Abstract
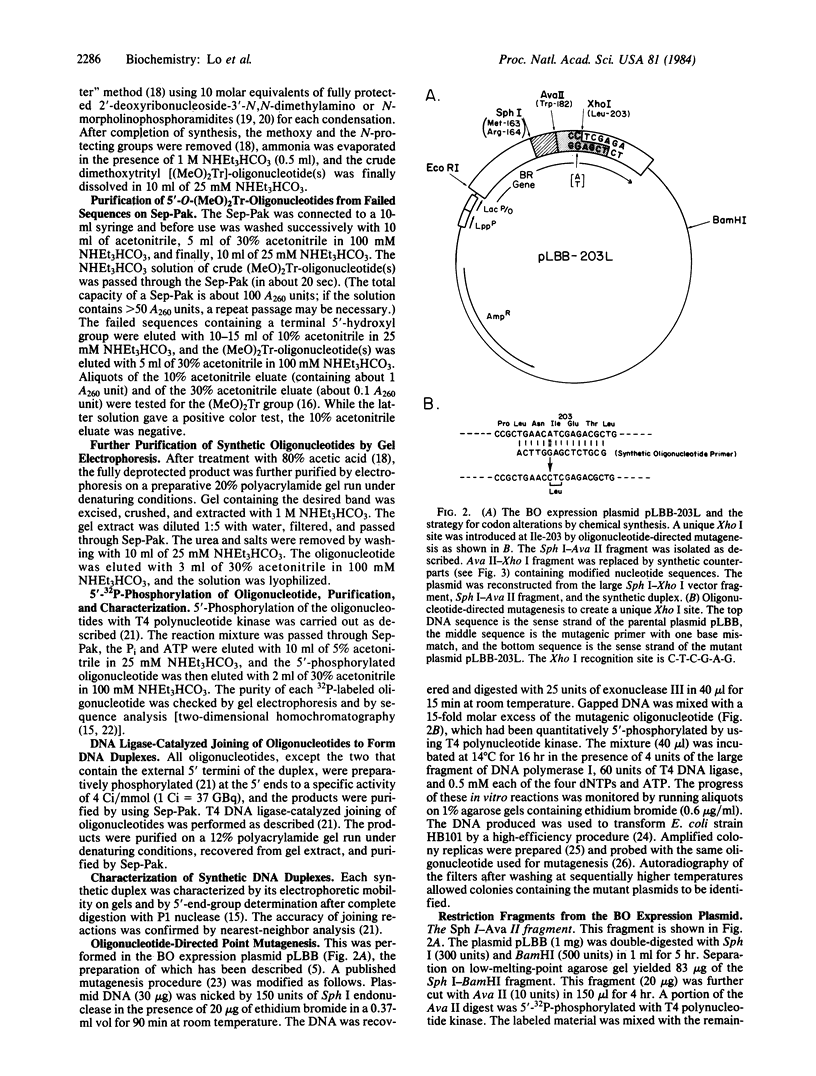
To study the mechanism of light-dependent proton translocation by bacteriorhodopsin, we have introduced single-codon changes in the gene so as to produce the following specific amino acid substitutions in the protein: Tyr-185 to Phe, Pro-186 to Leu, Trp-189 to Phe, Ser-193 to Ala, and Glu-194 to Gln. The strategy involved replacement of a 62-base-pair restriction fragment by synthetic DNA duplexes containing the modified nucleotide sequences. This required a unique restriction site (Xho I) at Ile-203 which was created by oligonucleotide-directed point mutagenesis. The six DNA duplexes corresponding to the modified native and mutant restriction fragments were all prepared by DNA ligase-catalyzed joining of chemically synthesized deoxyribooligonucleotides. The bacterioopsin expression plasmids reconstructed by using the synthetic DNA fragments were characterized by restriction analysis and DNA sequence determination. An extremely rapid, efficient, and general method for purification of the synthetic oligonucleotides and of DNA fragments was developed.
Keywords: purple membrane, proton pump, oligonucleotide purification, site-specific mutagenesis, T4 DNA ligase
Full text
PDF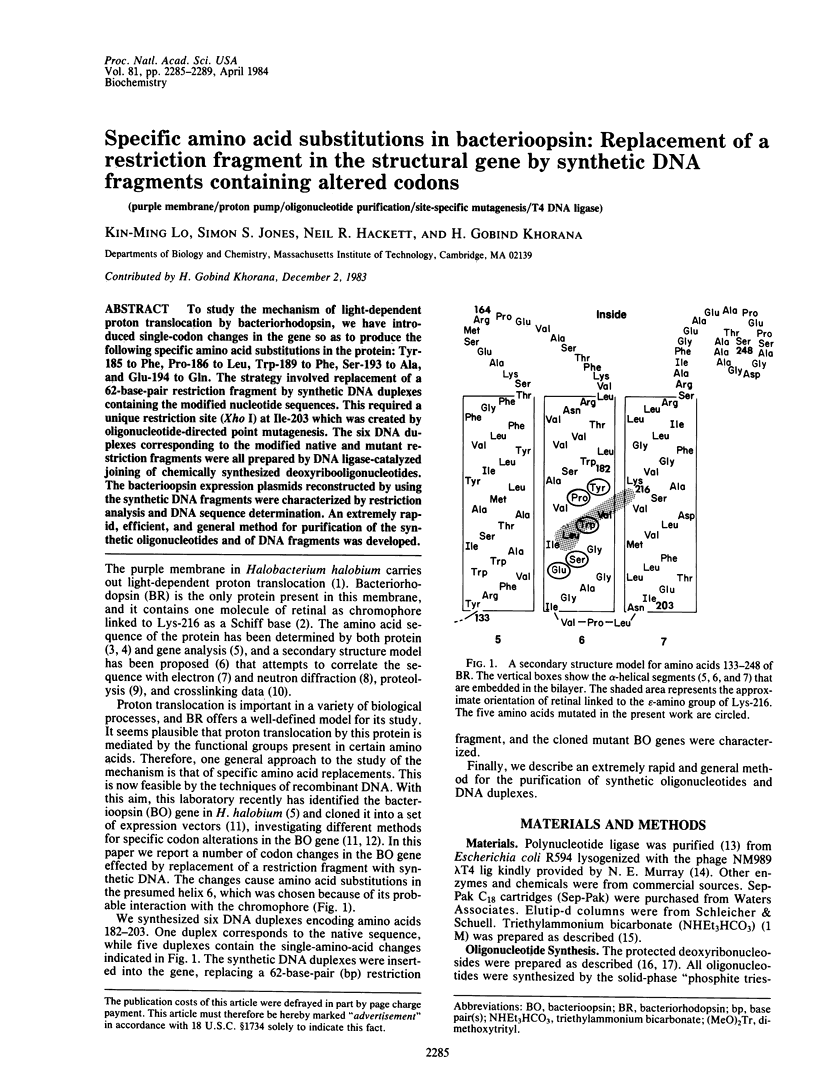


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayley H., Huang K. S., Radhakrishnan R., Ross A. H., Takagaki Y., Khorana H. G. Site of attachment of retinal in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2225–2229. doi: 10.1073/pnas.78.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. L., Belagaje R., Ryan M. J., Khorana H. G. Chemical synthesis and cloning of a tyrosine tRNA gene. Methods Enzymol. 1979;68:109–151. doi: 10.1016/0076-6879(79)68010-2. [DOI] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Cohen L. W., Riggs A. D., Morin C., Itakura K., Richards J. H. Oligonucleotide-directed mutagenesis as a general and powerful method for studies of protein function. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6409–6413. doi: 10.1073/pnas.79.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A. K. A proton motive force transducer and its role in proton pumps, proton engines, tobacco mosaic virus assembly and hemoglobin allosterism. J Theor Biol. 1982 Jul 7;97(1):95–127. doi: 10.1016/0022-5193(82)90281-8. [DOI] [PubMed] [Google Scholar]
- Dunn R., McCoy J., Simsek M., Majumdar A., Chang S. H., Rajbhandary U. L., Khorana H. G. The bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6744–6748. doi: 10.1073/pnas.78.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Henderson R., McLachlan A. D., Wallace B. A. Path of the polypeptide in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2023–2027. doi: 10.1073/pnas.77.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Zaccai G. Bacteriorhodopsin is an inside-out protein. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5894–5898. doi: 10.1073/pnas.77.10.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber G. E., Gray C. P., Wildenauer D., Khorana H. G. Orientation of bacteriorhodopsin in Halobacterium halobium as studied by selective proteolysis. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5426–5430. doi: 10.1073/pnas.74.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975 Sep 4;257(5521):28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Huang K. S., Radhakrishnan R., Bayley H., Khorana H. G. Orientation of retinal in bacteriorhodopsin as studied by cross-linking using a photosensitive analog of retinal. J Biol Chem. 1982 Nov 25;257(22):13616–13623. [PubMed] [Google Scholar]
- Khorana H. G., Gerber G. E., Herlihy W. C., Gray C. P., Anderegg R. J., Nihei K., Biemann K. Amino acid sequence of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5046–5050. doi: 10.1073/pnas.76.10.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- McCoy J. M., Khorana H. G. Introduction and characterization of amber mutations in the bacteriorhodopsin gene. J Biol Chem. 1983 Jul 10;258(13):8456–8461. [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- Nagle J. F., Morowitz H. J. Molecular mechanisms for proton transport in membranes. Proc Natl Acad Sci U S A. 1978 Jan;75(1):298–302. doi: 10.1073/pnas.75.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Abdulaev N. G., Feigina M. Y., Kiselev A. V., Lobanov N. A. The structural basis of the functioning of bacteriorhodopsin: an overview. FEBS Lett. 1979 Apr 15;100(2):219–224. doi: 10.1016/0014-5793(79)80338-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T., Besmer P., Takeya T., Khorana H. G. Total synthesis of the structural gene for the precursor of a tyrosine suppressor transfer RNA from Escherichia coli. 7. Enzymatic joining of the chemically synthesized segments to form a DNA duplex corresponding to the nucleotide sequence 1-26. J Biol Chem. 1976 Feb 10;251(3):634–641. [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Stoeckenius W., Bogomolni R. A. Bacteriorhodopsin and related pigments of halobacteria. Annu Rev Biochem. 1982;51:587–616. doi: 10.1146/annurev.bi.51.070182.003103. [DOI] [PubMed] [Google Scholar]
- Tu C. P., Jay E., Bahl C. P., Wu R. A reliable mapping method for sequence determination of oligodeoxyribonucleotides by mobility shift analysis. Anal Biochem. 1976 Jul;74(1):73–93. doi: 10.1016/0003-2697(76)90311-0. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]