Abstract
Many biotechnology applications require the evolution of enhanced protein stability. Using polymerase chain reaction-based recovery of engineered clones during the screen enrichment phase, we describe a yeast display method capable of yielding engineered proteins having thermal stability that substantially exceeds the viability threshold of the yeast host. To this end, yeast-enhanced green fluorescent protein destabilized by dual-loop insertion was engineered to possess a substantially enhanced resistance to thermal denaturation at 70°C. Stabilized proteins were secreted, purified and found to have three- to six-fold increased resistance to thermal denaturation. The validated method enables yeast display-based screens in previously inaccessible regions of the fitness landscape.
Keywords: protein engineering, thermal stability, yeast surface display
Introduction
Yeast surface display is a powerful platform for engineering protein properties (Gai and Wittrup, 2007; Pepper et al., 2008). As a few examples, yeast display has facilitated the evolution of a femtomolar affinity single-chain antibody fragment (Boder et al., 2000), the creation of a horseradish peroxidase with enhanced enantioselectivity (Lipovsek et al., 2007) and the identification of picomolar affinity binders using fibronectin as a scaffold (Hackel et al., 2008). Thermal stability has also been engineered using yeast surface display either by evolving the protein of interest using a restrictive induction temperature or by screening mutants for slowed denaturation at an elevated temperature (Shusta et al., 2000; Hackel et al., 2010; Jones et al., 2011). Typically when using yeast surface display, yeast that display engineered proteins with the desired properties are isolated using a cell sorter and are regrown to amplify the recovered clones while maintaining the genotype–phenotype linkage. Because of this requirement, thermal stability screens have been performed at temperatures where the yeast retain at least some viability (<50°C), and thus only apply for low-to-moderate stability proteins. One way to overcome the viability temperature limitation is to evolve the protein of interest in clonal assays in microtiter plates, but these approaches suffer from the disadvantage that a limited number of clones and hence, limited protein sequence diversity, can be analyzed in a given screen. Thus, in order to take advantage of the high throughput and quantitative nature of yeast surface display for stability engineering with elevated screening temperatures, we have developed a methodology that instead employs PCR-based recovery of selected clones after each round of library enrichment (Fig. 1). As a specific test case, we evolved a destabilized form of monomeric yeast-enhanced green fluorescent protein (GFPM) for improved resistance to thermal denaturation at 70°C, well beyond temperatures where yeast remains viable.
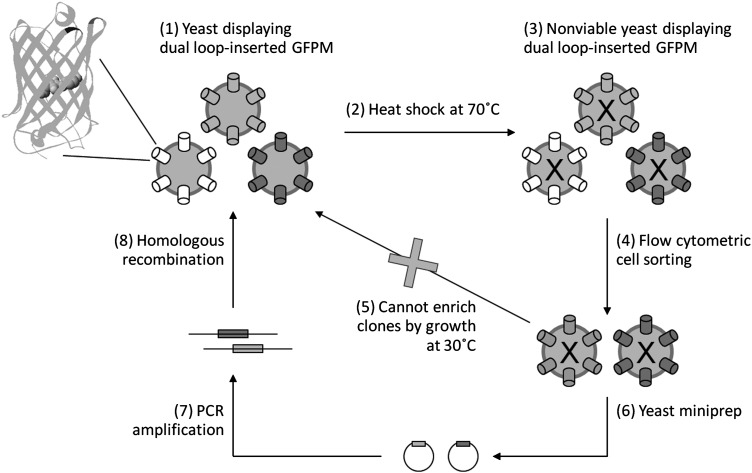
Fig. 1.
Schematic of the PCR-based procedure to engineer thermally stable proteins using yeast display. Dual-loop-inserted GFPM consists of yeast-enhanced GFP with amino acid insertions at 101–102 (black) and 172–173 (dark gray). The chromophore at center of β-barrel is shown in spacefill. Standard GFP structure pdb ID = 1EMA was used for this rendering. A library of yeast cells expressing dual-loop-inserted GFPM proteins on their surface (1) is subjected to thermal denaturation (2). The nonviable cells are then labeled with anti-c-myc antibody to measure expression, and retention of GFP fluorescence was used to monitor resistance to thermal denaturation (3). Yeast-harboring dual-loop-inserted GFPM mutants that are well expressed and resistant to thermal denaturation are recovered by flow cytometric cell sorting (4). However, the recovered yeast cells are dead and cannot be amplified by regrowth at 30°C as in traditional yeast display screens (5). Instead, plasmid DNA is recovered from the pool of sorted cells by yeast miniprep (6) and used as template for PCR-based amplification of the open reading frames of the dual-loop-inserted GFPM fragments (7). These recovered open reading frames are then used in a homologous recombination reaction to reconstruct a library enriched in thermally stable variants (8). The process is then repeated until the resultant pool is substantially enriched in dual-loop-inserted GFPM fragments that retain fluorescence after thermal denaturation.
Materials and methods
Strains and media
Yeast surface display was performed using the EBY100 yeast strain and compatible pCT-ESO-GFPM series of display plasmids (Pavoor et al., 2009). Saccharomyces cerevisiae strain BJ5464 and the pRS316-GFP-based expression vector were used to secrete proteins (Huang and Shusta, 2005). Minimal synthetic dextrose - casamino acids (SD-CAA) medium (20 g/l dextrose, 6.7 g/l yeast nitrogen base, 5 g/l casamino acids, 10.19 g/l Na2HPO4•7H2O, 8.56 g/l NaH2PO4•H2O) was used for yeast culture. Proteins were subsequently displayed or secreted using SG-CAA medium (20 g/l galactose replacing dextrose) to activate the GAL1-10 promoter. For secretion experiments, yeast cells were grown for 72 h in SD-CAA at 30°C. The medium was switched to SG-CAA supplemented with 1 g/l bovine serum albumin as a nonspecific carrier for 72 h at 20°C. Western blots, and nickel-nitrilotriacetic acid-based protein purification were performed as described earlier (Huang and Shusta, 2005). The Escherichia coli strains XL1-Blue (Stratagene, La Jolla, CA, USA) and DH5α (Invitrogen, Carlsbad, CA, USA) were used for cloning. Luria-Bertani (LB) medium (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl and pH 7.5, 50 μg/ml ampicillin) was used for bacteria growth and plasmid amplification.
Library creation, sorting and PCR-based mutant recovery
A mutagenic library of the 37-2-7 and 37-2-1 templates (Table I) was created such that the inserted loops were held constant and the remaining GFPM scaffold was randomly mutagenized. The strategy for introduction of low levels of mutagenesis employed PCR with nucleoside analogs and was identical to that previously described (Pavoor et al., 2009). The mutated and amplified open reading frames were then used in a homologous recombination reaction to create the dual-loop-inserted yeast display library (Pavoor et al., 2009), and the resultant library size determined by counting the number of yeast transformants was 6.6 × 106 clones. All flow cytometric sorting of the library was performed on a Becton Dickinson FACSVantage SE sorter at the University of Wisconsin Comprehensive Cancer Center. Yeast cells were grown in SD-CAA medium at 30°C to an OD600= 1.0 and subsequently display was induced in SG-CAA medium at 20°C for 14 h. Yeast cells (2 × 106) were resuspended in 500 μl of PBS plus 1 mg/ml BSA (PBS/BSA) and incubated at 70°C for 30 min. The cells were harvested by centrifugation and labeled with anti-c-myc antibody (1 : 100 dilution) (Covance, CA, USA) followed by anti-mouse phycoerythrin (PE, 1 : 40 dilution) (Sigma Aldrich, St Louis, MO, USA). The top 1% of the cells having both GFP fluorescence and full length expression (PE fluorescence), and at least 105 total yeast cells, were recovered in 1 ml of PBS/BSA. The cells were collected by centrifugation at 14 000 × g for 5min. To test the generalizable nature of the thermal denaturation and labeling methodologies reported here, several additional primary and secondary reagents were tested for nonspecific labeling after 70°C heat shock. Primary detection reagents included biotinylated TrkB receptor tyrosine kinase, biotinylated glyceraldehyde-3-phosphate dehydrogenase (GAPDH), recombinant human transferrin receptor (TfR), mouse anti-c-myc and rabbit anti-c-myc antibodies. The tested secondary reagents included streptavidin-PE (TrkB and GAPDH detection), anti-human alexa 647 (TrkB detection), mouse anti-TfR and anti-mouse Alexa 647 (TfR detection), anti-mouse Alexa 488 (anti-c-myc detection), anti-rabbit FITC (anti-c-myc detection). None of the tested reagents resulted in increases in nonspecific labeling upon heat shock (data not shown).
Table I.
Mutations in dual-loop-inserted GFPM variants
| Mutations | 37-2-7 | 37-2-1 | 70C-3 | 70C-6 | 70C-9 | 70C-6(−C) | 37-2-7(+C) |
|---|---|---|---|---|---|---|---|
| G10D | x | ||||||
| D19N | x | x | x | x | x | x | x |
| Y39H | x | x | x | x | x | x | |
| F46C | x | x | |||||
| I47V | x | ||||||
| F64L | x | x | x | x | x | x | x |
| A87T | x | x | x | x | x | x | x |
| I123V | x | x | |||||
| K140R | x | x | |||||
| N144S | x | x | x | ||||
| Y151C | x | x | |||||
| V163A | x | x | x | x | x | x | x |
| E172K | x | x | x | x | x |
The plasmids were recovered from the sorted yeast pellet using the yeast miniprep Zymoprep II kit (Zymo Research, Orange, CA, USA). The plasmid-containing elution resulting from the Zymoprep kit was further washed of potential contaminants using a PCR purification kit (Qiagen Valencia, CA, USA). The PCR purification elution volume was 30 μl and about 4 μl of this elution was used as template for each of seven parallel 30-cycle PCR reactions so as to utilize the entire plasmid preparation resulting from the sort. The reaction mixture was as follows: 1 μl of 10 μM dNTP mix (Invitrogen, Carlsbad, CA, USA), 5 μl of 10x buffer (–MgCl2), 1.5 μl of 50 mM MgCl2, 1 μl of Platinum Taq (Invitrogen, Carlsbad, CA, USA), 1 μl 100 μM PNL6 Forward primer (5′-GTA CGA GCT AAA AGT ACA GTG-3′), and 1 μl of 100 μM PNL6 reverse primer (5′-TAG ATA CCC ATA CGA CGT TC-3′) to total volume of 50 μl using water. Whereas the PNL primers are efficient for PCR amplification, a second set of primers (ESO) was used for final PCR amplification as they are longer and lead to more efficient homologous recombination for recreation of the yeast library. To this end, 2 μl of each of the seven PNL-based PCR products was utilized for a similar 30-cycle PCR reaction except that the ESO forward (5′-GTG GAG GAG GCT CTG GTG GAG GCG GTA GCG GAG GCG GAG GGT CGG CTA GC-3′) and ESO reverse (5′-TAT CAG ATC TCA GCT ATT ACA AGT CCT CTT CAG AAA TAA GCT TTT GTT C-3′) primers were employed. All the PCR products were combined, concentrated using Pellet Paint (Novagen, Madison, WI, USA) as per the manufacturer's instructions and resuspended in sterile water to a final concentration of 5 µg/µl. The enriched pool for the next round was then created using the resultant PCR-amplified dual-loop-inserted GFPM open reading frames and pCT-ESO-GFPM acceptor vector, predigested with NheI and BamHI restriction enzymes, in a homologous recombination reaction.
Individual dual-loop-inserted GFPM clones were evaluated using a Becton Dickinson FACSCalibur benchtop flow cytometer after being labeled exactly as described above for cell sorting. For measurement of resistance to thermal denaturation, individual clones were first subjected to 70°C for 30 min in PBS/BSA and then evaluated for retained GFP fluorescence. The GFP fluorescence measured in this study is contributed to by the GFP fluorescence from proteins inside the cell as well as from those being displayed on the surface of yeast. After measuring the total GFP fluorescence, the cells were treated with 0.5 M dithiothreitol (DTT) (Sigma Aldrich, St Louis, MO, USA) for 90 min to remove fluorescence associated with surface localized GFPM. After DTT treatment, the remaining internal GFP fluorescence was measured again using a flow cytometer and the GFP fluorescence resulting from the externally displayed protein calculated (external GFP fluorescence = total−internal).
Measurement of purified protein thermal denaturation rates
The denaturation rate of dual-loop-inserted GFPM protein was determined as previously described (Dai et al., 2007). Equal amounts of purified protein in a final volume of 50 μl were subjected to thermal denaturation. Using a BioRad iCycler compensated using external well factor solution (BioRad, Hercules, CA, USA), GFP fluorescence of the samples was measured every 30 s for 3.75 h at 70°C. After an initial adsorption phase, the remaining temporal fluorescence profile was representative of a single exponential decay and used to compute the denaturation rate constant and half-life for each protein at 70°C.
Results and discussion
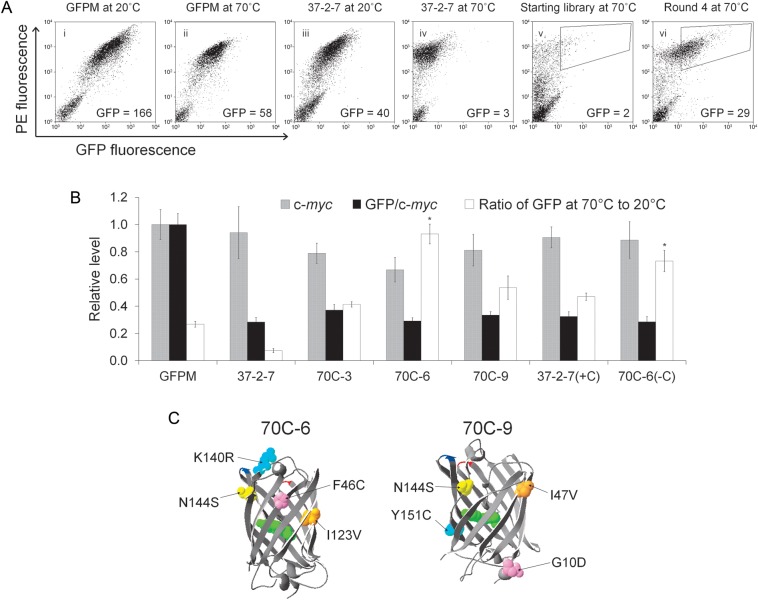
The target of stability engineering was monomeric, yeast-enhanced GFPM having destabilizing dual-loop insertion (Cormack et al., 1997; Pavoor et al., 2009). In previous work, we demonstrated that a GFPM variant possessing a 13 amino acid loop inserted between Asp102 and Asp103 (LKQHFWSTPRTTS) and a 12 amino acid loop inserted between Glu172 and Asp173 (SRERDYRLDYTR) of GFPM (Fig. 1), was not expressed or fluorescent (Pavoor et al., 2009). In addition, a dual-loop-inserted mutant known as 37-2-7 was identified (Pavoor et al., 2009) that could recover yeast surface expression levels (Fig. 2A panels i and iii—PE fluorescence and 2B—c-myc) and some fluorescence per molecule (Fig. 2A panels i and iii and 2B—GFP/c-myc). Since GFP fluorescence requires properly folded protein (Reid and Flynn, 1997), fractional retention of fluorescence after heat shock was used to evaluate 37-2-7 resistance to thermal denaturation in comparison with the non-loop-inserted GFPM parent molecule. When subjected to denaturation at 70°C for 30 min, yeast-displayed 37-2-7 (7% fluorescence retained) was not nearly as stable as non-loop-inserted GFPM (27% fluorescence retained) (Fig. 2A panels ii and iv and 2B), indicating that 37-2-7 would be a good protein template to validate our PCR-based stability engineering protocol (Fig. 1).
Fig. 2.
Directed evolution of dual-loop-inserted GFPM mutants that resist thermal denaturation. (A) Flow cytometric evaluation of resistance to thermal denaturation for non-loop-inserted GFPM (i and ii) and dual-loop-inserted mutant 37-2-7 (iii and iv). Full-length expression was monitored using the C-terminal c-myc epitope (PE fluorescence). Expression and GFP fluorescence were monitored both at the permissive temperature of 20°C and after 30 min at the denaturing temperature of 70°C. For panels i–iv, inset GFP values are the geometric mean GFP fluorescence of the entire displaying population. Also depicted are flow cytometric plots of the mutagenic dual-loop-inserted library after denaturation for 30 min at 70°C (v) and after four rounds of enrichment (vi) using a sorting gate similar to that depicted on the panels to recover mutants with template-like expression (high PE) and improved resistance to thermal denaturation (high GFP). Insets indicate the GFP mean of the entire high PE population. (B) Quantified flow cytometric data denoting full-length expression (c-myc) and yeast cell surface-localized GFP fluorescence per molecule (GFP/c-myc). It is important to note the GFP fluorescence comprises both intracellular and yeast cell surface contributions. For the GFP/c-myc quantity, only the cell surface GFP contribution is quantified so that per molecule fluorescence properties (per cell surface c-myc) can be monitored during the evolution process. The resistance to denaturation is presented as the ratio of GFP fluorescence after 30 min at 70°C to that at the permissive temperature of 20°C. Data are derived from triplicate yeast transformants and normalized to non-loop-inserted GFPM for comparison. (C) GFP structure denoting newly added mutations found in the evolved, denaturation resistant dual-loop-inserted GFPM molecules. Mutant 70C-3 only carries the Y151C mutation denoted in 70C-9 structure. Loop insertion sites are indicated at positions 101–102 (red) and 172–173 (blue).
The initial mutagenic library construction began with a mixture of two templates, 37-2-7 and a highly related mutant 37-2-1 (Table I), having similar expression and fluorescence characteristics (Pavoor et al., 2009). Mutagenesis was restricted to the GFP scaffold leaving the inserted loops free of mutation to help ensure that any variant would gain stability as a result of changes in the GFP scaffold and not in the destabilizing loop insertions. The resultant library had 6.6 × 106 clones, with roughly 20% of the clones having expression and fluorescence levels similar to those of 37-2-7 at the permissive 20°C temperature. After 30 min of 70°C heat shock (Fig. 1.2), GFP fluorescence of the library dropped dramatically as expected based on the comparative performance of 37-2-7 (Fig. 2A, panels iv and v). Using retention of GFP fluorescence as the proxy screening variable for increased resistance to thermal denaturation, improved clones were recovered by flow cytometry (Fig. 2A, panel v). At this point, the conventional procedure for directed evolution using yeast surface display would call for the sorted cells to be grown at 30°C so as to enrich the positive pool for further purification and/or screening rounds (Fig. 1(5)). Whereas it has been shown that yeast cells can be preconditioned to partially survive heat shocks of up to 50°C for 1 h (de Marañón et al., 1999; Guyot et al., 2005), yeast could not be similarly preconditioned to survive 70°C for 30 min (data not shown). Instead, the plasmids encoding the mutants collected in each sorting round were recovered by yeast miniprep for subsequent PCR-based amplification (Fig. 1(6) and 1(7)). Previous approaches focused on recursive mutagenesis strategies employed yeast minipreps and PCR-based recovery of improved mutants from sorted yeast surface display pools (Hackel et al., 2008). Importantly, since these previous screens were performed using viable yeast, the yeast cells were grown post-sort for amplification and 108 yeast cells were subjected to yeast miniprep recovery of clones (Hackel et al., 2008). Since the thermal denaturation screens performed here resulted in nonviable yeast, the number of yeast sorted is the number available for gene recovery and 108 yeast cells would be an unrealistic sorting burden. Instead, we found that we could efficiently PCR amplify the dual-loop-inserted GFPM open reading frames from the yeast minipreps starting with as few as 105 yeast cells. Therefore, in each sort round we collected the top 1% of the population maintaining GFP fluorescence after heat shock and to reach the benchmark of 105 recovered yeast, we sorted at least 107 yeast in each round, a very manageable flow cytometric sorting burden. This approach resulted in oversampling library clonal diversity 3-fold in round 1, but between 50- and 150-fold in subsequent rounds to help ensure maintenance of clone diversity. Additionally, while flow cytometry was used for every sorting round in this study, magnetic bead-based separations could be performed in the early rounds to more significantly oversample library diversity if there are concerns about loss of rare clones (Ackerman et al., 2009). Employing homologous recombination with the PCR-recovered mutant GFP open reading frames, enriched yeast display libraries of thermal denaturation resistant dual-loop-inserted GFPM clones were created for the next round of sorting (Fig. 1.8). After four rounds of sorting enrichment, a pool of dual-loop-inserted GFPM mutants that retain a significant portion of their GFP fluorescence after thermal denaturation was obtained (Fig. 2A, panels v and vi). In this study, additional mutagenesis was not introduced during the PCR-based recovery process that followed each sorting round, and thus, the resultant pool was the product of a single round of directed evolution. However, recursive mutagenesis would be directly compatible with these approaches (Hackel et al., 2008).
From the enriched, fourth round pool, ten individual clones were isolated and analyzed in surface display format. The three clones exhibiting the most improved resistance to thermal denaturation were 70C-3, 70C-6 and 70C-9 (Fig. 2C, Table I). Whereas the 37-2-7 (7%) template retains only a small amount of initial fluorescence after heat shock of 70°C for 30 min, clones 70C-6 (93%), 70C-9 (54%) and 70C-3 (41%) even outperformed non-loop-inserted GFPM (27%) in terms of resisting thermal denaturation (Fig. 2B). Although thermal stability increased, it is important to note that the mutations in 70C-3, 70C-6 and 70C-9 did not confer increased cell surface expression, nor did it substantially change the GFP fluorescence per molecule compared with the starting dual-loop-inserted template (Fig. 2B). These data indicate that at least in the surface displayed format, the mutations mainly confer increased resistance to thermal denaturation.
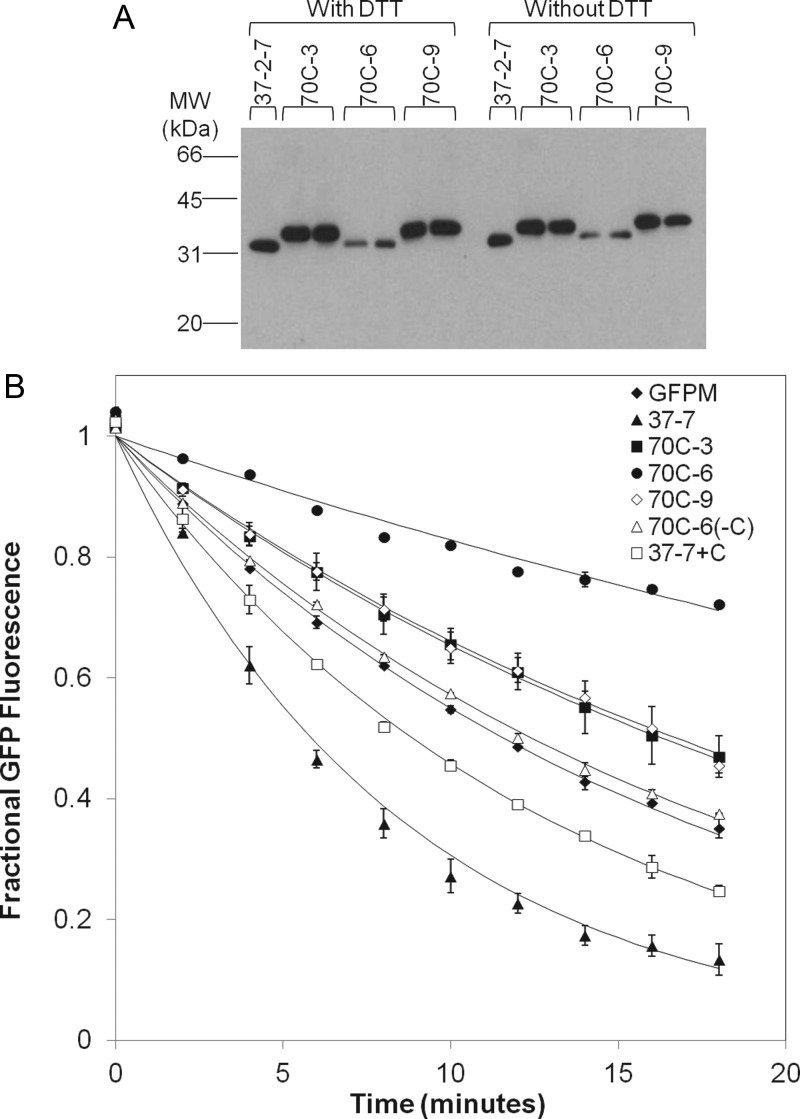
The stabilized dual-loop-inserted GFPM variants were secreted as soluble products from yeast at levels similar to that of the 37-2-7 template (∼1–2 mg/l, Table II, Fig. 3A), although secretion titers for the most stable clone, 70C-6, were decreased by 50%. Thus, the increased thermal stability of the clones isolated in this study did not correlate with increased secretion as has been observed previously (Shusta et al., 1999; Shusta et al., 2000). However, in contrast to these earlier studies that utilized the secretory machinery as a screening filter for improved folding, processing and surface display of protein, we instead used a post-display thermal denaturation screen without a direct linkage to effects on secretory processing and expression. Next, the resistance to thermal denaturation was also measured for the secreted and purified proteins to determine if yeast cell surface stability improvements translated to properties of the soluble protein. GFP fluorescence was monitored at 70°C as a function of time to generate thermal denaturation rate constants. Clones 70C-3, 70C-6 and 70C-9 maintained the same trend with respect to thermal stability as was observed on the surface of yeast: 70C6 > 70C-9 = 70C-3 > GFPM > 37-2-7 (Fig. 3B, Table II). Thus, the proteins engineered for resistance to high temperature thermal denaturation on the surface of yeast using the PCR-based enrichment methodology retain the stabilizing attributes as soluble proteins.
Table II.
Secretion and stability properties of purified dual-loop-inserted GFPM proteins
| Protein | Secretion level (mg/l) | Denaturation rate constant (×104 min−1) | Half-life (min) |
|---|---|---|---|
| GFPM | 3.0 ± 0.1 | 600 ± 20 | 12 |
| 37-2-7 | 1.4 ± 0.1 | 1180 ± 80 | 6 |
| 70C-3 | 1.6 ± 0.1 | 430 ± 40 | 16 |
| 70C-6 | 0.7 ± 0.1 | 190 ± 20 | 36 |
| 70C-9 | 1.8 ± 0.2 | 410 ± 10 | 17 |
| 70C-6(−C) | 1.4 ± 0.2 | 560 ± 10 | 13 |
| 37-2-7(+C) | 1.2 ± 0.4 | 780 ± 20 | 9 |
Fig. 3.
Properties of the soluble dual-loop-inserted GFPM proteins. (A) Western blot of yeast supernatants containing secreted dual-loop-inserted GFPM proteins. The western blot was performed with and without reducing agent (DTT) with no changes in migration. The slightly larger size for mutants 70C-3 and 70C-9 is a result of differential retention of the pro leader region that is part of secretion system used (Huang and Shusta, 2006). Whereas the western blot gives a qualitative idea of comparative secretion levels, quantitative western blotting with triplicate samples was performed to generate the secretion titers denoted in Table II. (B) Thermal denaturation rates of secreted and purified dual-loop-inserted GFPM proteins. GFP fluorescence was monitored as a function of time during denaturation at 70°C. The loss of fluorescence was fit to an exponential decay model to derive denaturation rate constants reported in Table II. Data points ± SD represent duplicate purified samples and the solid lines are the exponential curve fits.
Table I lists the mutations conferring resistance to thermal denaturation in clones 70C-3, 70C-6 and 70C-9 and these mutations are illustrated in Fig. 2C. Of interest, 70C-6 (F46C), 70C-3 (Y151C) and 70C-9 (Y151C) all had the introduction of cysteine residues with 70C-3 having the Y151C mutation as the only addition to those found in the dual-loop-inserted starting templates and thus likely to be responsible for the increase in stability. Moreover, clone 70C-6 with the F46C mutation was extremely stable (Fig. 3B, Table II). Evaluation of the soluble mutant protein sizes by western blotting indicated no shift in migration in the presence or absence of reducing agent, ruling out the possibility of intermolecular disulfide-bonded dimers as the source of stabilization (Fig. 3A). Of course, intramolecular disulfide bonds with unpaired cysteine residues in the parent dual-loop-inserted GFPM molecule could play a stabilizing role in addition to the general change in physiochemical character provided by the Y151C and F46C mutations. Whereas such detailed mechanistic insights were beyond the scope of this study, we further investigated the functional contribution of the cysteine at position 46 in clone 70C-6, by creating a cysteine reversion mutant of 70C-6, denoted 70C-6(−C). The cysteine was also introduced into the 37-2-7 template (F46C) creating 37-2-7(+C) to evaluate the contribution of the cysteine mutation to the observed stability in the absence of the other mutations present in 70C-6. As expected, neither the 70C-6(−C) nor the 37-2-7(+C) mutants had altered expression (c-myc) or fluorescence properties (GFP/c-myc, Fig. 2B). In the yeast display format, the 70C-6(−C) mutant had a 20% decreased resistance to thermal denaturation compared to the 70C-6 parent indicating a partial role for F46C in conferring stability (Fig. 2B, P< 0.05). Moreover, the addition of cysteine at position 46 in the 37-2-7(+C) mutant allows for substantial gains in resistance to thermal denaturation compared with 37-2-7, but stability is not restored to the levels of 70C-6 (Fig. 2B). Analogous results were obtained with the solubly expressed and purified 70C-6(−C) and 37-2-7(+C) proteins, with cysteine reversion in 70C-6(−C) yielding a lowered resistance to thermal denaturation while additional mutation to cysteine at position 46 in 37-2-7(+C) led to increased resistance to thermal denaturation (Fig. 3B and Table II). Taken together, these data indicated an important, but not exclusive, stabilizing role for the F46C mutation and that additional mutations in 70C-6, I123V, K140R and N144S were also involved in the stabilization. In conclusion, incorporation of a PCR-based method for enriching screened yeast display clones allowed the engineering of proteins under conditions where the yeast cells are nonviable. Recently, a similar approach was also successful for the engineering of thermostable IgG1-Fc scaffolds (Traxlmayr et al., 2012). Thus, this adapted screening methodology could allow for the rapid isolation of engineered proteins under extreme conditions of temperature, detergents or chaotropes that would limit yeast surface display in its classical form.
Funding
This work was funded by the National Institutes of Health [grant number NS052649]; National Science Foundation [grant number CBET-0966014]; and the University of Wisconsin Graduate School. J.A.W. is the recipient of an NSF Graduate Research Fellowship and an NIH Biotechnology Training Grant Fellowship [grant number GM08349].
Footnotes
Edited by Frances Arnold
References
- Ackerman M., Levary D., Tobon G., Hackel B., Orcutt K.D., Wittrup K.D. Biotechnol. Prog. 2009;25:774–783. doi: 10.1002/btpr.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder E.T., Midelfort K.S., Wittrup K.D. Proc. Natl. Acad. Sci. USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B.P., Bertram G., Egerton M., Gow N.A.R., Falkow S., Brown A.J.P. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Dai M., Fisher H.E., Temirov J., Kiss C., Phipps M.E., Pavlik P., Werner J.H., Bradbury A.R.M. Protein Eng. Des. Sel. 2007;20:69–79. doi: 10.1093/protein/gzl056. [DOI] [PubMed] [Google Scholar]
- de Marañón I.M., Chaudanson N., Joly N., Gervais P. Biotechnol. Bioeng. 1999;65:176–181. [PubMed] [Google Scholar]
- Gai S.A., Wittrup K.D. Curr. Opin. Struct. Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyot S., Ferret E., Gervais P. Biotechnol. Bioeng. 2005;92:403–409. doi: 10.1002/bit.20600. [DOI] [PubMed] [Google Scholar]
- Hackel B.J., Ackerman M.E., Howland S.W., Wittrup K.D. J. Mol. Biol. 2010;401:84–96. doi: 10.1016/j.jmb.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel B.J., Kapila A., Wittrup K.D. J. Mol. Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Shusta E.V. Biotechnol. Prog. 2005;21:349–357. doi: 10.1021/bp0497482. [DOI] [PubMed] [Google Scholar]
- Huang D., Shusta E.V. Appl. Environ. Microbiol. 2006;72:7748–7759. doi: 10.1128/AEM.01403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.S., Tsai P.-C., Cochran J.R. Proc. Natl. Acad. Sci. USA. 2011;108:13035–13040. doi: 10.1073/pnas.1102561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsek D., Antipov E., Armstrong K.A., Olsen M.J., Klibanov A.M., Tidor B., Wittrup K.D. Chem. Biol. 2007;14:1176–1185. doi: 10.1016/j.chembiol.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Pavoor T.V., Cho Y.K., Shusta E.V. Proc. Natl. Acad. Sci. USA. 2009;106:11895. doi: 10.1073/pnas.0902828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper L.R., Cho Y.K., Boder E.T., Shusta E.V. Comb. Chem. High Throughput Screen. 2008;11:127–134. doi: 10.2174/138620708783744516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B.G., Flynn G.C. Biochemistry (Mosc) 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- Shusta E.V., Holler P.D., Kieke M.C., Kranz D.M., Wittrup K.D. Nat. Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- Shusta E.V., Kieke M.C., Parke E., Kranz D.M., Wittrup K.D. J. Mol. Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- Traxlmayr M.W., Faissner M., Stadlmayr G., Hasenhindl C., Antes B., Rüker F., Obinger C. Biochim. Biophys. Acta. 2012;1824:542–549. doi: 10.1016/j.bbapap.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]