Abstract
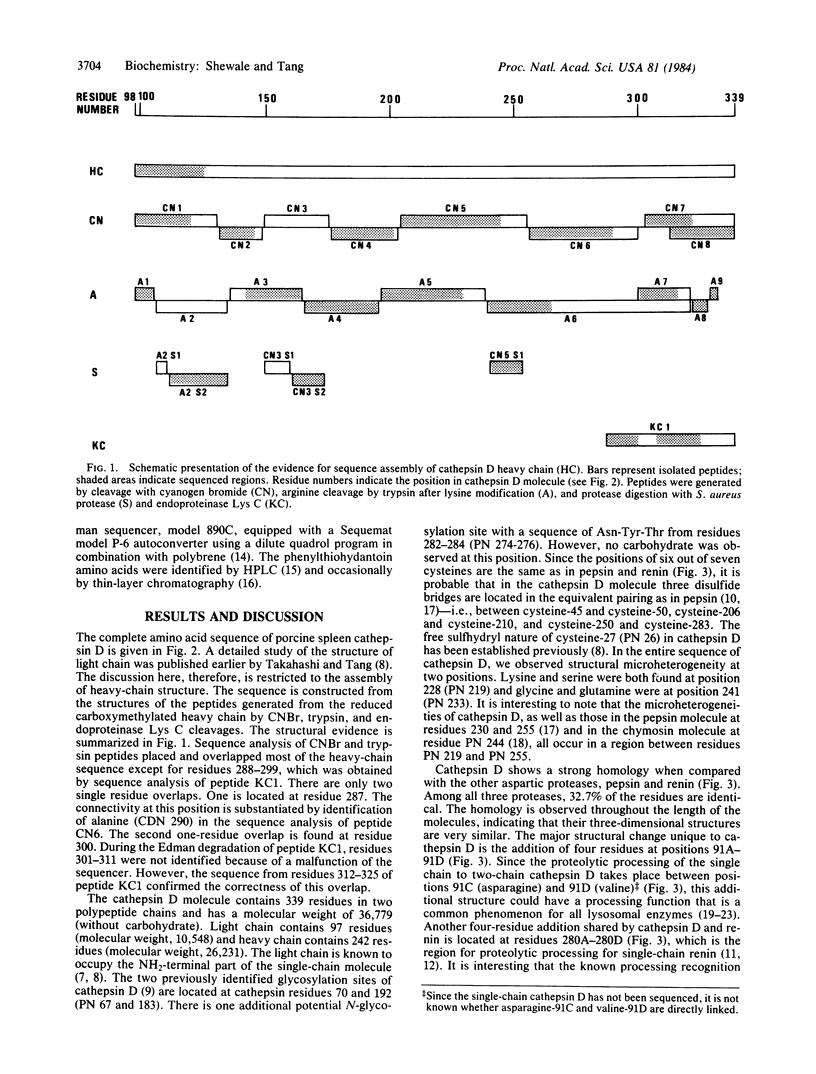
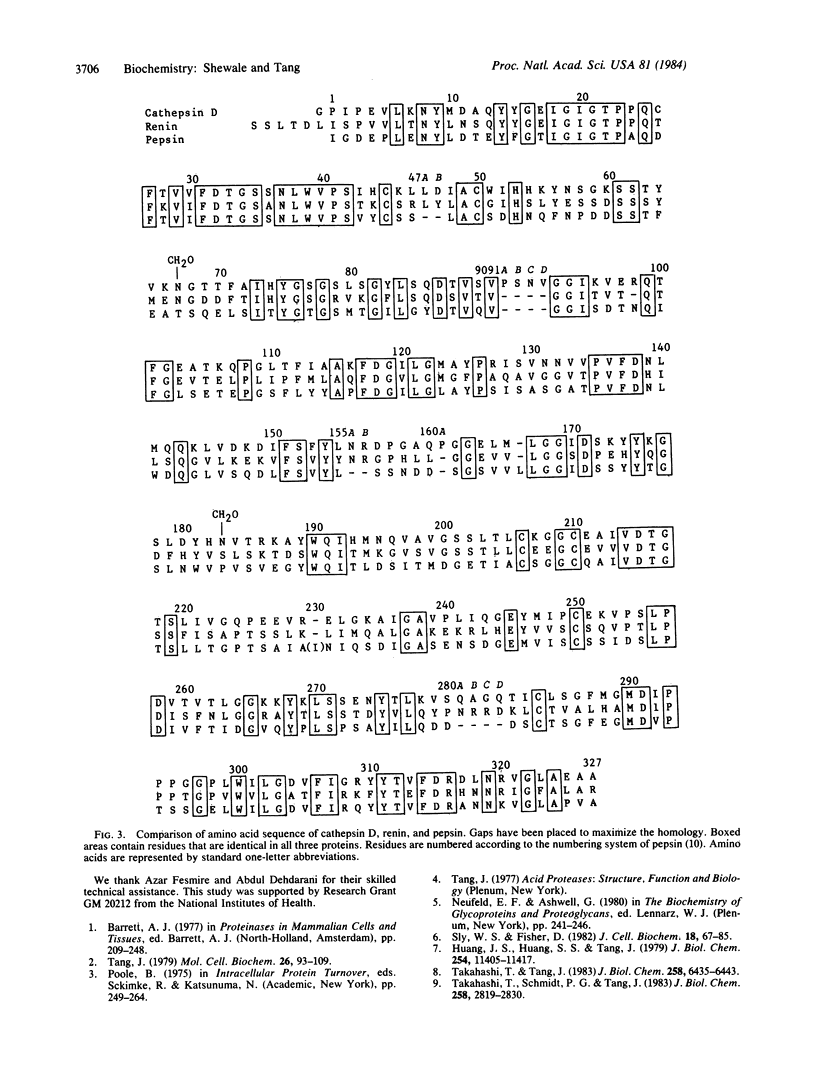
The amino acid sequence of porcine spleen cathepsin D heavy chain has been determined and, hence, the complete structure of this enzyme is now known. The sequence of heavy chain was constructed by aligning the structures of peptides generated by cyanogen bromide, trypsin, and endo-proteinase Lys C cleavages. The structure of the light chain has been published previously. The cathepsin D molecule contains 339 amino acid residues in two polypeptide chains: a 97-residue light chain and a 242-residue heavy chain, with a combined Mr of 36,779 (without carbohydrate). There are two carbohydrate units linked to asparagine residues 70 and 192. The disulfide bond arrangement in cathepsin D is probably similar to that of pepsin, because the positions of six half-cystine residues are conserved. The active site aspartyl residues, corresponding to aspartic acid-32 and -215 of pepsin, are located at residues 33 and 224 in the cathepsin D molecule. The amino acid sequence around these aspartyl residues is strongly conserved. Cathepsin D shows a strong homology with other acid proteases. When the sequence of cathepsin D, renin, and pepsin are aligned, 32.7% of the residues are identical. The homology is observed throughout the length of the molecules, indicating that three-dimensional structures of all three molecules are similar.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brauer A. W., Margolies M. N., Haber E. The application of 0.1 M quadrol to the microsequence of proteins and the sequence of tryptic peptides. Biochemistry. 1975 Jul;14(13):3029–3035. doi: 10.1021/bi00684a036. [DOI] [PubMed] [Google Scholar]
- Cunningham M., Tang J. Purification and properties of cathepsin D from porcine spleen. J Biol Chem. 1976 Aug 10;251(15):4528–4536. [PubMed] [Google Scholar]
- Erickson A. H., Conner G. E., Blobel G. Biosynthesis of a lysosomal enzyme. Partial structure of two transient and functionally distinct NH2-terminal sequences in cathepsin D. J Biol Chem. 1981 Nov 10;256(21):11224–11231. [PubMed] [Google Scholar]
- Ferguson J. B., Andrews J. R., Voynick I. M., Fruton J. S. The specificity of cathepsin D. J Biol Chem. 1973 Oct 10;248(19):6701–6708. [PubMed] [Google Scholar]
- Foltmann B., Pedersen V. B., Jacobsen H., Kauffman D., Wybrandt G. The complete amino acid sequence of prochymosin. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2321–2324. doi: 10.1073/pnas.74.6.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruton J. S. The mechanism of the catalytic action of pepsin and related acid proteinases. Adv Enzymol Relat Areas Mol Biol. 1976;44:1–36. doi: 10.1002/9780470122891.ch1. [DOI] [PubMed] [Google Scholar]
- Hasilik A., Neufeld E. F. Biosynthesis of lysosomal enzymes in fibroblasts. Synthesis as precursors of higher molecular weight. J Biol Chem. 1980 May 25;255(10):4937–4945. [PubMed] [Google Scholar]
- Henderson L. E., Copeland T. D., Oroszlan S. Separation of amino acid phenylthiohydantoins by high-performance liquid chromatography on phenylalkyl support. Anal Biochem. 1980 Feb;102(1):1–7. doi: 10.1016/0003-2697(80)90307-3. [DOI] [PubMed] [Google Scholar]
- Hsu I. N., Delbaere L. T., James M. N., Hofmann T. Penicillopepsin from Penicillium janthinellum crystal structure at 2.8 A and sequence homology with porcine pepsin. Nature. 1977 Mar 10;266(5598):140–145. doi: 10.1038/266140a0. [DOI] [PubMed] [Google Scholar]
- Huang J. S., Huang S. S., Tang J. Cathepsin D isozymes from porcine spleens. Large scale purification and polypeptide chain arrangements. J Biol Chem. 1979 Nov 25;254(22):11405–11417. [PubMed] [Google Scholar]
- James M. N., Hsu I. N., Delbaere L. T. Mechanism of acid protease catalysis based on the crystal structure of penicillopepsin. Nature. 1977 Jun 30;267(5614):808–813. doi: 10.1038/267808a0. [DOI] [PubMed] [Google Scholar]
- Keilová H. Inhibition of cathepsin D by diazoacetylnorleucine methyl ester. FEBS Lett. 1970 Feb 25;6(4):312–314. doi: 10.1016/0014-5793(70)80086-2. [DOI] [PubMed] [Google Scholar]
- Myerowitz R., Neufeld E. F. Maturation of alpha-L-iduronidase in cultured human fibroblasts. J Biol Chem. 1981 Mar 25;256(6):3044–3048. [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Newton A. Isolation and expression of cloned hook protein gene from Caulobacter crescentus. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4863–4867. doi: 10.1073/pnas.79.16.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panthier J. J., Foote S., Chambraud B., Strosberg A. D., Corvol P., Rougeon F. Complete amino acid sequence and maturation of the mouse submaxillary gland renin precursor. Nature. 1982 Jul 1;298(5869):90–92. doi: 10.1038/298090a0. [DOI] [PubMed] [Google Scholar]
- Sepulveda P., Marciniszyn J., Jr, Liu D., Tang J. Primary structure of porcine pepsin. III. Amino acid sequence of a cyanogen bromide fragment, CB2A, and the complete structure of porcine pepsin. J Biol Chem. 1975 Jul 10;250(13):5082–5088. [PubMed] [Google Scholar]
- Skudlarek M. D., Swank R. T. Biosynthesis of two lysosomal enzymes in macrophages. Evidence for a precursor of beta-galactosidase. J Biol Chem. 1979 Oct 25;254(20):9939–9942. [PubMed] [Google Scholar]
- Sly W. S., Fischer H. D. The phosphomannosyl recognition system for intracellular and intercellular transport of lysosomal enzymes. J Cell Biochem. 1982;18(1):67–85. doi: 10.1002/jcb.1982.240180107. [DOI] [PubMed] [Google Scholar]
- Summers M. R., Smythers G. W., Oroszlan S. Thin-layer chromatography of sub-nanomole amounts of phenylthiohydantoin (PTH) amino acids on polyamide sheets. Anal Biochem. 1973 Jun;53(2):624–628. doi: 10.1016/0003-2697(73)90114-0. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Schmidt P. G., Tang J. Oligosaccharide units of lysosomal cathepsin D from porcine spleen. Amino acid sequence and carbohydrate structure of the glycopeptides. J Biol Chem. 1983 Mar 10;258(5):2819–2830. [PubMed] [Google Scholar]
- Takahashi T., Tang J. Amino acid sequence of porcine spleen cathepsin D light chain. J Biol Chem. 1983 May 25;258(10):6435–6443. [PubMed] [Google Scholar]
- Takio K., Towatari T., Katunuma N., Teller D. C., Titani K. Homology of amino acid sequences of rat liver cathepsins B and H with that of papain. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3666–3670. doi: 10.1073/pnas.80.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. Evolution in the structure and function of carboxyl proteases. Mol Cell Biochem. 1979 Jul 31;26(2):93–109. doi: 10.1007/BF00232887. [DOI] [PubMed] [Google Scholar]
- Tang J., Sepulveda P., Marciniszyn J., Jr, Chen K. C., Huang W. Y., Tao N., Liu D., Lanier J. P. Amino-acid sequence of porcine pepsin. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3437–3439. doi: 10.1073/pnas.70.12.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]


