Abstract
Hepatitis D is a liable reason of mortality and morbidity worldwide. It is caused by an RNA virus known as Hepatitis Delta Virus (HDV). Genetic studies of HDV have shown that delta antigen protein is responsible for replication of genome and play a foremost role in viral infection. Therefore, delta antigen protein may be used as suitable target for disease diagnosis. Viral activity can be restrained through RNA interference (RNAi) technology, an influential method for post transcriptional gene silencing in a sequence specific manner. However, there is a genetic variability in different viral isolates; it is a great challenge to design potential siRNA molecules which can silence the respective target genes rather than any other viral gene simultaneously. In current study two effective siRNA molecules for silencing of HDV were rationally designed and validated using computational methods, which may lead to knockdown the activity of virus. Thus, this approach may provide an insight for the chemical synthesis of antiviral RNA molecule for the treatment of hepatitis D, at genome level.
Keywords: Antiviral, RNAi, siRNA, Thermodynamics
Background
Hepatitis Delta virus (HDV) is a human sub viral pathogen which is able to infect only such individuals who were previously or are simultaneously infected with Hepatitis B virus [1]. According to Kuo et al. HDV is able to replicate its RNA within cells in the absence of hepatitis B virus but require hepatitis B antigen for packaging and release of HDV virion [2]. HDV currently infects about 20 million people worldwide [3] and is most common among populations using inject-able drugs particularly in countries bordering the Mediterranean Sea. It is least common in Eastern Asia, but is also present in Taiwan, China and India. Most children with HDV have been identified in Italy and Greece, with a few in Northern Africa. Genetic diversity of HDV is related to the geographic origin of the various isolates. Apart from HDV-1, which is ubiquitous, each virus clad is geographically localized: HDV-2 (previously labeled HDV-IIa) is found in Japan [4], Taiwan [5], and Yakutia, Russia [6]; HDV-4 (previously labeled HDV-IIb) in Taiwan [7] and Japan [8, 9]; HDV-3 in the Amazonian region [10] and HDV-5, HDV-6, and HDV- 7 in Africa. The studies of Radjef et al. shows that more than seven clades exist and characterized a sequence (dFr644) that was not strongly affiliated to any of the seven HDV clades [11]. Frédéric et al. described two HDV isolates (dFr2072 and dFr2736) that have strong phylogenetic relation to dFr644 reported by Radjef et al., and proposed as 8th clade [12]. The whole genome of HDV was cloned and sequenced in 1986 [13], which is a single stranded circular RNA of about 1,700nt [14]. Genome replication accumulates three RNA species [15, 16]. The genome and its exact complement, the antigenome, are with unit length and exist primarily in a circular conformation but also in a linear conformation [17]. The third RNA species with relatively lower amounts of an 800-nt polyadenylated RNA (antigenome polarity), is translated to produce a 195-amino-acid protein, known as the delta antigen.
Delta antigen is essential for HDV genome replication [18]. Clinical association of HDV with hepatitis B infection is due to the fact that the outer coat of HDV consists of hepatitis B virus surface glycoprotein [19]. Therefore immune prophylaxis against HDV is achieved by vaccination against hepatitis B virus. This mode of prevention is effective only in case of coinfections in hepatitis B virus susceptible individuals [3] as it fails to show any significant effect in case of super-infection, which is more serious state of health [3]. So there is an urgent need to identify the suitable therapeutic molecule for the treatment of HDV.
RNA interference (RNAi), an evolutionary conserved gene silencing mechanism, uses short double-stranded RNA (dsRNA) to trigger degradation or translation repression of homologous RNA targets in a sequence-specific manner. This has been used as alternative antiviral therapy [20]. RNAi therapy has been successful in many pathogenic infection as well as genetic disorders, which interferes with expression of disease causing genes. RNAi was first discovered in Caenorhabditis elegans [21] and plants, [22], but it can also be used to induce gene silencing in a diverse range of organisms including fungi, protozoans and metazoan animals.
Small interfering RNAs (siRNAs), double-stranded RNAs typically of length between 19 and 25 with 2 nucleotide overhangs on the 3' ends, that are capable of inducing sequencespecific, post-transcriptional gene silencing. Naturally occurring siRNAs are cleavage products of long double-stranded RNAs (dsRNAs) by Dicer, a ribonuclease III enzyme [23, 24]. The siRNA-induced mRNA degradation is a complicated process involving multiple steps, initiated by the binding of siRNA with RNA induced silencing complex (RISC), followed by RISC's activation, resulting in the recognition of the target mRNA and its degradation [23, 25, 26]. As a gene knockdown tool used in labs, siRNAs can also be chemically synthesized and introduced into the cells by direct transfection [27, 28] or delivered into the cells in forms of hairpin precursors through plasmid or viral vectors [29, 30]. The siRNA-based gene knock-down techniques are preferred by several investigators because of their ability to disrupt individual gene's function without affecting related genes [31]. The role of RNAi in mammalian innate immunity is poorly understood and relatively little data is available. However, the existences of viruses that encode genes are able to suppress the RNAi response in mammalian cells may be evidence in favor of an RNAi-dependent mammalian immune response [32, 33]. However, this hypothesis of RNAi-mediated immunity in mammals has been challenged as poorly substantiated [34]. But Elbashir et al. in their study found that these techniques are successful for gene silencing studies in mammalian cells because unlike longer double-stranded RNAs, siRNAs do not trigger interferon responses which lead to nonspecific mRNA degradation [27]. The RNAi technique is being successfully used as antiviral therapy in various known viral diseases [35–37]. Currently siRNA gene silencing technology is one of the significant aspects to block the expression of disease causing gene in many biological systems and helpful for gene level treatment. However, HDV genomic and antigenomic RNAs can fold into an unbranched rod-like structure with 74% of the bases paired [02], and this folding might interfere with siRNA action. The delta antigen protein has the ability to bind double stranded RNA [38] and thus might also interfere. Chang and Taylor study with nonreplicating HDV RNA sequences support the interpretation that neither the potential for intramolecular rod-like RNA folding nor the presence of the delta protein conferred resistance to siRNA. However, the genomic and antigenomic RNAs are resistant to siRNA action. The antigenome is localized in the nucleus and so could be inaccessible for RNAi action [39]. But the reason for the resistance of genomic RNA is unclear since a large amount of genomic RNA is cytoplasmic [40, 41]. Chang and Taylor subsequently showed that the delta antigen mRNA can be successfully targeted by siRNAs in cell culture and also verified that siRNA cannot target the replicating viral RNA transcripts directly but only indirectly via action on the viral mRNA species [39]. Thus, delta antigen protein coding mRNA of HDV is obligatory target to inhibit the RNA processing and may be suitable for antiviral therapy. Therefore, in the current study an attempt has been made to identify potential siRNA molecules for silencing of delta antigen coding mRNA or gene in HDV using computational approach.
Methodology
Data collection and analysis
Sixty eight genome isolate sequences of HDV were retrieved from viral GenBank database, available at http://www.ncbi.nlm.nih.gov/. The viral database contains all experimentally identified widespread genome isolates of HDV which were further used for siRNA designing Table 2 (see supplementary material).
Target identification and rational siRNA molecule designing:
siDirect 2.0 [42] (http://siDirect2.RNAi.jp/) tool, was used for target identification and designing of potential siRNA molecules. It utilized mixed rule approach of Ui-Tei, Amarzguioui and Reynolds rules [43] and melting temperature (Tm) below 21.5°C for siRNA duplex, as parameter. For further verification of predicted molecules GeneScript siRNA Target Finder (http://www.genescript.com/index.html), tool was also applied. Besides these other parameters were taken on the concept of algorithms given in Table 1 (see supplementary material).
Similarity search:
Blast tool (http://www.ncbi.nlm.nih.gov/blast) [44] was used to identify any off target sequence similarity in other non targeted organism's genome against whole Genebank datasets by applying expected threshold value 10 and BLOSUM 62 matrix as parameter. The target sites having similarity of more than 16 adjoining base pair with any other organism were excluded from the consideration.
GC calculation and siRNA secondary structure prediction:
GC calculator tool www.genomicsplace.com/gc_calc.html was used to calculate the GC content for selected siRNA molecule while secondary structure and free energy of folding was computed through Mfold server http://mfold.rna.albany.edu/?q=mfold/download-mfold.
Thermodynamics calculation of RNA-RNA interaction:
RNAup program (www.tbi.univie.ac.at/~ulim/RNAup) at Vienna web suit [45] was used to study the thermodynamics of interactions between target gene and predicted siRNA molecules. It works on extension of the standard partition function approach to RNA secondary structures that compute energetic of RNA-RNA interactions [46]. Flow chart shows complete methodology used for screening of effective siRNA molecules in this study (Figure 1).
Figure 2.
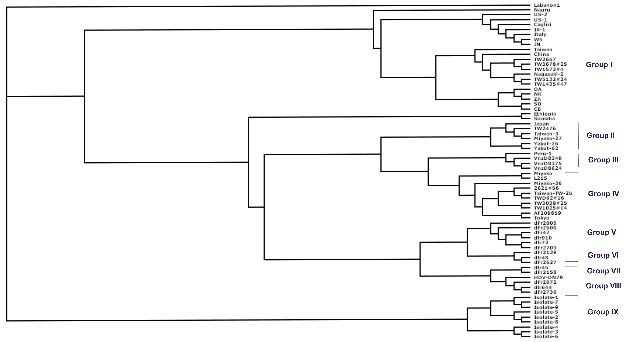
Phylogenetic tree for genome isolates of HDV available at NCBI Genebank database showing nine major clades or groups. Eight major clades of HDV were already known but in current analysis ninth one was proposed after phylogenetic study.
Discussion
The HDV genome exists as an enveloped negative sense, singlestranded, closed circular RNA. Because of a nucleotide sequence that is 70% self-complementary, the HDV genome forms a partially double stranded RNA structure that is described as rod-like. With a genome of approximately 1700 nucleotides, HDV is the smallest “virus” known to infect animals. On the basis of geographical origin of various isolates of HDV eight major clade were identified till date [12]. Sixty eight genome isolates of HDV are available in NCBI Genebank database which used in current study Table 2 (see supplementary material). Out of these sixty eight, nineteen sequences did not belong to any of the above described clade or groups. After phylogenic study of these nineteen sequences, five sequences were found to have strong similarity with group I; four sequences had similarity with group IV, while only one sequence had similarity to group VIII. Rest nine sequences out of these nineteen did not come under any group, but they had strong similarity with each other and so a new group or clad i.e. group IX (Figure 2) has been proposed. It has been earlier reported that delta antigen protein is responsible for virus replication and RNA processing [47]. Hence, this protein coding mRNA or gene from all genome isolates have immense potential for molecular diagnosis and used as target for siRNA molecule designing. There are diverse computational algorithms and tools existing for rational designing of siRNA molecules to knock down the activity of genes Table 1 (see supplementary material). siDirect 2.0 tool was used in current study to provide functional, target-specific siRNA molecules, which significantly reduces off-target silencing. To avoid offtarget effect, Tm for the seed-target duplex was calculated using the nearest neighbor model and the thermodynamic parameters for the formation of RNA duplex were also studied [48]. The formula for calculating Tm is:
Tm = {(1000 × ΔH) / (A + ΔS + R ln (CT/4))} - 273.15 + 16.6 log [Na+] ⇗ (Equation 1)
Where ΔH (kcal/ mol) is the sum of the nearest neighbor enthalpy change, A is the helix initiation constant (-10.8), ΔS are the sum of the nearest neighbor entropy change [49]. R is the gas constant (1.987 cal/deg/mol), and CT is the total molecular concentration of the strand (100 µM). [Na+] was fixed at 100 mM. Apart from it, to check the accuracy of result Gene script target Finder was also applied and usage statistical modeling method.
In present study three hundred fifty three siRNA targets were identified for small delta antigen of HDV and potential siRNA molecules against these targets were obtained using mixed rule approach i.e. Ui-Tei, Amarzguioui and Reynolds rule. Out of three hundred fifty three predicted siRNA targets, only sixty were following all three rules. Hence, these sixty siRNA targets were filtered out for further study and considered possible candidates. Consequently these sixty targets were subjected to NCBI Blast tool. Out of theses sixty target only forty eight were selected on the basis of low off target similarity Table 3 (see supplementary material). No acceptable siRNA molecule was identified for coding sequences of selected eight isolates (highlighted in Table 2 (see supplementary material). All the forty eight effective siRNA molecules selected on the basis of low off target similarity were assessed for target site convenience that can be suitable to knockdown the activity of HDV. MSA of these selected forty eight siRNA targets were depicted that forty one targets have precise similarity (Figure 3) while only seven sequences are least similar to them. Subsequently a consensus sequence was executed using EMBOSS tool for these forty one targets and siRNA molecules were designed against these consensus target. Therefore single siRNA might be used to knock down the activity of forty one isolates of HDV while rest seven molecules also create another consensus target and siRNA molecules against that target were also designed. After this analysis only two molecules were found appropriate for silencing of HDV delta antigen gene. However, there are the incompatible results regarding the effect of GC content and secondary structure on siRNA efficiency. Therefore, these parameters cannot be preferred as a primary determinant of siRNA efficiency. Still, it is recommended to choose sequences with low GC content (31- 58%) [50–52], in the present study both predicted siRNA molecules having recommended range of GC content. Furthermore, the possible folding of predicted siRNA molecules for HDV was done with the online MFold package. Mfold follows most widely used algorithms for RNA secondary structure prediction, which are based on a search for the minimal free energy state [52]. Here, one siRNA molecule is having more than zero free energy of folding at 370C Table 4 (see supplementary material). Earlier studies have recommended that an RNA molecule should have minimum free energy of folding for their stability. Therefore, the molecule with positive energy may be more accessible for target site and have high potential to bind with target and lead to in effective gene silencing. While other molecule is also having less than -1 kcal frees energy of folding.
Figure 3.
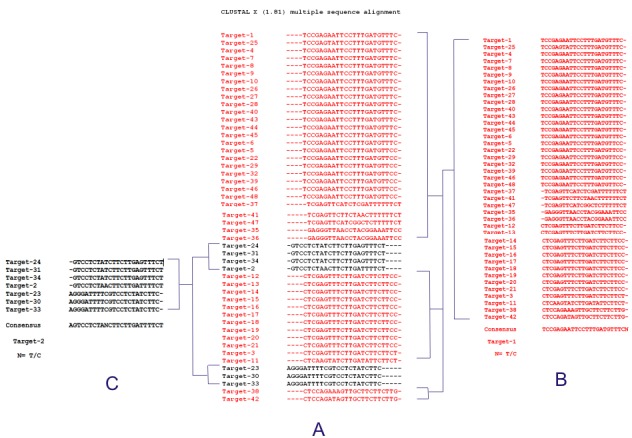
Multiple sequence alignment of predicted siRNA target sequences. (A) All aligned siRNA target sequence. (B) Forty one siRNA targets showing exact similarity and a consensus sequence of them executed using EMBOSS tool. (C) Seven target sequences with dissimilarity to forty one sequences, while exact similar to each other and create another good consensus.
Apart from this, a variety of biologically important RNAs were used for prediction of their function by interacting with other RNA molecules. Thus, thermodynamics study of RNA-RNA interactions may be an important aspect for siRNA molecule efficiency. The predicted siRNA molecules were subjected to RNA-RNA interaction study with their respective targets. The Vienna web suite is a comprehensive collection of tool that offers state of the art algorithms for RNA folding, comparison and prediction of RNA–RNA interactions. RNAup one of the important tools of Vienna web suite was used to predict free energy of RNA-RNA interactions. It models the binding energy for the interaction at a particular site as
(BE) ΔGbinding = ΔGuA + ΔGuB + ΔGh → (Equation 2)
Where ΔGuAB (ΔGuA + ΔGuB) is the free energy required to make the binding region in molecule A (target) or B (siRNA) accessible by removing intra-molecular structure. While ΔGh denotes the free energy gained from forming the intermolecular duplex by the partition function over all structures where the short RNA binds to target region. Calculation of the free energy of interaction (binding) between a siRNA molecule and its target was performed by using equation (2) Table 4 (see supplementary material).
Schubert et al. in their study analyzed that RNAi efficiency correlates well with the binding energies of siRNAs to their respective mRNA target [53]. Similarly, Mueckstein et al. in their study have chosen the target sites provided by Schubert et al. and agreed with the experimental results. Optimal free energy of binding (BE) is highly favorable and the siRNA will bind almost exclusively to the intended target site. The stepwise decrease of the target accessibility is directly correlated to a poor optimal BE and decreased silencing efficiency [46]. In present analysis, siRNA molecule for consensus target-1 was found to be more effective with BE. Therefore, it may qualify as high quality candidate for silencing the delta antigen gene and be used to cure the delta hepatitis.
RNAi approach is successfully exploited in various cases such as hepatitis B infection [36] silencing of endonuclease Argonaute 2 in Drosophila melanogaster [54]. RNAi utilized in HIV-1 infection in human peripheral blood mononuclear cells via best env-specific siRNAs, E7145 targeted to the central region of the V3 loop and E7490 targeted to the CD4 binding site of conserved regions on gp120, significantly inhibited the HIV-1 gene expression. Furthermore, E7145 and E7490 were effective against HIV-1NL4-3 replication in PBMCs for a relatively long time (14 days) [37]. In experimental brain cancer pegylated immunoliposomes (PIL) carrying short hairpin RNA expression plasmids driven by the U6 RNA polymerase promoter and directed to target EGFR expression by RNAi. The PIL is comprised of a mixture of known lipids containing polyethyleneglycol (PEG), which stabilizes the PIL structure in vivo in circulation. The tissue target specificity of PILs is given by conjugation of ~1% of the PEG residues to monoclonal antibodies (mAbs) that bind to specific endogenous receptors (i.e., insulin and transferrin receptors) located in the brain vascular endothelium [55]. Trypanosoma cruzi gp83 ligand, a cell surface trans-sialidase-like molecule that the parasite uses to attach to host cells, increases the level of laminin Υ-1 transcript and its expression in mammalian cells, leading to an increase in cellular infection. Stable RNA interference (RNAi) with host cell laminin Υ-1 knocks down the levels of laminin Υ-1 transcript and protein expression in mammalian cells, causing a dramatic reduction in cellular infection by Trypanosoma cruzi [56]. This approach was found to be successful in targeting bovine prion gene PRNP in livestock [57], carcinoma of the breast [58] and crown gall tumorigenesis in plants [59]. This technique was also used for silencing of capsid genes of Flavivirus using computational methods [60]. However, siRNA is the most influential means to control over gene expression in various organisms and showing antiviral activity too. Therefore, rational siRNA has provided the advancement in the development of experiment based approaches to prevent the HDV infections via gene silencing mechanism.
Conclusion
Using RNAi technology a number of siRNA molecules may be designed for silencing of significant genes in various biological systems. Further their interactions with target can also be calculated, computationally. Therefore, in this study two siRNA molecules were predicted against delta antigen protein as effective candidate using computational approaches. These molecules may lead to a novel antiviral therapy against HDV. Study outcome would also provide a basis to the researchers and pharma industry persons to develop the antiviral therapeutics at genomic level, experimentally.
Supplementary material
Figure 1.
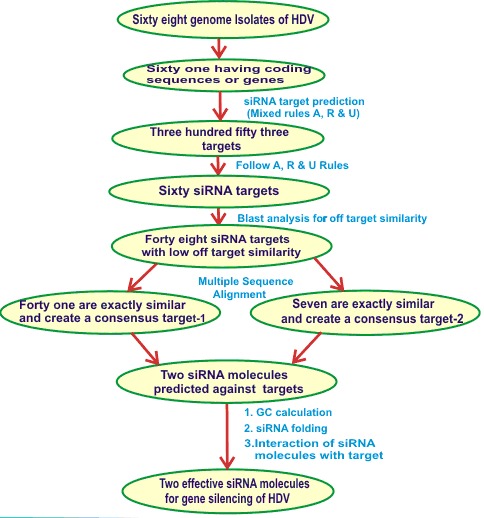
Flow chart showing complete methodology used for screening of effective siRNA molecules in this study
Acknowledgments
The support of Department of Biotechnology, Ministry of Science and Technology, Government of India, to Bioinformatics Centre at Biotech Park Lucknow is gratefully acknowledged. The authors would also like to thank the Department of Pharmacology & Therapeutics, Chhatrapati Shahuji Maharaj Medical University, Lucknow, India for its great support and providing the facilities for this work. Authors acknowledge the kind support of Dr. Sangeeta Saxena, Head, Department of Biotechnology, Babasaheb Bhimrao Ambedkar University (BBAU), Lucknow for technical review of the manuscript.
Footnotes
Citation:Singh et al, Bioinformation 8(16): 749-757 (2012)
References
- 1.M Rizzetto, et al. Proc Natl Acad Sci U S A. 1980;77:6124. [Google Scholar]
- 2.MYP Kuo, et al. J Virol. 1988;62:1855. [Google Scholar]
- 3.SJ Hadziyannis, et al. J Gastroenterol Hepatol. 1997;12:289. doi: 10.1111/j.1440-1746.1997.tb00424.x. [DOI] [PubMed] [Google Scholar]
- 4.F Imazeki, et al. J Virol. 1990;64:5594. doi: 10.1128/jvi.64.11.5594-5599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.JC Wu, et al. Herpetology. 1995;22:1656. [Google Scholar]
- 6.V Ivaniushina, et al. J Gen Virol. 2001;82:2709. doi: 10.1099/0022-1317-82-11-2709. [DOI] [PubMed] [Google Scholar]
- 7.JC Wu, et al. J Gen Virol. 1998;79:1105. doi: 10.1099/0022-1317-79-5-1105. [DOI] [PubMed] [Google Scholar]
- 8.H Sakugawa, et al. J Med Virol. 1999;58:366. doi: 10.1002/(sici)1096-9071(199908)58:4<366::aid-jmv8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 9.H Watanabe, et al. J Gen Virol. 2003;84:3275. doi: 10.1099/vir.0.19499-0. [DOI] [PubMed] [Google Scholar]
- 10.JL Casey, et al. Proc Natl Acad Sci U S A. 1993;90:9016. [Google Scholar]
- 11.N Radjef, et al. J Virol. 2004;78:2537. doi: 10.1128/JVI.78.5.2537-2544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LG Frédéric, et al. Emerg Infect Dis. 2006;12:1447. doi: 10.3201/eid1209.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.KS Wang. Nature. 1986;323:508. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 14.JM Taylor, et al. Annu Rev Microbiol. 1992;46:253. doi: 10.1146/annurev.mi.46.100192.001345. [DOI] [PubMed] [Google Scholar]
- 15.PJ Chen, et al. Proc Natl Acad Sci U S A. 1986;83:8774. [Google Scholar]
- 16.MM Lai. Annual Rev Biochem. 1995;64:259. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 17.J Chang, et al. Viral. 2000;74:9889. [Google Scholar]
- 18.MYP Kuo, et al. J Virol. 1989;63:1945. [Google Scholar]
- 19.C Cunha, et al. RNA. 1998;4:680. [Google Scholar]
- 20.M Yan, et al. World J Gastroenterol. 2007;13:5169. doi: 10.3748/wjg.v13.i39.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A Fire, et al. Nature. 1998;391:806. [Google Scholar]
- 22.RA Jorgensen, et al. Plant Mol Biol. 1996;31:957. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- 23.SM Elbashir, et al. Genes Dev. 2001;15:188. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.PD Zamore, et al. Cell. 2000;101:25. [Google Scholar]
- 25.E Bernstein, et al. Nature. 2001;409:363. [Google Scholar]
- 26.SM Hammond, et al. Nature. 2000;404:293. [Google Scholar]
- 27.SM Elbashir, et al. Nature. 2001;411:494. [Google Scholar]
- 28.NJ Caplen, et al. Proc Natl Acad Sci U S A. 2001;98:9742. [Google Scholar]
- 29.TR Brummelkamp, et al. Science. 2002;296:550. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 30.DA Rubinson, et al. Nat Genet. 2003;33:401. [Google Scholar]
- 31.MT McManus, PA Sharp. Nat Rev Genet. 2002;3:737. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 32.B Berkhout, J Haasnoot. FEBS Lett. 2006;580:2896. doi: 10.1016/j.febslet.2006.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S Schütz, P Sarnow. Virology. 2006;344:151. doi: 10.1016/j.virol.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 34.B Cullen. Nat Immunol. 2006;7:563. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 35.J Xu, et al. Antiviral Res. 2007;73:126. doi: 10.1016/j.antiviral.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Y Chen, et al. Pharm Res. 2008;25:72. [Google Scholar]
- 37.WS Park, et al. Nucleic Acids Res. 2002;30:4830. doi: 10.1093/nar/gkf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M Chao, et al. Virol. 1991;65:4057. [Google Scholar]
- 39.J Chang, et al. J Virol. 2003;77:9728. doi: 10.1128/JVI.77.17.9728-9731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.S Gudima, et al. J Virol. 2002;76:3709. doi: 10.1128/JVI.76.8.3709-3719.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.TB Macnaughton, MMC Lai. J Virol. 2002;76:3928. doi: 10.1128/JVI.76.8.3928-3935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Y Naito, et al. BMC Bioinformatics. 2009;10:392. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.SK Gupta, et al. J Compur Scie & Syst Biol. 2010;3:1. doi:10.4172/jcsb.1000048. [Google Scholar]
- 44.SF Altschul, et al. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 45.AR Gruber, et al. Nucleic Acids Res. 2008;36:W70. [Google Scholar]
- 46.U Mueckstein, et al. Bioinformatics. 2006;22:1177. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 47.JP Monjardino, JA Saldanha. Br Med Bull. 1990;46:399. doi: 10.1093/oxfordjournals.bmb.a072406. [DOI] [PubMed] [Google Scholar]
- 48.K Ui-Tei, et al. Nucleic Acids Res. 2008;36:7100. doi: 10.1093/nar/gkn902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.SM Freier, et al. Proc Natl Acad Sci U S A. 1986;83:9373. [Google Scholar]
- 50.M Amarzguioui, H Prydz. Biochem Biophys Res commun. 2004;316:1050. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 51.A Reynolds, et al. Nat biotech. 2004;22:326. [Google Scholar]
- 52.M Zuker, et al. Science. 1989;244:48. [Google Scholar]
- 53.SJ Schubert, et al. Mol Biol. 2005;348:883. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 54.RP Van Rii, et al. Genes Dev. 2006;20:2985. [Google Scholar]
- 55.RJ Boado. NeuroRx. 2005;2:139. [Google Scholar]
- 56.PN Nde, et al. Infect immun. 2006;74:1643. doi: 10.1128/IAI.74.3.1643-1648.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MC Golding, et al. Proc Natl Acad Sci U S A. 2006;103:5285. [Google Scholar]
- 58.MA Stoff-Khalili, et al. Cancer Gene Ther. 2006;13:633. doi: 10.1038/sj.cgt.7700929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MA Escobar, et al. Proc Natl Acad Sci U S A. 2001;98:13437. [Google Scholar]
- 60.P Somvanshi, et al. Interdiscip Sci. 2009;1:298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


