Abstract
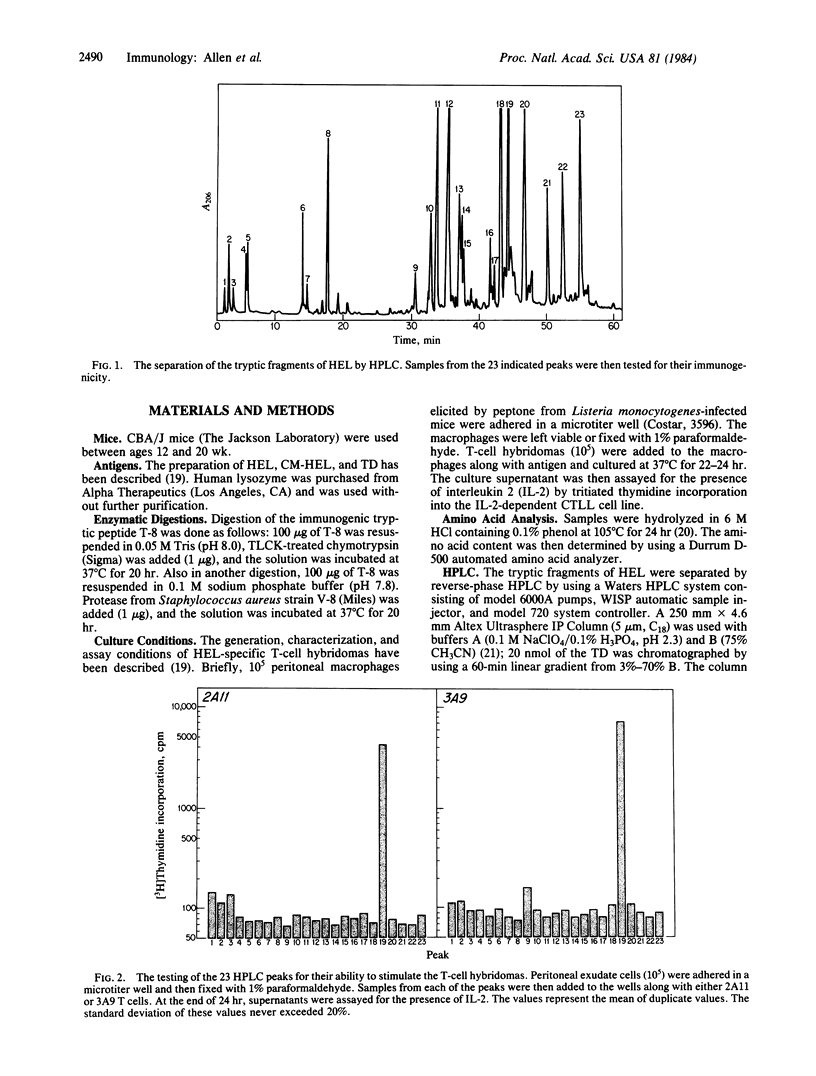
The purpose of this study was to identify the fragment of the hen egg-white lysozyme (HEL) molecule presented by macrophages to helper T cells. This was investigated by using T-cell hybridomas and macrophages prefixed in paraformaldehyde. We previously had shown that such prefixed macrophages could present a tryptic digest of HEL. The tryptic peptides were separated by HPLC and tested for their ability to stimulate the T-cell hybridomas. Only one tryptic peptide was found to be immunogenic. This immunogenic peptide was identified as the tryptic peptide T-8, containing amino acids 46-61. The precise determinant on the peptide T-8 being recognized was further defined by testing the response of the two T-cell hybridomas to human lysozyme. Neither clone responded to human lysozyme. From the amino acid sequence of human lysozyme, the determinant was localized to the four amino-terminal residues. Cleavage of the immunogenic peptide with either chymotrypsin or protease V-8 completely abolished the immunogenicity. This suggested that the T-cell determinant is located in the hydrophilic amino-terminal residues and that it must be associated with a hydrophobic stretch of amino acids, which allows the peptide to associate with the macrophage plasma membrane.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adorini L., Harvey M. A., Miller A., Sercarz E. E. Fine specificity of regulatory T cells. II. Suppressor and helper T cells are induced by different regions of hen egg-white lysozyme in a genetically nonresponder mouse strain. J Exp Med. 1979 Aug 1;150(2):293–306. doi: 10.1084/jem.150.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen P. M., Unanue E. R. Differential requirements for antigen processing by macrophages for lysozyme-specific T cell hybridomas. J Immunol. 1984 Mar;132(3):1077–1079. [PubMed] [Google Scholar]
- Atassi M. Z. Precise determination of the entire antigenic structure of lysozyme: molecular features of protein antigenic structures and potential of "surface-simulation" synthesis--a powerful new concept for protein binding sites. Immunochemistry. 1978 Dec;15(12):909–936. doi: 10.1016/0161-5890(78)90126-8. [DOI] [PubMed] [Google Scholar]
- CANFIELD R. E. PEPTIDES DERIVED FROM TRYPTIC DIGESTION OF EGG WHITE LYSOZYME. J Biol Chem. 1963 Aug;238:2691–2697. [PubMed] [Google Scholar]
- Canfield R. E., Kammerman S., Sobel J. H., Morgan F. J. Primary structure of lysozymes from man and goose. Nat New Biol. 1971 Jul 7;232(27):16–17. doi: 10.1038/newbio232016a0. [DOI] [PubMed] [Google Scholar]
- Chesnut R. W., Colon S. M., Grey H. M. Requirements for the processing of antigens by antigen-presenting B cells. I. Functional comparison of B cell tumors and macrophages. J Immunol. 1982 Dec;129(6):2382–2388. [PubMed] [Google Scholar]
- Chesnut R. W., Endres R. O., Grey H. M. Antigen recognition by T cells and B cells: recognition of cross-reactivity between native and denatured forms of globular antigens. Clin Immunol Immunopathol. 1980 Mar;15(3):397–408. doi: 10.1016/0090-1229(80)90051-3. [DOI] [PubMed] [Google Scholar]
- GELL P. G., BENACERRAF B. Studies on hypersensitivity. II. Delayed hypersensitivity to denatured proteins in guinea pigs. Immunology. 1959 Jan;2(1):64–70. [PMC free article] [PubMed] [Google Scholar]
- Goetzl E. J., Peters J. H. Immunologic alterations in bovine serum albumin resulting from partial or complete reduction and alkylation. J Immunol. 1972 Mar;108(3):785–799. [PubMed] [Google Scholar]
- Gordon S., Todd J., Cohn Z. A. In vitro synthesis and secretion of lysozyme by mononuclear phagocytes. J Exp Med. 1974 May 1;139(5):1228–1248. doi: 10.1084/jem.139.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaka K., Okudaira H., King T. P. Immunogenic properties of modified antigen E. II. Ability of urea-denatured antigen and alpha-polypeptide chain to prime T cells specific for antigen E. J Immunol. 1975 Jan;114(1 Pt 1):110–115. [PubMed] [Google Scholar]
- Lee C. L., Atassi M. Z. Conformation and immunochemistry of methylated and carboxymethyulated derivatives of lysozyme. Biochemistry. 1973 Jul 3;12(14):2690–2695. doi: 10.1021/bi00738a022. [DOI] [PubMed] [Google Scholar]
- Lee K. C., Wong M., Spitzer D. Chloroquine as a probe for antigen processing by accessory cells. Transplantation. 1982 Sep;34(3):150–153. doi: 10.1097/00007890-198209000-00008. [DOI] [PubMed] [Google Scholar]
- Maizels R. M., Clarke J. A., Harvey M. A., Miller A., Sercarz E. E. Epitope specificity of the T cell proliferative response to lysozyme: proliferative T cells react predominantly to different determinants from those recognized by B cells. Eur J Immunol. 1980 Jul;10(7):509–515. doi: 10.1002/eji.1830100705. [DOI] [PubMed] [Google Scholar]
- Meek J. L. Prediction of peptide retention times in high-pressure liquid chromatography on the basis of amino acid composition. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1632–1636. doi: 10.1073/pnas.77.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G. D. Prediction of chain turns in globular proteins on a hydrophobic basis. Nature. 1978 Apr 13;272(5654):586–590. doi: 10.1038/272586a0. [DOI] [PubMed] [Google Scholar]
- Rosenthal A. S., Shevach E. M. Function of macrophages in antigen recognition by guinea pig T lymphocytes. I. Requirement for histocompatible macrophages and lymphocytes. J Exp Med. 1973 Nov 1;138(5):1194–1212. doi: 10.1084/jem.138.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANGER F., THOMPSON E. O. Halogenation of tyrosine during acid hydrolysis. Biochim Biophys Acta. 1963 May 14;71:468–471. doi: 10.1016/0006-3002(63)91108-9. [DOI] [PubMed] [Google Scholar]
- Schirrmacher V., Wigzell H. Immune responses against native and chemically modified albumins in mice. I. Analysis of non-thymus-processed (B) and thymus-processed (T) cell responses against methylated bovine serum albumin. J Exp Med. 1972 Dec 1;136(6):1616–1630. doi: 10.1084/jem.136.6.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. H., Paul W. E. T-lymphocyte-enriched murine peritoneal exudate cells. II. Genetic control of antigen-induced T-lymphocyte proliferation. J Exp Med. 1976 Mar 1;143(3):529–540. doi: 10.1084/jem.143.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimonkevitz R., Kappler J., Marrack P., Grey H. Antigen recognition by H-2-restricted T cells. I. Cell-free antigen processing. J Exp Med. 1983 Aug 1;158(2):303–316. doi: 10.1084/jem.158.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unanue E. R. The regulatory role of macrophages in antigenic stimulation. Part Two: symbiotic relationship between lymphocytes and macrophages. Adv Immunol. 1981;31:1–136. doi: 10.1016/s0065-2776(08)60919-0. [DOI] [PubMed] [Google Scholar]
- Ziegler H. K., Unanue E. R. Decrease in macrophage antigen catabolism caused by ammonia and chloroquine is associated with inhibition of antigen presentation to T cells. Proc Natl Acad Sci U S A. 1982 Jan;79(1):175–178. doi: 10.1073/pnas.79.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K., Unanue E. R. Identification of a macrophage antigen-processing event required for I-region-restricted antigen presentation to T lymphocytes. J Immunol. 1981 Nov;127(5):1869–1875. [PubMed] [Google Scholar]
- Ziegler K., Unanue E. R. The specific binding of Listeria monocytogenes-immune T lymphocytes to macrophages. I. Quantitation and role of H-2 gene products. J Exp Med. 1979 Nov 1;150(5):1143–1160. doi: 10.1084/jem.150.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]



