Abstract
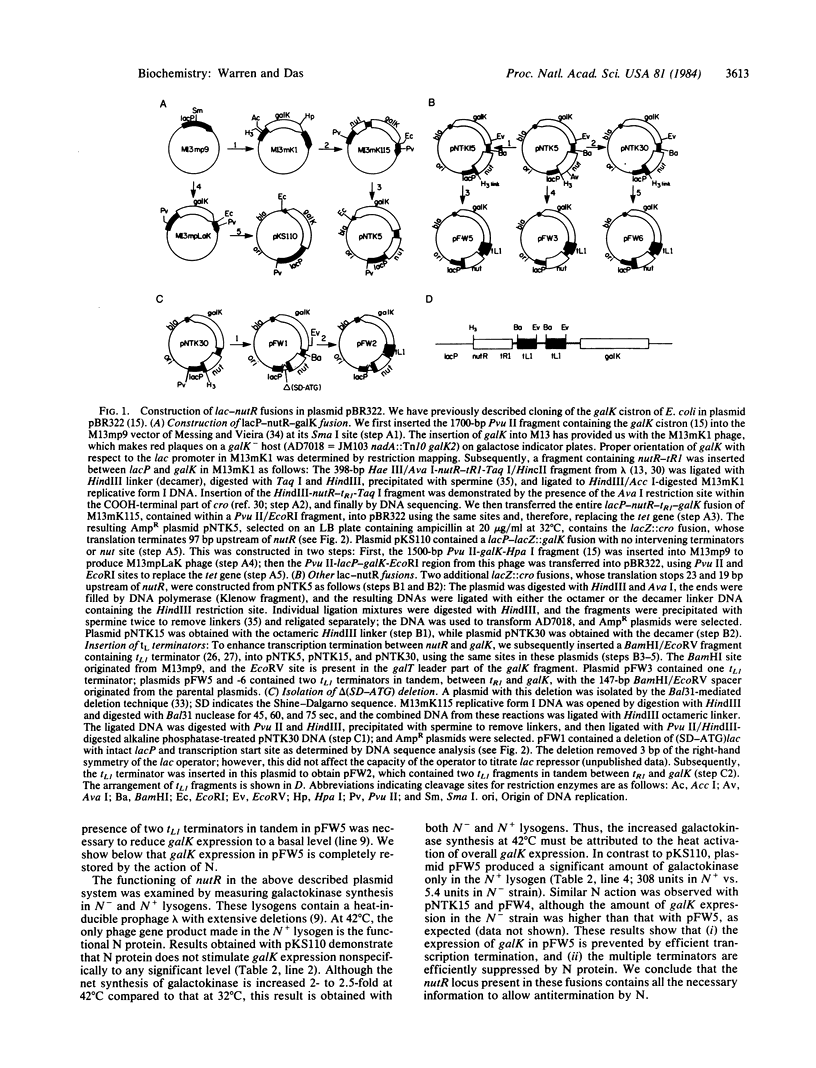
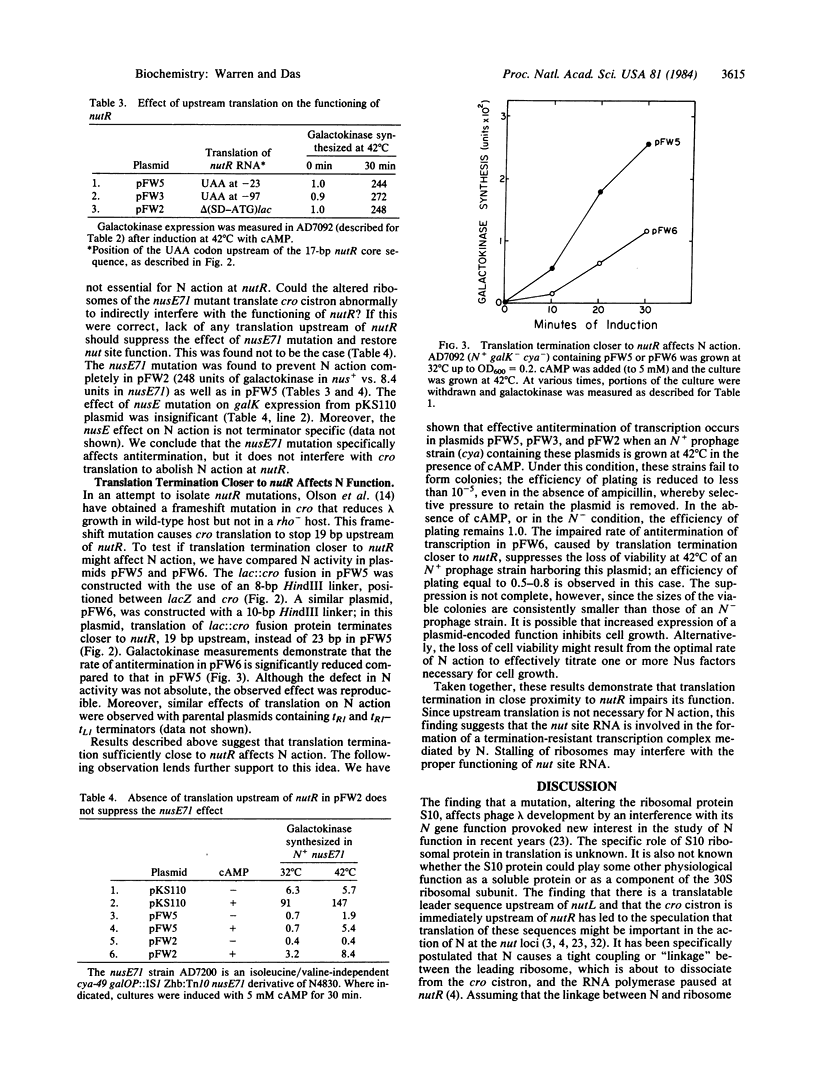
Transcription antitermination by lambda N gene product is affected in a mutant Escherichia coli with altered ribosomal protein S10, caused by the nusE71 mutation. To study the role of translation in antitermination, we have fused the phage nutR locus, the site of action of N, with the lac regulatory region. We have monitored N action by measuring galactokinase, whose synthesis depends on suppression of terminators located between nutR and the galK cistron. We show that a deletion removing potential ribosome binding signals and AUG codons from the upstream region of nut site does not affect N action. Moreover, the lack of translation upstream of nutR does not overcome the antitermination defect caused by nusE mutation. When the upstream region is translated, however, N action is impaired if translation terminates 19 base pairs upstream of nutR . Termination of translation at further upstream sites, such as 23 or 97 base pairs upstream, does not interfere with N action. Our results suggest that the S10 ribosomal protein is required for N action without involving translation. These results also suggest that the nut site RNA itself plays an important role in the formation of a termination-resistant transcription complex.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Adhya S., Gottesman M., De Crombrugghe B. Release of polarity in Escherichia coli by gene N of phage lambda: termination and antitermination of transcription. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2534–2538. doi: 10.1073/pnas.71.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann M. F., Friedman D. I. Cooperative effects of bacterial mutations affecting lambda N gene expression. II. Isolation and characterization of mutations in the rif region. Virology. 1976 Aug;73(1):128–138. doi: 10.1016/0042-6822(76)90067-2. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Gottesman M. E., Wardwell J., Trisler P., Gottesman S. lambda mutation in the Escherichia coli rho gene that inhibits the N protein activity of phage lambda. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5530–5534. doi: 10.1073/pnas.80.18.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin N. C. Altered reading of genetic signals fused to the N operon of bacteriophage lambda: genetic evidence for modification of polymerase by the protein product of the N gene. J Mol Biol. 1974 Oct 15;89(1):33–48. doi: 10.1016/0022-2836(74)90161-2. [DOI] [PubMed] [Google Scholar]
- Franklin N. C., Bennett G. N. The N protein of bacteriophage lambda, defined by its DNA sequence, is highly basic. Gene. 1979 Dec;8(1):107–119. doi: 10.1016/0378-1119(79)90011-8. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Baumann M., Baron L. S. Cooperative effects of bacterial mutations affecting lambda N gene expression. I. Isolation and characterization of a nusB mutant. Virology. 1976 Aug;73(1):119–127. doi: 10.1016/0042-6822(76)90066-0. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Schauer A. T., Baumann M. R., Baron L. S., Adhya S. L. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1115–1118. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Wilgus G. S., Mural R. J. Gene N regulator function of phage lambda immun21: evidence that a site of N action differs from a site of N recognition. J Mol Biol. 1973 Dec 25;81(4):505–516. doi: 10.1016/0022-2836(73)90519-6. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. P., Swindle J., Keppel F., Ballivet M., Bisig R., Eisen H. Studies on the E. coli groNB (nusB) gene which affects bacteriophage lambda N gene function. Mol Gen Genet. 1980;179(1):55–61. doi: 10.1007/BF00268446. [DOI] [PubMed] [Google Scholar]
- Ghysen A., Pironio M. Relationship between the N function of bacteriophage lambda and host RNA polymerase. J Mol Biol. 1972 Mar 28;65(2):259–272. doi: 10.1016/0022-2836(72)90281-1. [DOI] [PubMed] [Google Scholar]
- Gottesman M. E., Adhya S., Das A. Transcription antitermination by bacteriophage lambda N gene product. J Mol Biol. 1980 Jun 15;140(1):57–75. doi: 10.1016/0022-2836(80)90356-3. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J. The nusA gene protein of Escherichia coli. Its identification and a demonstration that it interacts with the gene N transcription anti-termination protein of bacteriophage lambda. J Mol Biol. 1981 Mar 25;147(1):11–23. doi: 10.1016/0022-2836(81)90076-0. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Malnoe P., Li J. Purification of the gene N transcription anti-termination protein of bacteriophage lambda. J Biol Chem. 1980 Feb 25;255(4):1465–1470. [PubMed] [Google Scholar]
- Herskowitz I., Hagen D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- Hoopes B. C., McClure W. R. Studies on the selectivity of DNA precipitation by spermine. Nucleic Acids Res. 1981 Oct 24;9(20):5493–5504. doi: 10.1093/nar/9.20.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani M., Orosz L., Busby S., Taniguchi T., Adhya S. Cyclic AMP-dependent constitutive expression of gal operon: use of repressor titration to isolate operator mutations. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4775–4779. doi: 10.1073/pnas.80.15.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. E. coli lactose operon ribosome binding site. Nature. 1974 Jun 14;249(458):647–649. doi: 10.1038/249647b0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B., Crapo L., Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968 Apr;59(4):1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. R., Flamm E. L., Friedman D. I. Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell. 1982 Nov;31(1):61–70. doi: 10.1016/0092-8674(82)90405-6. [DOI] [PubMed] [Google Scholar]
- Salstrom J. S., Szybalski W. Coliphage lambdanutL-: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol. 1978 Sep 5;124(1):195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Scherer G., Hobom G., Kössel H. Nucleotide sequence of cro, cII and part of the O gene in phage lambda DNA. Nature. 1978 Mar 30;272(5652):410–414. doi: 10.1038/272410a0. [DOI] [PubMed] [Google Scholar]
- Sternberg N. A class of rifR RNA polymerase mutations that interferes with the expression of coliphage lambda late gene. Virology. 1976 Aug;73(1):139–154. doi: 10.1016/0042-6822(76)90068-4. [DOI] [PubMed] [Google Scholar]
- Strauch M., Friedman D. I. Identification of the nusB gene product of Escherichia coli. Mol Gen Genet. 1981;182(3):498–501. doi: 10.1007/BF00293941. [DOI] [PubMed] [Google Scholar]
- Ward D. F., Gottesman M. E. Suppression of transcription termination by phage lambda. Science. 1982 May 28;216(4549):946–951. doi: 10.1126/science.6281888. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Mudryj M., DiLauro R., Gottesman M. Specificity of the bacteriophage lambda N gene product (pN): nut sequences are necessary and sufficient for antitermination by pN. Cell. 1979 Dec;18(4):1145–1151. doi: 10.1016/0092-8674(79)90227-7. [DOI] [PubMed] [Google Scholar]



