Abstract
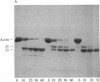
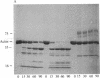
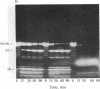
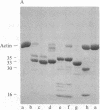
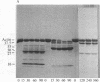
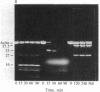
Under standard conditions, G-actin has been submitted to nine proteases of varying specificity, and in each case the pattern of fragments produced has been studied by NaDodSO4 gel electrophoresis. The results suggest that the actin monomer consists of a large region (ca. 33 kilodaltons) and a small, easily degraded region (ca. 9 kilodaltons). The COOH terminus is in the large region. Consideration of primary sequence homologies, medium resolution maps of actin crystals, and certain reactions of actin suggests that the NH2 terminus is in the small region, as is the negative sequence to which a divalent metal cation is normally chelated, but that the nucleotide-binding site is on the large region near the junction between the regions. From analysis of these results, numerous properties of actin are understandable.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asai H., Tawada K. Enzymic nature of F-actin at high temperature. J Mol Biol. 1966 Sep;20(2):403–417. doi: 10.1016/0022-2836(66)90071-4. [DOI] [PubMed] [Google Scholar]
- Barden J. A., Cooke R., Wright P. E., dos Remedios C. G. Proton nuclear magnetic resonance and electron paramagnetic resonance studies on skeletal muscle actin indicate that the metal and nucleotide binding sites are separate. Biochemistry. 1980 Dec 9;19(25):5912–5916. doi: 10.1021/bi00566a038. [DOI] [PubMed] [Google Scholar]
- Benyamin Y., Roustan C., Boyer M. Induction by chemically modified actin derivatives of antibody specificity. A relation between modified sites and antibody interactions with monomeric and filamentous actins. FEBS Lett. 1983 Aug 22;160(1-2):41–45. doi: 10.1016/0014-5793(83)80932-6. [DOI] [PubMed] [Google Scholar]
- Brauer M., Sykes B. D. Effects of manganous ion on the phosphorus-31 nuclear magnetic resonance spectrum of adenosine triphosphate bound to nitrated G-actin: proximity of divalent metal ion and nucleotide binding sites. Biochemistry. 1982 Nov 9;21(23):5934–5939. doi: 10.1021/bi00266a032. [DOI] [PubMed] [Google Scholar]
- Burtnick L. D., Chan K. W. Protection of actin against proteolysis by complex formation with deoxyribonuclease I. Can J Biochem. 1980 Dec;58(12):1348–1354. doi: 10.1139/o80-183. [DOI] [PubMed] [Google Scholar]
- Collins J. H., Elzinga M. The primary structure of actin from rabbit skeletal muscle. Three cyanogen bromide peptides that are insoluble at neutral pH. J Biol Chem. 1975 Aug 10;250(15):5906–5914. [PubMed] [Google Scholar]
- Collins J. H. Homology of myosin DTNB light chain with alkali light chains, troponin C and parvalbumin. Nature. 1976 Feb 26;259(5545):699–700. doi: 10.1038/259699a0. [DOI] [PubMed] [Google Scholar]
- Curmi P. M., Barden J. A., dos Remedios C. G. Conformational studies of G-actin containing bound lanthanide. Eur J Biochem. 1982 Feb;122(2):239–244. doi: 10.1111/j.1432-1033.1982.tb05872.x. [DOI] [PubMed] [Google Scholar]
- De Couet H. G. Studies on the antigenic sites of actin: a comparative study of the immunogenic crossreactivity of invertebrate actins. J Muscle Res Cell Motil. 1983 Aug;4(4):405–427. doi: 10.1007/BF00711947. [DOI] [PubMed] [Google Scholar]
- Egelman E. H., Francis N., DeRosier D. J. F-actin is a helix with a random variable twist. Nature. 1982 Jul 8;298(5870):131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- Eisenberg E., Kielley W. W. Troponin-tropomyosin complex. Column chromatographic separation and activity of the three, active troponin components with and without tropomyosin present. J Biol Chem. 1974 Aug 10;249(15):4742–4748. [PubMed] [Google Scholar]
- Fagraeus A., Norberg R., Biberfeld G. Anti-actin antibodies of human and rabbit origin. Ann Immunol (Paris) 1978 Feb-Mar;129(2-3):245–254. [PubMed] [Google Scholar]
- Jacobson G. R., Rosenbusch J. P. ATP binding to a protease-resistant core of actin. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2742–2746. doi: 10.1073/pnas.73.8.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P., Wester P. J., Hikida R. S. Protein-protein interactions of proteolytic fragments of actin. Biochim Biophys Acta. 1979 May 23;578(1):253–257. doi: 10.1016/0005-2795(79)90133-8. [DOI] [PubMed] [Google Scholar]
- Kagawa H. The reaction of a positively-charged N-ethylmaleimide derivative with actin and myosin subfragment-1. Int J Biochem. 1981;13(7):871–873. doi: 10.1016/0020-711x(81)90109-9. [DOI] [PubMed] [Google Scholar]
- Kassab R., Mornet D., Pantel P., Bertrand R., Audemard E. Structural aspects of actomyosin interaction. Biochimie. 1981 Apr;63(4):273–289. doi: 10.1016/s0300-9084(81)80116-2. [DOI] [PubMed] [Google Scholar]
- Korn E. D. Actin polymerization and its regulation by proteins from nonmuscle cells. Physiol Rev. 1982 Apr;62(2):672–737. doi: 10.1152/physrev.1982.62.2.672. [DOI] [PubMed] [Google Scholar]
- Kretsinger R. H., Nockolds C. E. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973 May 10;248(9):3313–3326. [PubMed] [Google Scholar]
- MARTONOSI A., GOUVEA M. A., GERGELY J. Studies on actin. III. G-F transformation of actin and muscular contraction (experiments in vivo). J Biol Chem. 1960 Jun;235:1707–1710. [PubMed] [Google Scholar]
- MARTONOSII A., MOLINO C. M., GERGELY J. THE BINDING OF DIVALENT CATIONS TO ACTIN. J Biol Chem. 1964 Apr;239:1057–1064. [PubMed] [Google Scholar]
- Mornet D., Bertrand R. U., Pantel P., Audemard E., Kassab R. Proteolytic approach to structure and function of actin recognition site in myosin heads. Biochemistry. 1981 Apr 14;20(8):2110–2120. doi: 10.1021/bi00511a007. [DOI] [PubMed] [Google Scholar]
- Mornet D., Bertrand R., Pantel P., Audemard E., Kassab R. Structure of the actin-myosin interface. Nature. 1981 Jul 23;292(5821):301–306. doi: 10.1038/292301a0. [DOI] [PubMed] [Google Scholar]
- Mornet D., Ue K., Morales M. F. Proteolysis and the domain organization of myosin subfragment 1. Proc Natl Acad Sci U S A. 1984 Feb;81(3):736–739. doi: 10.1073/pnas.81.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszbek L., Gladner J. A., Laki K. The fragmentation of actin by thrombin. Isolation and characterization of the split products. Arch Biochem Biophys. 1975 Mar;167(1):99–103. doi: 10.1016/0003-9861(75)90445-2. [DOI] [PubMed] [Google Scholar]
- Muszbek L., Laki K. Cleavage of actin by thrombin. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2208–2211. doi: 10.1073/pnas.71.6.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offer G., Moos C., Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J Mol Biol. 1973 Mar 15;74(4):653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- Rich S. A., Estes J. E. Detection of conformational changes in actin by proteolytic digestion: evidence for a new monomeric species. J Mol Biol. 1976 Jul 15;104(4):777–792. doi: 10.1016/0022-2836(76)90181-9. [DOI] [PubMed] [Google Scholar]
- Selden L. A., Estes J. E., Gershman L. C. The tightly bound divalent cation regulates actin polymerization. Biochem Biophys Res Commun. 1983 Oct 31;116(2):478–485. doi: 10.1016/0006-291x(83)90548-x. [DOI] [PubMed] [Google Scholar]
- Suck D., Kabsch W., Mannherz H. G. Three-dimensional structure of the complex of skeletal muscle actin and bovine pancreatic DNAse I at 6-A resolution. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4319–4323. doi: 10.1073/pnas.78.7.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh K. Identification of myosin-binding sites on the actin sequence. Biochemistry. 1982 Jul 20;21(15):3654–3661. doi: 10.1021/bi00258a020. [DOI] [PubMed] [Google Scholar]
- Takashi R. Fluorescence energy transfer between subfragment-1 and actin points in the rigor complex of actosubfragment-1. Biochemistry. 1979 Nov 13;18(23):5164–5169. doi: 10.1021/bi00590a021. [DOI] [PubMed] [Google Scholar]
- Taylor D. L., Reidler J., Spudich J. A., Stryer L. Detection of actin assembly by fluorescence energy transfer. J Cell Biol. 1981 May;89(2):362–367. doi: 10.1083/jcb.89.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeds A. G., McLachlan A. D. Structural homology of myosin alkali light chains, troponin C and carp calcium binding protein. Nature. 1974 Dec 20;252(5485):646–649. doi: 10.1038/252646a0. [DOI] [PubMed] [Google Scholar]
- Weeds A. G., Taylor R. S. Separation of subfragment-1 isoenzymes from rabbit skeletal muscle myosin. Nature. 1975 Sep 4;257(5521):54–56. doi: 10.1038/257054a0. [DOI] [PubMed] [Google Scholar]
- West J. J., Nagy B., Gergely J. Free adenosine diphosphate as an intermediary in the phosphorylation by creatine phosphate of adenosine diphosphate bound to actin. J Biol Chem. 1967 Mar 25;242(6):1140–1145. [PubMed] [Google Scholar]
- Yanagida T., Nakase M., Nishiyama K., Oosawa F. Direct observation of motion of single F-actin filaments in the presence of myosin. Nature. 1984 Jan 5;307(5946):58–60. doi: 10.1038/307058a0. [DOI] [PubMed] [Google Scholar]
- Yanagida T., Oosawa F. Conformational changes of F-actin-epsilon-ADP in thin filaments in myosin-free muscle fibers induced by Ca2+. J Mol Biol. 1980 Jun 25;140(2):313–320. doi: 10.1016/0022-2836(80)90108-4. [DOI] [PubMed] [Google Scholar]