Abstract
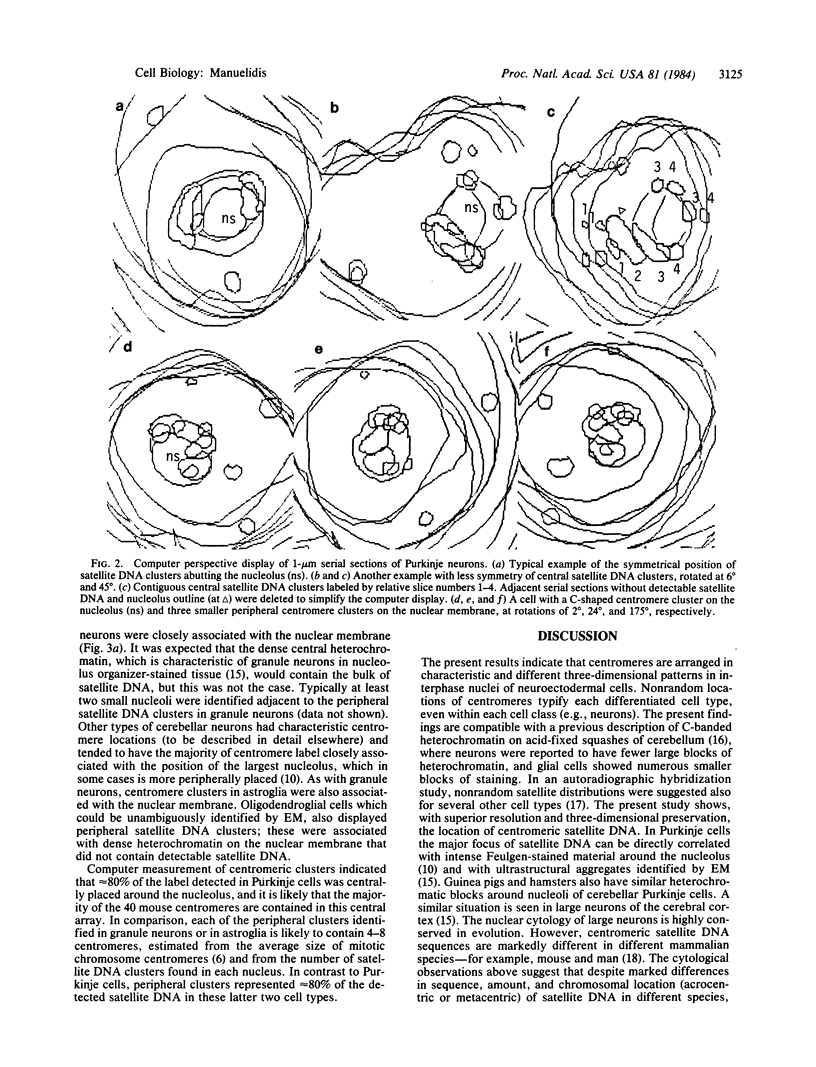
Paraformaldehyde-fixed tissue from mouse cerebellum was hybridized with biotin-labeled satellite DNA for identification of centromeres. By using avidin-peroxidase conjugates, it was possible to define the nuclear position of centromeres at the ultrastructural level. Three-dimensional analysis of well-resolved centromere arrays were aided by computer reconstruction of serial sections. Different cell types displayed distinct, nonrandom centromere locations. In Purkinje neurons, the majority of detected sequences were clustered together around the central nucleolus, whereas in granule neurons, more numerous, dispersed centromere clusters were associated with the nuclear membrane. In Purkinje cells, peroxidase-labeled regions corresponded to dense heterochromatic aggregates were detected in Purkinje cells of several different species. These observations suggest that in these highly differentiated cells, the nuclear position of centromeres is maintained in evolution despite species differences in centromeric DNA sequence. Such defined ordering of centromeres may be integral to specific functional capacities.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avivi L., Feldman M. Arrangement of chromosomes in the interphase nucleus of plants. Hum Genet. 1980;55(3):281–295. doi: 10.1007/BF00290206. [DOI] [PubMed] [Google Scholar]
- Brigati D. J., Myerson D., Leary J. J., Spalholz B., Travis S. Z., Fong C. K., Hsiung G. D., Ward D. C. Detection of viral genomes in cultured cells and paraffin-embedded tissue sections using biotin-labeled hybridization probes. Virology. 1983 Apr 15;126(1):32–50. doi: 10.1016/0042-6822(83)90460-9. [DOI] [PubMed] [Google Scholar]
- Comings D. E. The rationale for an ordered arrangement of chromatin in the interphase nucleus. Am J Hum Genet. 1968 Sep;20(5):440–460. [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Pardue M. L. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Natl Acad Sci U S A. 1969 Jun;63(2):378–383. doi: 10.1073/pnas.63.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. C., Cooper J. E., Mace M. L., Jr, Brinkley B. R. Arrangement of centromeres in mouse cells. Chromosoma. 1971;34(1):73–87. doi: 10.1007/BF00285517. [DOI] [PubMed] [Google Scholar]
- Hutchison N. J., Langer-Safer P. R., Ward D. C., Hamkalo B. A. In situ hybridization at the electron microscope level: hybrid detection by autoradiography and colloidal gold. J Cell Biol. 1982 Nov;95(2 Pt 1):609–618. doi: 10.1083/jcb.95.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John H. A., Birnstiel M. L., Jones K. W. RNA-DNA hybrids at the cytological level. Nature. 1969 Aug 9;223(5206):582–587. doi: 10.1038/223582a0. [DOI] [PubMed] [Google Scholar]
- Kurnit D. M., Maio J. J. Subnuclear redistribution of DNA species in confluent and growing mammalian cells. Chromosoma. 1973 May 14;42(1):23–36. doi: 10.1007/BF00326328. [DOI] [PubMed] [Google Scholar]
- Langer P. R., Waldrop A. A., Ward D. C. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. Consensus sequence of mouse satellite DNA indicates it is derived from tandem 116 basepair repeats. FEBS Lett. 1981 Jun 29;129(1):25–28. doi: 10.1016/0014-5793(81)80746-6. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Langer-Safer P. R., Ward D. C. High-resolution mapping of satellite DNA using biotin-labeled DNA probes. J Cell Biol. 1982 Nov;95(2 Pt 1):619–625. doi: 10.1083/jcb.95.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae M. M., Franke W. W. The interphase distribution of satellite DNA-containing heterochromatin in mouse nuclei. Chromosoma. 1972;39(4):443–456. doi: 10.1007/BF00326177. [DOI] [PubMed] [Google Scholar]
- Sedat J., Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- Singer R. H., Ward D. C. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7331–7335. doi: 10.1073/pnas.79.23.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Manuelidis L. Sequence definition and organization of a human repeated DNA. J Mol Biol. 1980 Sep 25;142(3):363–386. doi: 10.1016/0022-2836(80)90277-6. [DOI] [PubMed] [Google Scholar]