid changes in protein complex. The identification of active site amino acids present in WSP complex structure was predicted using CastP. A greater number of variable active sites are present in these organisms and these protein structures can be used for drug binding sites. In WSP, there is less recombination impact and divergence of amino acids distribution in D. melanogaster and A. tabida but more diverged in B. malayi.
The complex protein shows possibility of polymorphism as its significant feature of evolution. However, it has been shown that recombination and selection have played a key role in adaptive evolution [11]. This variation may be the effect of mutation and recombination. However, we conclude the remote possibility of polymorphism in WSP due to positive selection [4], further such variation in course of host pathogen interactions having variable antigenic property, goodwill to sexual selection, and as potential drug targets has been proved. The host pathogen interaction and immune response is supported by positive selection, although it needs further confirmed by the replication of the results of our study.
Conclusion
Generally, most of the nematode depends on this bacterium Wolbachia for their survival. For e.g. Brugia malayi, Wuchereria bancrofti, Onchocerca volvulus, etc., nematode which cause Lymphatic filariasis and River Blindness in humans are so much dependent on Wolbachia that in the absence leads to reduced growth rates followed by eventual death. The initial work on Wolbachia surface protein as shown that it may function to inhibit apoptosis of host, antigenic role and it shows strong immune response against human who are infected with filariasis.
The Biophysical characterization of the protein would provide the clues to its biological functions, physiological role. Although work has been iniated in this regard, the comparison between different sources of Wolbachia or has not been undertaken. Here we predict the structure of Wolbachia surface protein from three different sources which behaves as parasitic in D.melanogaster, mutalistic in B.malayi, both parasitic and mutalistic in A.tabida. The comparative analysis will highlight the multiple roles of Wolbachia surface protein which extend to combating pests, major vectors specifically through gene drive strategies. Further the function of WSP would facilitate, and is a prerequisite for the development of new drug targets.
Supplementary material
Acknowledgments
The authors would like to thank to Bangalore University and DAE Board of research in Nuclear Sciences (BRNS) Grant No 2012/37B/BRNS/No.953 for their financial assistance during the work. We would like to thank our team members for their contribution and support. First author would like to thank Dr. Manish Chandra Patak, IISER Bhopal, ITI (Gas Rahat) Building, Govindpura, Bhopal 462023 India for his review and also would like to thank Mr.S.Sampath Kumar, Mr. C.H. Huchesh, Mr. H. Ravikumar, Dr. N.S.Surendra, Mr. Kunal A and ChethanaV.C for their support during the study.
Footnotes
Citation:Uday & Puttaraju, Bioinformation 8(15): 711-715 (2012)
References
- 1.F Vavre, et al. Adv Parasitol. 2009;70:299. doi: 10.1016/S0065-308X(09)70012-0. [DOI] [PubMed] [Google Scholar]
- 2.HR Braig, et al. J Bacteriol. 1998;180:2373. doi: 10.1128/jb.180.9.2373-2378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang, Haibin Veterinary Biosciences. 2006 [Google Scholar]
- 4.FM Jiggins, et al. Mol Biol Evol. 2002;19:1341. doi: 10.1093/oxfordjournals.molbev.a004195. [DOI] [PubMed] [Google Scholar]
- 5.Y Zheng, et al. Nucleic Acids Res. 2004;32:6347. doi: 10.1093/nar/gkh935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WC Wimley. Curr Opin Struct Biol. 2003;13:404. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 7.L Baldo, et al. J Bacteriol. 2005;187:5406. doi: 10.1128/JB.187.15.5406-5418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DA Haake, et al. J Bacteriol. 2004;186:2818. doi: 10.1128/JB.186.9.2818-2828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FM Jiggins. J Mol Evol. 2006;62:99. doi: 10.1007/s00239-005-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.B Malorny, et al. J Bacteriol. 1998;180:1323. doi: 10.1128/jb.180.5.1323-1330.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L Baldo, et al. BMC Evol Biol. 2010;10:48. doi: 10.1186/1471-2148-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.H Hong, et al. J Biol chem. 2006;281:7568. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


