Abstract
Background
Anaplasma phagocytophilum infects a wide variety of hosts and causes granulocytic anaplasmosis in humans, horses and dogs and tick-borne fever in ruminants. Infection with A. phagocytophilum results in the modification of host gene expression and immune response. The objective of this research was to characterize gene expression in pigs (Sus scrofa) naturally and experimentally infected with A. phagocytophilum trying to identify mechanisms that help to explain low infection prevalence in this species.
Results
For gene expression analysis in naturally infected pigs, microarray hybridization was used. The expression of differentially expressed immune response genes was analyzed by real-time RT-PCR in naturally and experimentally infected pigs. Results suggested that A. phagocytophilum infection affected cytoskeleton rearrangement and increased both innate and adaptive immune responses by up regulation of interleukin 1 receptor accessory protein-like 1 (IL1RAPL1), T-cell receptor alpha chain (TCR-alpha), thrombospondin 4 (TSP-4) and Gap junction protein alpha 1 (GJA1) genes. Higher serum levels of IL-1 beta, IL-8 and TNF-alpha in infected pigs when compared to controls supported data obtained at the mRNA level.
Conclusions
These results suggested that pigs are susceptible to A. phagocytophilum but control infection, particularly through activation of innate immune responses, phagocytosis and autophagy. This fact may account for the low infection prevalence detected in pigs in some regions and thus their low or no impact as a reservoir host for this pathogen. These results advanced our understanding of the molecular mechanisms at the host-pathogen interface and suggested a role for newly reported genes in the protection of pigs against A. phagocytophilum.
Keywords: Anaplasmosis, Genetics, Pig, Wild boar, Genomics, Immune response
Background
Anaplasma phagocytophilum (Rickettsiales: Anaplasmataceae) is a tick-borne pathogen that infects a wide range of hosts including humans and wild and domestic animals [1,2]. A. phagocytophilum is the causative agent of human, equine and canine granulocytic anaplasmosis and tick-borne fever in ruminants [1,3,4]. In Europe, A. phagocytophilum is the most widespread tick-borne infection in animals with an increasing incidence in humans [5-10]. A. phagocytophilum is transmitted by Ixodes spp., but other tick species may subsequently also prove to be vectors [11,12]. Evidence suggests that persistent infections occur in domestic and wild ruminants, which can then serve as reservoir hosts [1,9]. The broad geographic distribution and the clinical and host tropism diversity of A. phagocytophilum strains suggest the presence of complex infection-transmission networks that may influence the epizootiology of the disease [13].
A. phagocytophilum has been reported with low prevalence in wild pigs (Sus scrofa) in the Czech Republic [14] and Slovenia [15]. Recently, 12% prevalence of was detected in wild boar in Poland [16]. In Slovenia and Poland, the A. phagocytophilum gene sequences found in wild pigs were identical to that found in humans and I. ricinus ticks [15,16]. In Sicily, evidence suggested that A. phagocytophilum infection might occur in pigs [17]. In south-central Spain, where I. ricinus are scarce [18], Anaplasma spp. has not been reported in wild boar [13,19,20], although other tick species feeding on wild boar were positive for A. phagocytophilum DNA [12]. Recently, 16S rDNA but not p44/msp2 genotypes identical to A. phagocytophilum were found with low prevalence in wild boar in Japan [21] but a survey in Mississippi, United States, failed to detect pathogen DNA in feral pigs [22]. These results suggested that wild pigs might play a role in the epizootiology of A. phagocytophilum by serving as a natural reservoir host in some regions only.
Infection with A. phagocytophilum has been shown to modify the host cell gene expression. The gene expression profile has been characterized in human cells [23-28] and sheep [29] infected with A. phagocytophilum. As shown by recent studies in sheep [29], gene expression profile in response to A. phagocytophilum infection may differ between human cells and ruminant hosts. These differences may be the result of species-specific differences and/or the effect of different pathogen strains [2,29].
The objective of this study was to characterize gene expression profiles emphasizing on immune response genes in wild and domestic pigs in response to A. phagocytophilum using a combination of microarray hybridization and real-time RT-PCR. These results will expand current information on the mammalian host response to A. phagocytophilum infection and contribute to the overall understanding of the molecular mechanisms involved in pathogen infection, multiplication and persistence.
Materials and methods
Experimental design and rationale
The finding of wild pigs naturally infected with A. phagocytophilum in Slovenia suggested that this pathogen might also infect pigs, thus probably affecting gene expression in this species. The genes differentially expressed in response to A. phagocytophilum infection were first characterized in wild pigs naturally infected with A. phagocytophilum by microarray hybridization and real-time RT-PCR. The differentially expressed immune response genes were then further characterized in domestic pigs experimentally infected with A. phagocytophilum under controlled experimental conditions.
Wild pigs and sample preparation
Buffy coats were prepared from blood samples collected from adult (≥1 year-old) wild pig males hunter-killed during 2007 in Kočevje–Šubičeva and Kostel–Delač, Slovenia. Total DNA and RNA were extracted using MagneSil KF genomic DNA (Promega, Madison, WI, USA) and TRIzol Reagent (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA), respectively according to manufacturer’s instructions. The DNA was used to test for A. phagocytophilum infection using 16S rDNA and groESL PCRs and sequence analysis as previously reported [15]. Three of the 113 pigs analyzed tested positive for the presence of A. phagocytophilum DNA and were selected for further analysis. Control Buffy coats were prepared from uninfected adult wild pig males hunter-killed in south-central Spain where pigs are not infected with Anaplasma spp. [13,19,20]. Control animals tested negative in the A. phagocytophilum 16S rDNA and groESL PCRs. All animals tested negative for other pathogens commonly found in wild pigs such as Mycobacterium bovisBrucella suis, Aujeszky´s Disease Virus (ADV) and porcine circovirus type 2 (PCV2).
Microarray hybridization and analysis
Total RNA from wild pigs was characterized using the Experion™ Automated Electrophoresis System (Bio-Rad, Hercules, CA, USA) in order to evaluate the quality and integrity of RNA preparations. One RNA sample from infected animals did not have the quality required for microarray hybridization. Therefore, two samples from infected animals were selected for microarray hybridization analysis together with three RNA samples from uninfected control animals. To obtain a comprehensive gene expression profile in response to A. phagocytophilum infection, the GeneChip® Porcine Genome Array was used, which contains 23,937 probe sets that interrogate approximately 23,256 transcripts from 20,201 S. scrofa genes (Affymetrix, Santa Clara, CA, USA; http://www.affymetrix.com/products_services/arrays/specific/porcine.affx). Two μg total RNA were labeled using the GeneChip® HT IVT Labeling Kit (Affymetrix). The images were processed with Microarray Analysis Suite 5.0 (Affymetrix). Raw expression values obtained directly from CEL files were preprocessed using the RMA method [30], a three-step process which integrates background correction, normalization and summarization of probe values. Standard quality controls based on Affymetrix original methods including average background, scale factor, number of genes called present, 3´ to 5´ ratios computed from the MAS 5.0 algorithm and probe-level models (PLM) based on fitting a model for probe values and analyzing its residuals (Relative Log Expression and Normal Unscaled Standard Error) were performed. Arrays that did not show the minimum acceptable quality based on these standard quality controls were discarded. After quality control, the mean expression of each probe set in controls was compared with that of the infected samples to summarize the comparison between controls and the infected samples. Microarray data analysis was done using the free statistical language R and the libraries developed by the Bioconductor Project (http://www.bioconductor.org). In order to deal with the multiple testing issues derived from the fact that many tests (one per gene) were performed simultaneously, p-values were adjusted to obtain strong control over the false discovery rate using the Benjamini and Hochberg method [31]. All the microarray data were deposited at the NCBI Gene Expression Omnibus (GEO) under the platform accession number GPL3533 and the series number GSE15766.
Sequence analysis
Gene ontology (GO) assignments were retrieved from the GeneChip® Porcine Genome Array (Affymetrix) and verified by searching the Entrez (http://www.ncbi.nlm.nih.gov/sites/gquery) and Gene ontology (http://www.geneontology.org/) databases. The gene ontology (GO) enrichment analysis was performed with GOstats package [32]. For each GO category of interest, entries in the array were compared with results of differentially expressed genes by χ2-test (p = 0.01).
Domestic pigs and sample preparation
Six 9-weeks-old pathogen-free male pigs were randomly distributed into two experimental groups with three animals each, infected and uninfected. Pigs were experimentally infected with A. phagocytophilum by intravenous inoculation (iv) of ISE6 tick cell cultures infected with the human NY-18 isolate of A. phagocytophilum[33,34]. Pigs were each inoculated with one T-25 flask of A. phagocytophilum-infected ISE6 tick cells (11-15% infection, as determined by detection of intracellular morulae in stained cytospin cell smears; Hema-3 Stain, Fisher Scientific, Middletown, VA, USA) at days 0 and 36 of the experiment. Control pigs were inoculated with control uninfected tick cells. Uninfected and infected cultures were centrifuged at 1,000 x g for 5 min and resuspended in L-15B medium without fetal bovine serum and antibiotics in a final iv dose of 1 x 107 cells/2 ml. All pigs were monitored for infection by recording clinical signs, PCR of blood samples, examination of stained blood films and by Anaplasma serology at days 0 (before first inoculation), 7, 15, 36 (before second inoculation), 47 and 62. At day 62, pigs were euthanized by a licensed veterinarian and subjected to gross necropsy examination. Animals were cared for in accordance with standards specified in the Guide for Care and Use of Laboratory Animals and approved by ethical committee for animal care and experimentation (No. 10/397354.9/11).
Detection of A. phagocytophilum in experimentally infected pigs by PCR
DNA was extracted from pig blood samples using TriReagent (Sigma, St. Louis, MO, USA) following manufacturer’s recommendations. A. phagocytophilum infection levels were characterized by msp4 PCR using the iQ5 thermal cycler (Bio-Rad, Hercules, CA, USA) as described previously using oligonucleotide primers MAP4AP5: 5’-ATGAATTACAGAGAATTGCTTGTAGG-3’ and MSP4AP3: 5’-TTAATTGAAAGCAAATCTTGCTCCTATG-3’) in a 50-μl volume PCR (1.5mMMgSO4, 0.2mM dNTP, 5XGoTaq reaction buffer, 5u GoTaqDNA polymerase) (Promega, Madison, WI, USA) [2]. Negative control reactions were performed with the same procedures, but adding water instead of DNA to monitor contamination of the PCR. PCR products were electrophoresed on 1% agarose gels to check the size of amplified fragments by comparison to a DNA molecular weight marker (1 kb DNA Ladder, Promega). Amplified fragments were resin purified (Invitrogen, Carlsbad, CA, USA) for sequencing both strands by double-stranded dye-termination cycle sequencing (Secugen SL, Madrid, Spain). The msp4 coding region was used for sequence alignment. Multiple sequence alignment was performed using the program DNA Baser (Heracle BioSoft S.R.L., Pitesti, Romania).
Detection of anti-A. phagocytophilum antibodies in experimentally infected pigs by ELISA
Serum samples were tested for IgG antibodies by means of an in-house indirect ELISA using the A. phagocytophilum (NY-18) recombinant MSP4 protein as antigen and protein G horseradish peroxidase as a conjugate using the protocol described by Araújo et al. [35] with some modifications. Briefly, 96-well plates (MaxiBinding, SPL Life sciences, Korea) were coated overnight at 4°C with 0.4 μg/ml of MSP4, diluted in carbonate-bicarbonate phosphate buffer. Plates were blocked for 1 hr at 37°C with 140 μl/well of a solution containing 5% skim milk with phosphate buffered saline and 0.05% Tween-20 (PBST). Sera were added directly on plate (100 μl/well) at a dilution of 1:100 in PBST and incubated for 1 hr at 37°C. Plates were then washed five times with PBST, and Protein G (Sigma Aldrich, Saint Louis, USA) was added (100 μl/well) at a dilution of 1:1,000 in PBST and incubated at 37°C for 1 hr. After five washes with PBST, the chromogen/substrate o-phenylene diamine dihydrochloride (OPD; Sigma)/H2O2 was added. The reaction was stopped with 50 μl/well of sulphuric acid (H2SO4; 3N), and the optical density (OD) was measured in a spectrophotometer at 450 nm. White-tailed deer and cattle sera positive to Anaplasma were included as controls. Antibody titers in experimentally infected and control pigs were expressed as the OD450nm (ODpig sera - ODPBS control) and compared between inoculated and control groups by ANOVA test (P = 0.05).
Detection of TNF-alpha, IL-1 beta and IL-8 in experimentally infected pigs by ELISA
The serum levels of TNF-alpha, IL-1 beta and IL-8 (ELISA kits; RayBiotech Inc., Norcross, GA, USA) were determined at days 0, 7, 15, 33, 36, 47 and 62 in infected and control pigs following manufacturer’s recommendations. Briefly, 100 μl of each standard recombinant porcine cytokine (RayBiotech) and pig serum samples were added to 96-well plates (MaxiBinding, SPL Life sciences, Korea) and incubated for 2.5 hrs. Plates were washed 4 times with 1x wash solution, inverted and cleaned with paper towels. Then, 100 μl of biotinylated anti-pig antibody (RayBiotech) were added to each well, incubated for 1 hr and washed as described before. Hundred μl of horseradish peroxidase labeled streptavidin solution (RayBiotech) were added to each well, incubated for 45 min and washed as before. Finally, 100 μl of 3,3’,5,5’-tetramethylbenzidine (TMB) One-Step substrate reagent (RayBiotech) were added to each well, incubated for 30 min in the dark and 50 μl of stop solution (0.2 M sulfuric acid) were added to each well before reading the plate at OD450nm immediately. All incubations were done at room temperature with gentle shaking. The mean OD450nm was calculated for each set of duplicate standards, controls and samples and the average zero standard OD450nm was subtracted. The standard curve was plotted and used to calculate serum cytokine concentrations in infected and control pigs. Infected to uninfected ratios were compared between infected and control pigs by Student’s t-test (P = 0.05).
Buffy coat cell composition in experimentally infected pigs
Buffy coat was obtained by centrifugation of 10 ml of heparin-treated blood at 200xg for 10 min. The Buffy coat was removed and resuspended in 5 ml PBS. Cell suspension (100 μl) was then treated with 1 ml of BD-FACS lysing solution (Becton Dickinson, Madrid, Spain), centrifuged and resuspended in 500 μl PBS. The analyses of cell subpopulations were performed using a FACScalibur (Becton Dickinson) flow cytometer. Subpopulation were gated and counted by their characteristic forward and side scatter. Results were compared between inoculated and control pigs by ANOVA test (P = 0.05).
Analysis of mRNA levels by real-time RT-PCR analysis
Real-time RT-PCR was performed on RNA samples from naturally and experimentally infected and uninfected pigs with gene specific primers (Table 1) using the iScript One-Step RT-PCR Kit with SYBR Green and the iQ5 thermal cycler (Bio-Rad, Hercules, CA, USA) following manufacturer's recommendations. A dissociation curve was run at the end of the reaction to ensure that only one amplicon was formed and that the amplicon denatured consistently in the same temperature range for every sample [36]. The mRNA levels were normalized against porcine cyclophlilyn, beta-actin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using the genNorm method (ddCT method as implemented by Bio-Rad iQ5 Standard Edition, Version 2.0) [37]. In all cases, the mean of the duplicate values was used and data from infected and uninfected animals were compared using the Student’s t-test (P = 0.05) or ANOVA test (P = 0.05) for wild and experimental pigs, respectively.
Table 1.
Primer sets and real-time PCR conditions used for analysis of differentially expressed genes
| Gene description | Genbank accession number | Upstream/downstream primer sequences (5´-3´) | PCR conditionsa |
|---|---|---|---|
| Interleukin 1 receptor accessory protein-like 1 (IL1RAPL1) |
NG_008292 CN163387 |
IL1-L: GTTGTCATTTCGCCAAACCT IL1-R: GCCTATGACCGATGGCTTTA |
58°C, 30 sec/72 °C, 30 sec |
| T-cell receptor alpha chain (TCR-alpha) |
AB087958.1 |
TcellR-L: TTCTGACCCTGGGGACTATG TcellR-R: GAGAAGCCATGCTGTTGGT |
58°C, 30 sec/72°C, 30 sec |
| Gap junction protein alpha 1 (GJA1) |
BC105464.1 CK465005 |
GAP-L: TGCAATGAAGCTGAACATGA GAP-R: TGGAATGCAAGAGAGGTTGA |
58°C, 30 sec/72°C, 30 sec |
| Thrombospondin 4 (TSP-4) |
XM_001926236 BM190304 |
TROMB4-L: GGGCAAGGTTTTTGTTCTGA TROMB4-R: TCATAGGGGTCCAGCACTTC |
60°C, 30 sec/72°C, 30 sec |
| Beta-actin |
DQ845171 |
SusBetActin-L: GACATCCGCAAGGACCTCTA SusBetActin-R: ACACGGAGTACTTGCGCTCT |
60°C, 30 sec/72°C, 30 sec |
| Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) |
AF069649 |
GADPHSus-L: CCAGAACATCATCCCTGCTT GADPHSus-R: GTCCTCAGTGTAGCCCAGGA |
60°C, 30 sec/72 °C, 30 sec |
| Cyclophilin | AY008846 | SSCYCLOPHILIN-L: AGCACTGGGGAGAAAGGATT SSCYCLOPHILIN-R: CTTGGCAGTGCAAATGAAAA | 55 °C, 30 sec/72 °C, 30 sec |
aPCR conditions are shown as annealing/extension in real-time RT-PCR analysis.
Results
Gene expression in pigs naturally infected with A. phagocytophilum
All infected wild pigs contained a single A. phagocytophilum 16S rDNA and groESL genotype. The 16S rDNA sequence was identical to the sequence of the USG3 strain [GenBank: AY055469] originally isolated from a dog infected by feeding infected I. scapularis ticks, as well as to strains obtained from patients diagnosed with human granulocytic anaplasmosis (HGA) [38]. The sequence of the groESL locus was identical to that identified previously in wild boar, human and I. ricinus samples in Slovenia [GenBank: AF033101 and EU246961] [15].
Of the 20,201 S. scrofa genes that were analyzed in the microarray, 942 showed significant (P < 0.05) differences between infected and control samples (936 upregulated and 6 down regulated) and 61 of them had >2 fold changes in expression in wild pigs (Table 2). Of these genes, 56 were upregulated and 5 were down regulated in infected animals (Table 2).
Table 2.
Gene ontology and description of significant differentially expressed genes (P < 0.05; > 2 fold change)
| Affymetrix ID1 | Genbank accession number | Fold Change2 | SD3 | P-value4 | Description5 | GO Molecular funtion6 | GO Biological process7 |
|---|---|---|---|---|---|---|---|
| Ssc.30381.1.A1_at |
CO991016 |
361.988 |
84.148 |
0.039 |
Unknown |
||
| Ssc.17891.1.A1_at |
CF175823 |
29.073 |
8.518 |
0.029 |
Unknown |
||
| Ssc.13408.1.A1_at |
BI405159 |
19.229 |
5.72 |
0.030 |
Unknown |
||
| Ssc.10537.1.A1_at |
BF711416 |
15.897 |
1.497 |
0.002 |
Unknown |
||
| Ssc.29577.1.A1_at |
CO940471 |
14.279 |
0.259 |
0.006 |
Unknown |
||
| Ssc.24631.1.S1_at |
CK461650 |
11.635 |
1.792 |
0.007 |
Formin 1 |
Protein binding |
Cell adhesion |
| Ssc.31062.1.S1_at |
AJ663560 |
8.047 |
1.593 |
0.021 |
Unknown |
||
| Ssc.28701.1.S1_at |
BG895814 |
8.008 |
0.889 |
0.015 |
Sorbin and SH3 domain isoform 2, transcript variant 14 |
Receptor activity |
Signaling pathway |
| Ssc.29538.1.A1_at |
CO941727 |
7.984 |
0.567 |
0.020 |
Unknown |
||
| Ssc.10128.1.A1_at |
BI399899 |
7.217 |
1.986 |
0.039 |
similar to H. sapiens SIX homebox 4 |
Unknown |
Unknown |
| Ssc.16289.1.A1_at |
U15437.1 |
5.927 |
1.945 |
0.047 |
Ig heavy chain variable VDJ region |
Protein binding |
Immune response |
| Ssc.16269.1.S1_at |
U15523.1 |
2.919 |
0.594 |
0.047 |
Ig heavy chain variable VDJ region |
Protein binding |
Immune response |
| Ssc.15942.5.A1_x_at |
U38202.1 |
2.571 |
0.398 |
0.044 |
|
|
|
| Ssc.17872.1.A1_at |
CF175649 |
5.731 |
1.428 |
0.047 |
COUP transcription factor 1 (COUP-TF1) |
Transcription factor |
Signaling pathway |
| Ssc.31126.1.A1_at |
CO942136 |
5.391 |
0.792 |
0.035 |
Unknown |
||
| Ssc.29622.1.A1_at |
CO942607 |
4.96 |
0.634 |
0.009 |
Unknown |
||
| Ssc.17942.1.A1_at |
CF176409 |
4.605 |
0.287 |
0.020 |
Unknown |
||
| Ssc.31069.1.A1_s_at |
BF712013 |
4.578 |
0.478 |
0.014 |
DAZ interacting protein 3, zinc finger |
Protein binding |
Ubiquitin-dependent protein catabolic process |
| Ssc.6157.1.A1_at |
BQ597772 |
3.782 |
0.555 |
0.021 |
Zinc finger protein 521 |
Unknown |
Unknown |
| Ssc.1411.1.S1_at |
BM190304 |
3.586 |
0.152 |
0.013 |
Thrombospondin 4 (TSP-4) |
Cation binding, protein binding |
Cell adhesion |
| Ssc.7524.1.A1_at |
BQ599075 |
3.397 |
0.719 |
0.033 |
Sk/Dkk-1 protein precursor |
Protein binding |
Signaling pathway |
| Ssc.8931.1.A1_at |
BI398736 |
3.336 |
0.818 |
0.037 |
Angiopoietin-like protein 2 (Angptl2) |
Unknown |
Signaling pathway |
| Ssc.13693.1.A1_at |
BQ603203 |
3.313 |
0.589 |
0.026 |
Unknown |
||
| Ssc.4707.1.A1_at |
BI118246 |
3.271 |
0.917 |
0.049 |
H. sapiens kit ligand (KITLG) |
Protein binding |
Cell adhesion |
| Ssc.13265.1.A1_at |
BQ605073 |
3.236 |
0.602 |
0.029 |
Unknown |
||
| Ssc.7967.1.A1_at |
BQ599891 |
3.153 |
0.663 |
0.033 |
Unknown |
||
| Ssc.8871.2.A1_at |
CK457442 |
2.929 |
0.711 |
0.043 |
Cyclin-dependent kinase inhibitor 1C (CDKN1C) |
Protein binding |
Signaling pathway |
| Ssc.20473.2.S1_at |
CK456061 |
2.909 |
0.139 |
0.001 |
Unknown |
||
| Ssc.20452.1.S1_at |
BX670488 |
2.890 |
0.616 |
0.035 |
Keratin associated protein 26-1 |
Protein binding |
Cell structure |
| Ssc.29030.1.S1_at |
CO988330 |
2.838 |
0.499 |
0.032 |
Unknown |
||
| Ssc.26632.1.S1_at |
CN155689 |
2.813 |
0.333 |
0.030 |
Tripartite motif protein 32 |
Protein binding |
Cell differentiation, ubiquitin-dependent protein catabolic process |
| Ssc.24221.2.A1_at |
BI181166 |
2.805 |
0.302 |
0.007 |
NADH-ubiquinone oxidoreductase |
Enzymatic activity |
Cell metabolism |
| |
|
|
|
|
18 kDa subunit |
|
|
| Ssc.428.10.S1_at |
AB087975.1 |
2.767 |
0.181 |
0.027 |
T cell receptor alpha chain (TCR-alpha) |
Receptor activity |
Immune response |
| Ssc.17790.1.S1_at |
AB087958.1 |
2.334 |
0.369 |
0.031 |
T cell receptor alpha chain (TCR-alpha) |
Receptor activity |
Immune response |
| Ssc.18884.1.A1_at |
CF365209 |
2.67 |
0.083 |
0.008 |
Unknown |
||
| Ssc.25538.1.S1_at |
BX918287 |
2.597 |
0.377 |
0.033 |
Zinc finger protein 502 |
Transcription factor |
Transcription |
| Ssc.26587.1.A1_at |
CN154795 |
2.592 |
0.334 |
0.030 |
Unknown |
||
| Ssc.20172.1.A1_at |
BX676733 |
2.547 |
0.347 |
0.027 |
Tumor endothelial marker 8 isoform 3 |
Protein binding, receptor activity |
Cell adhesion |
| Ssc.7090.1.A1_at |
NM_214233.1 |
2.465 |
0.435 |
0.026 |
Thioltransferase (GLRX1) |
Enzymatic activity |
Stress |
| Ssc.13474.1.A1_at |
BQ602423 |
2.454 |
0.405 |
0.022 |
Unknown |
||
| Ssc.8511.1.A1_at |
BF703957 |
2.449 |
0.275 |
0.009 |
Sus scrofa mRNA,clone: OVRM10011A06, expressed in ovary |
Unknown |
Unknown |
| Ssc.30148.1.A1_at |
CO987207 |
2.341 |
0.208 |
0.048 |
Rho-related BTB domain containing 3 (RHOBTB3) |
Protein binding, receptor activity |
Signaling pathway, ubiquitin-dependent protein catabolic process |
| Ssc.22336.1.S1_at |
CF793417 |
2.302 |
0.366 |
0.023 |
Homeobox protein Hox-B7 (Hox-2C) |
Transcription factor, protein binding |
Transcription |
| Ssc.1377.2.S1_at |
BI343023 |
2.264 |
0.19 |
0.016 |
Integrin alpha-8 (ITGA8) |
Cation binding, protein binding, receptor activity |
Cell differentiation, cell adhesion, signalling pathway |
| Ssc.29167.1.A1_at |
CO950916 |
2.198 |
0.272 |
0.030 |
Rho GTPase activating protein 5 |
Protein binding |
Cell adhesion |
| Ssc.13363.1.A1_at |
BI404946 |
2.188 |
0.31 |
0.038 |
Ubiquitin carboxyl-terminal hydrolase 24 |
Protein binding |
Ubiquitin-dependent protein catabolic process |
| Ssc.17370.1.A1_at |
BX665583 |
2.186 |
0.266 |
0.014 |
Adrenergic, alpha-1B-, receptor (ADRA1B) |
Protein binding, receptor activity |
Signaling pathway |
| Ssc.22210.2.S1_at |
CF788693 |
2.176 |
0.298 |
0.040 |
Unknown |
||
| Ssc.29565.1.A1_at |
CO942018 |
2.168 |
0.308 |
0.025 |
Unknown |
||
| Ssc.19407.1.A1_at |
CF359796 |
2.157 |
0.016 |
0.015 |
Unknown |
||
| Ssc.28265.1.A1_at |
CN025977 |
2.143 |
0.376 |
0.035 |
Unknown |
||
| Ssc.26179.1.S1_at |
BX922022 |
2.123 |
0.101 |
0.005 |
Midnolin (MIDN) |
Protein binding |
Transcription |
| Ssc.4848.1.S1_at |
CF789770 |
2.123 |
0.182 |
0.031 |
Calponin 3, acidic, transcript variant 1 |
Cation binding, protein binding |
Unknown |
| Ssc.20453.1.S1_at |
BX675824 |
2.092 |
0.228 |
0.012 |
Laminin receptor 1 |
Receptor activity |
Unknown |
| Ssc.14354.1.A1_at |
BQ601965 |
2.079 |
0.093 |
0.047 |
HHEX gene for hematopoietically expressed homeobox |
Transcription factor |
Cell differentiation, signaling pathway |
| Ssc.942.1.S1_at |
CK465005 |
2.061 |
0.165 |
0.012 |
Gap junction protein, alpha 1 (GJA1) |
Protein binding |
Cell adhesion, signaling pathway, immune response |
| Ssc.26933.1.S1_at |
CN163387 |
2.036 |
0.000 |
0.007 |
Interleukin 1 receptor accessory protein-like 1 (IL1RAPL1) |
Receptor activity |
Immune response |
| Ssc.30263.1.A1_at |
CO989398 |
2.033 |
0.300 |
0.039 |
Unknown |
||
| Ssc.16566.1.S1_at |
BF078197 |
2.025 |
0.271 |
0.025 |
Lactase phlorizinhydrolase |
Cation binding |
Cell metabolism |
| Ssc.9748.1.A1_at |
BI398784 |
−3.914 |
1.462 |
0.033 |
Unknown |
|
|
| Ssc.30189.1.A1_at |
CO987781 |
−4.078 |
1.800 |
0.038 |
Pig DNA sequence from clone CH242-94D11 on chromosome 7 |
Unknown |
Unknown |
| Ssc.8698.1.S1_at |
CN163671 |
−10.246 |
0.151 |
0.010 |
Cadherin 11, type 2, OB-cadherin (osteoblast) |
Cation binding, protein binding |
Cell adhesion |
| Ssc.18866.1.A1_at |
CF365015 |
−10.551 |
1.242 |
0.003 |
Unknown |
||
| Ssc.29259.1.A1_at | CO953119 | −14.935 | 2.417 | 0.048 | Zinc finger protein 567 | Transcription factor | Transcription |
1 Affymetrix ID identifies the probe based on Affymetrix accession number.
2 Fold change is the normalized average log2 ratio of infected/uninfected wild pigs (negative values denote dowregulation in infected animals).
3 SD is the standard deviation of the normalized average log2 ratio.
4 P-value determined from the average log2 ratio.
5 Description indicates a short description of the gene.
6 GO molecular function is the function of the gene according to GO entries.
7 GO biological process indicates the gene process according to GO entries.
Gene ontology (GO) could be assigned to 32 of the differentially expressed genes (Table 2). The differentially expressed genes in wild pigs infected with A. phagocytophilum included those with cation binding, protein binding, transcription factor, enzymatic activity and receptor activity protein function involved in cell differentiation, adhesion, metabolism and structure, signaling pathway, transcription, stress, immune response and catabolic processes (Table 2). The most frequently represented protein function and biological process GO assignments were significantly overrepresented among genes differentially expressed in wild pigs and contained genes upregulated in response to A. phagocytophilum infection (Table 3). Among them, the highest GO enrichment for molecular function and biological process occurred for protein binding and signaling pathway genes, respectively (Table 3).
Table 3.
Gene ontology enrichment analysis of genes differentially expressed in wild pigs naturally infected with A. phagocytophilum
| GO category | Represented on the microarray (%)a | Represented among differentially expressed genes (%)b |
|---|---|---|
|
Molecular function | ||
| Cation binding |
67 (2.4) |
5 (15.6)* |
| Protein binding |
14 (0.5) |
20 (62.5)* |
| Transcription factor |
10 (0.4) |
5 (15.6)* |
| Receptor activity |
10 (0.4) |
8 (25.0)* |
|
Biological process | ||
| Catabolic process |
108 (3.8) |
3 (9.4)* |
| Immune response |
23 (0.8) |
4 (12.5)* |
| Cell adhesion |
20 (0.7) |
8 (25.0)* |
| Signaling pathway |
13 (0.5) |
10 (31.2)* |
| Cell differentiation | 7 (0.2) | 3 (9.4)* |
aOf the 20,201 S. scrofa genes analyzed in the microarray, 2,840 had GO assignments and were used for GO enrichment analysis.
bOf the 61 genes that showed significant (P ≤ 0.05) ≥ 2 fold changes in expression in infected wild pigs, 32 had GO assignments and were used for GO enrichment analysis. For genes with multiple GO assignments, each category was included in the analysis. For each GO category of interest, entries in the array were compared with results of differentially expressed genes by χ2–test (*α < 0.01).
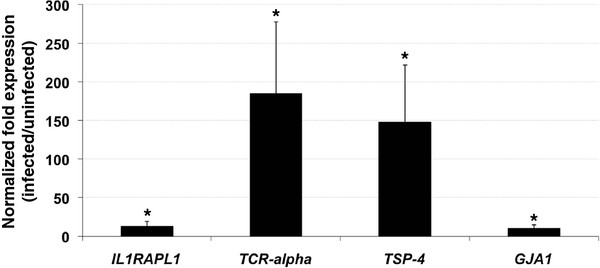
The immune response was among the biological processes significantly overrepresented in genes upregulated in response to A. phagocytophilum infection (Table 3). Thus, the immune response genes upregulated in response to A. phagocytophilum infection, interleukin 1 receptor accessory protein-like 1 (IL1RAPL1), T-cell receptor alpha chain (TCR-alpha), thrombospondin 4 (TSP-4) and Gap junction protein alpha 1 (GJA1), were selected for confirmation of microarray hybridization results by real-time RT-PCR. The real-time RT-PCR analysis confirmed the results of the microarray hybridization and demonstrated that the immune response genes IL1RAPL1, TCR-alpha, TSP-4 and GJA1 were upregulated in infected animals (Figure 1).
Figure 1.
Relative expression of immune response genes in naturally A. phagocytophilum -infected and uninfected wild pigs. The expression of selected genes was quantified by real-time RT-PCR in samples of infected (N = 3) and uninfected control pigs (N = 3). Amplification efficiencies were normalized against porcine cyclophlilyn, beta-actin and GAPDH and infected to uninfected average ± S.D. mRNA ratios determined. In all cases, the mean of duplicate values was used and data from infected and uninfected animals were compared using the Student`s t-test (*P < 0.05).
Gene expression in pigs experimentally infected with A. phagocytophilum
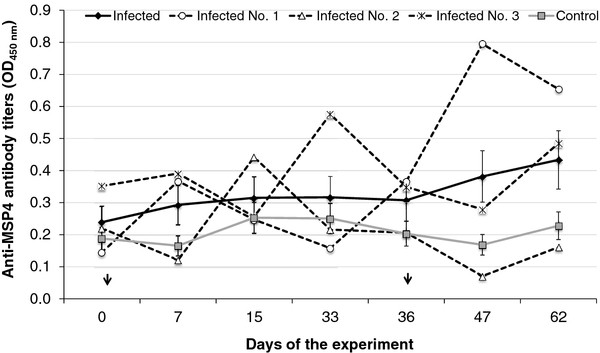
In experimentally infected pigs, A. phagocytophilum DNA was detected by msp4 PCR in blood samples collected at 15 (in all 3 pigs), 36 (before second inoculation in pigs No. 1 and No. 2), and 62 (in pig No. 1 only) days post-infection (dpi) in pigs inoculated with infected cells but not in control pigs. The A. phagocytophilum msp4 amplicons from pig blood were sequenced and corresponded to the NY-18 isolate sequence (Genbank accession number JQ522935). Infected and uninfected pigs did not show clinical signs or A. phagocytophilum morulae in stained blood films. Significant differences were not observed in anti-A. phagocytophilum MSP4 antibodies between pigs inoculated with infected cells and controls (P > 0.05; Figure 2). However, peaks in anti-MSP4 antibody titers were detected at 33 and 47 dpi in pigs No. 3 and No. 1, respectively (Figure 2).
Figure 2.
Detection of anti- A. phagocytophilum MSP4 antibodies in experimentally infected and control pigs. Antibody titers were determined by ELISA, expressed as the average ± S.E. OD450nm (ODpig sera - ODPBS control) and compared between infected and control pigs by ANOVA test (P > 0.05). OD450nm values for each infected pig are also shown. Arrows show time of pig inoculation with infected and uninfected tick cells.
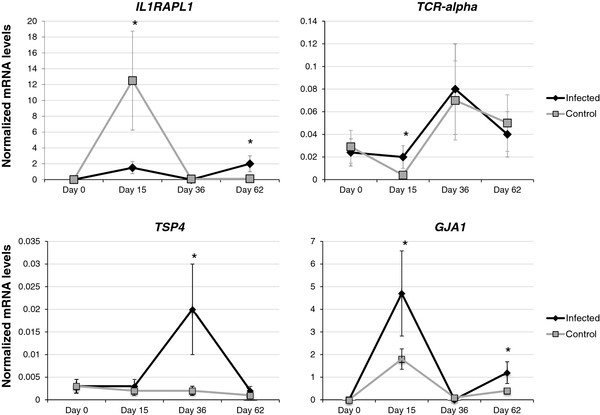
Buffy coat cell composition did not change during the experiment and was similar between infected (lymphocytes, 37.80 ± 0.13%; monocytes, 10.51 ± 0.02%; granulocytes, 49.56 ± 0.16%) and uninfected (lymphocytes, 36.16 ± 0.11; monocytes, 10.21 ± 0.01; granulocytes, 49.43 ± 0.13%) pigs (P > 0.4). The immune response genes upregulated in response to A. phagocytophilum infection in naturally infected pigs (IL1RAPL1, TCR-alpha, TSP-4 and GJA1) were selected to characterize the mRNA levels at different dpi by real-time RT-PCR in experimentally infected pigs (Figure 3). The results showed that TCR-alpha and GJA1 were upregulated in infected pigs at 15 dpi when compared to control animals. TSP-4 was upregulated at 36 dpi only while IL1RAPL1 and GJA1 were upregulated at 62 dpi (Figure 3). The highest mRNA levels for immune response genes at the end of the experimental infection (62 dpi) were found in the only infected pig in which pathogen DNA was detected by PCR (pig No. 1).
Figure 3.
Expression of immune response genes in experimentally A. phagocytophilum -infected and uninfected domestic pigs. The expression of selected genes was quantified by real-time RT-PCR in samples of infected (N = 3) and uninfected control pigs (N = 3). Amplification efficiencies were normalized against porcine cyclophlilyn, beta-actin and GAPDH and shown in arbitrary units as average ± S.D. mRNA levels. In all cases, the mean of duplicate values was used and data from infected and uninfected animals were compared using the ANOVA t-test (*P < 0.05).
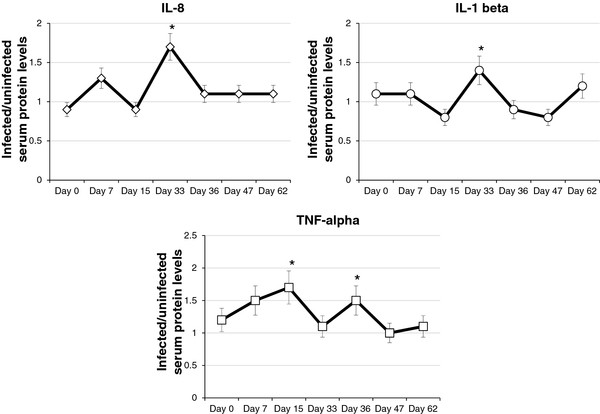
Serum IL-1 beta, IL-8 and TNF-alpha levels in pigs experimentally infected with A. phagocytophilum
Serum IL-1 beta, IL-8 and TNF-alpha levels were transiently higher in infected pigs when compared to uninfected controls (Figure 4). Significant (P < 0.05) infected to uninfected ratio for serum protein levels were obtained for IL-1 beta and IL-8 at 33 dpi and for TNF-alpha at 15 and 36 dpi (Figure 4). These protein levels were equivalent in infected animals to 3.73 ± 0.00 pg/ml (IL-1 beta), 2.18 ± 0.00 pg/ml (IL-8), 370.13 ± 0.00 pg/ml (TNF-alpha at 15 dpi) and 2.01 ± 0.00 pg/ml (TNF-alpha at 36 dpi). In uninfected control animals, protein levels at the same time points were bellow ELISA detection limits.
Figure 4.
Serum IL-8, IL-1 beta and TNF-alpha levels in experimentally infected pigs. Cytokine levels were determined by ELISA in the sera from infected and uninfected control pigs and infected to uninfected average ± S.D. ratios determined. Results were compared between infected and control pigs by Student’s t-test (*P ≤ 0.05).
Discussion
Molecular evidence suggested that wild pigs could be involved in the natural cycle of A. phagocytophilum in some regions [14-16,21]. The results of sequence analyses suggested that the A. phagocytophilum strain identified in wild pigs might be similar to those causing disease in dogs and humans, thus reinforcing the possible role of pigs in the epidemiology of HGA in these regions [15,38,39].
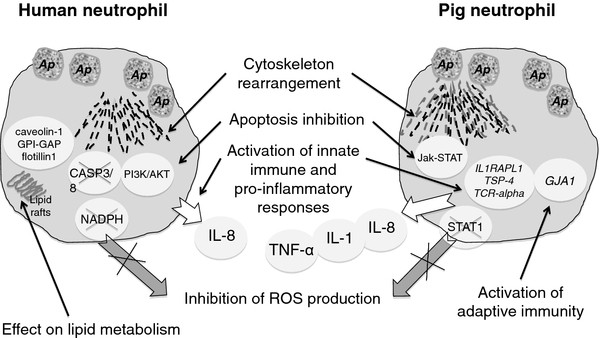
The overall effect of A. phagocytophilum on pig gene expression was low as only 4.7% (942/20,201) of the genes analyzed in the microarray were differentially expressed in pathogen-infected animals (P < 0.05) and only 61 genes (0.3%; 61/20,201) showed >2 fold difference between infected and control animals. Interestingly, 9 of the 61 (15%) differentially expressed genes in naturally infected pigs were related to cytoskeleton structure and function. Phagocytosis and autophagy are among the first lines of defense against bacterial infections and require a dramatic rearrangement of the cytoskeleton for internalization of invading microbes [40]. The expression of genes such as GJA1, integrin alpha-8, TSP-4, formin 1, Rho GTPase activating protein 5, keratin associated protein 26–1, calponin 3 and laminin receptor 1 was upregulated, while the expression of cadherin 11 was down regulated in A. phagocytophilum-infected wild pigs, thus suggesting an effect of pathogen infection on cytoskeleton rearrangement. It has been suggested that A. phagocytophilum affects actin reorganization to facilitate cell invasion but reduces neutrophil phagocytosis and subverts autophagy to establish intracellular infection and proliferation [41-43]. Furthermore, a recent study showed that Toll-like receptor signaling usurps components that are traditionally associated with autophagy to increase the efficiency of phagocytosis, thereby providing a link between these two microbial defense mechanisms [44]. Taken together, these results suggested that A. phagocytophilum infection of pigs impacted cytoskeleton rearrangement to promote phagocytosis and autophagy, thus resulting in effective pathogen clearance (Figure 5).
Figure 5.
Effect of A. phagocytophilum infection on host cells. A. phagocytophilum (Ap) infection causes cytoskeleton rearrangement required for infection, but in pigs it may also promote phagocytosis and autophagy for effective pathogen clearance. Ap delays the apoptotic death of neutrophils to increase infection, but different and complementary mechanisms may operate in human and pig cells. Pathogen infection stimulates innate immune and pro-inflammatory responses in both humans and pigs. IL-8 is likely secreted by infected neutrophils but monocytes, rather than neutrophils, are probably responsible for proinflammatory IL-1 beta and TNF-alpha cytokine production. The expression of genes involved in adaptive immunity was not impaired in pigs. ROS production is inhibited by pathogen infection of human neutrophils but although this mechanism was not found in pigs, upregulation of TGF-beta in infected pigs may inhibits NO production by suppressing STAT1 activation and accelerating iNOS protein degradation. The effect on lipid metabolism required for pathogen infection of human neutrophils was not found in pigs. Data for human neutrophils was obtained from the recent review by Severo et al. [43]
A. phagocytophilum infection has been shown to delay the apoptotic death of neutrophils [24,43,45,46]. The analysis of gene expression profile in naturally infected pigs did not show an effect on caspases 3 and 8 (CASP3/8) and the PI3K/AKT pathway, which have been linked to A. phagocytophilum-induced apoptosis inhibition in human neutrophils [43]. However, the activation of the Jak-STAT pathway that has been shown to occur in A. phagocytophilum-infected sheep and pigs may constitutes a new mechanism leading to delay in the apoptotic death of neutrophils in these species [47] (Figure 5). Reactive oxygen species (ROS) production is inhibited by A. phagocytophilum through modulation of NADPH oxidase assembly and/or regulation of gene expression in human cells [43], a mechanism that was not found in pigs. However, upregulation of TGF-beta in infected pigs [47] may inhibits NO production in neutrophils by suppressing STAT1 activation and accelerating iNOS protein degradation [47,48]. The effect of A. phagocytophilum on lipid metabolism required for pathogen infection of human neutrophils [25,43] was also not found in pigs. However, some of these discrepancies may be explained by the fact that results in pigs were obtained using RNA from Buffy coats and not purified neutrophils or cell cultures, which may produce a masking effect of other leukocyte mRNAs.
Our group is interested in the characterization of the host immune response to intracellular bacteria [29,49-52]. The infection with A. phagocytophilum has been shown to stimulate innate immune and pro-inflammatory responses [43,45,46,53]. However, experiments in mice have shown that A. phagocytophilum infection may be controlled, even in the absence of innate immune effectors [54,55]. In sheep and horses, evidence suggests that A. phagocytophilum infection triggers innate immune responses while impairing adaptive immunity [29,56], a factor that could contribute to pathogenicity in these species.
Analysis of gene expression in naturally and experimentally infected pigs suggested that A. phagocytophilum infection increased innate immunity by up regulation of IL1RAPL1TSP-4 and TCR-alpha (Figure 5). Furthermore, kinetics of mRNA levels in experimentally infected pigs showed an early, transient up regulation of immune response genes, probably coinciding with the first bacteremia of the acute infection phase [57]. Up regulation of IL1RAPL1 and TSP-4 may increase the innate immune proinflammatory response through improved signal transduction and secretion of IL-1 and IL-8, respectively [58,59]. T lymphocytes use their TCR as a pattern recognition receptor to sense the presence of infection and produce after activation proinflammatory cytokines such as TNF-alpha [60]. In experimentally inoculated pigs, IL-1 beta, IL-8 and TNF-alpha serum levels were transiently higher in infected animals when compared to controls, thus corroborating the stimulation of proinflammatory responses suggested by gene expression studies in A. phagocytophilum-infected pigs (Figure 5). IL-8 secretion in response to A. phagocytophilum infection in human cells leads to neutrophils recruitment [43]. Although IL-1 and TNF-alpha levels have not been found to be elevated in HGA patients, higher mRNA or serum levels have been observed in horses and sheep, for which A. phagocytophilum is also pathogenic [61]. In vitroA. phagocytophilum infection of human peripheral blood lymphocytes and monocytes induce transient mRNA expressions and protein secretion of IL-1 beta and TNF-alpha [61]. These studies suggested that although IL-8 is likely secreted by neutrophils, monocytes, rather than neutrophils, are responsible for proinflammatory IL-1 beta and TNF-alpha cytokine production [61,62]. The expression of genes involved in adaptive immunity was not impaired. In fact, the expression of GJA1, a member of the connexin gene family with a role in innate and adaptive immunity through the regulation of phagocytosis by macrophages and the host response to bacterial infection [63], was upregulated in infected pigs. The activation of the Jak-STAT pathway associated with A. phagocytophilum infection in sheep and pigs may results in immune development to aid in pathogen control [47].
The experimental infection with A. phagocytophilum demonstrated that pigs are susceptible to pathogen infection. The detection of bacterial DNA by PCR showed a prepatent period (calculated as the number of days from the time of pig inoculation with infected tick cells to the first day that blood samples were found to be A. phagocytophilum positive by PCR) of 15 days, similar to that found in sheep [64] and white-tailed deer [65] but lower than in mice [66] inoculated with A. phagocytophilum (NY-18) infected cells. At 36 dpi only two animals were PCR positive and by 47 dpi all animals were negative, suggesting duration of approximately 30 days for the primary bacteremia. However, although only one pig (No. 1) was PCR positive at 62 dpi after the second inoculation, recurrent bacteremias are possible [57]. The weak antibody response detected in infected animals supports a rapid control of pathogen infection. However, similar results were obtained in sheep experimentally inoculated with A. phagocytophilum infected cells [64]. The pigs used in this study for microarray analysis were naturally infected with A. phagocytophilum. Therefore, it was not possible to establish when animals were infected. Transient up regulation of immune response genes in experimentally infected pigs suggested that naturally infected pigs were also at early infection stages. However, we cannot exclude the possibility that, if pigs become persistently infected even at low infection levels, some of the gene expression profiles described in this study in naturally infected pigs may represent the response of persistently infected animals and may differ from the response during early infection. Persistent A. phagocytophilum infection has been documented in sheep [57] and horses [67] and previous studies have shown differences in gene expression profiles between acutely and chronically A. phagocytophilum-infected sheep [29].
Conclusions
These results suggested that pigs are susceptible to A. phagocytophilum but control infection, particularly through activation of innate immune responses and cytoskeleton rearrangement to promote phagocytosis and autophagy (Figure 5). Control of A. phagocytophilum infection in pigs may results in infection below PCR detection levels or infection clearance, thus contributing to the low percentage of infection prevalence detected for this species in most regions, with a low or no impact as a reservoir host for this pathogen [14,15,20]. The results reported here confirmed in pigs the activation of innate and adaptive immune pathways during A. phagocytophilum infection reported in humans and other species (Figure 5). However, this pathogen may uses other mechanisms to circumvent host-cell defenses and establish infection by dowregulating other adaptive immune response genes such as IL-2 and IL-4 and delaying the apoptotic death of neutrophils through activation of the Jak-STAT pathway [47]. These results further expand the existing information on the response of mammalian hosts to A. phagocytophilum infection and suggested a role for newly reported genes in the protection of pigs against A. phagocytophilum.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RCG performed microarray analysis and lab tests, NA, KSS, BB-B, MM and NG collected data and samples, NA, MB and JMP performed lab tests. RCG, NA, MB, JMP, CG and JF analyzed data and performed statistical analysis. JF, KMK and TA-Z conceived the study, JF designed the study, CG supervised part of study, RCG, KMK, TA-Z, CG and JF wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Ruth C Galindo, Email: ruthcecilia.galindo@uclm.es.
Nieves Ayllón, Email: Nieves.Ayllon@uclm.es.
Katja Strašek Smrdel, Email: katja.strasek@mf.uni-lj.si.
Mariana Boadella, Email: Mariana.Boadella@uclm.es.
Beatriz Beltrán-Beck, Email: Beatriz.Beltran@uclm.es.
María Mazariegos, Email: mazariegosm@vet.ucm.es.
Nerea García, Email: garcian@vet.ucm.es.
José M Pérez de la Lastra, Email: josemanuel.plastra@uclm.es.
Tatjana Avsic-Zupanc, Email: tatjana.avsic@mf.uni-lj.si.
Katherine M Kocan, Email: katherine.kocan@okstate.edu.
Christian Gortazar, Email: Christian.Gortazar@uclm.es.
José de la Fuente, Email: jose_delafuente@yahoo.com.
Acknowledgements
José A. Barasona (IREC), Laura Cuesta and Alejandro Navarro (VISAVET) are acknowledged for technical assistance. This research was supported by EU FP7, ANTIGONE project number 278976 and the CSIC intramural project 200830I249 to JF. N. Ayllón was funded by MINECO, Spain. Personnel at the Cap de la Unitat Cientificotécnica de Suport, Institut de Recerca Hospital Universitari Vall d´Hebron, Barcelona, Spain are acknowledged for technical assistance with microarray hybridization and analysis.
References
- Dumler JS, Barbet AC, Bekker CPJ, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of the genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions subjective synonyms of Ehrlichia phagocytophila. Int J Sys Evol Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Massung RF, Wong SJ, Chu FK, Lutz H, Meli M, von Loewenich FD, Grzeszczuk A, Torina A, Caracappa S, Mangold AJ, Naranjo V, Stuen S, Kocan KM. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J Clin Microbiol. 2005;43:1309–1317. doi: 10.1128/JCM.43.3.1309-1317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusterla N, Berger Pusterla J, Braun U, Lutz H. Serological, hematologic, and PCR studies of cattle in an area of Switzerland in which tick-borne fever (caused by Ehrlichia phagocytophila) is endemic. Clin Diagn Lab Immunol. 1998;5:325–327. doi: 10.1128/cdli.5.3.325-327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumler JS, Choi KS, Garcia-Garcia JC, Barat NS, Scorpio DG, Garyu JW, Grab DJ, Bakken JS. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg Infect Dis. 2005;11:1828–1834. doi: 10.3201/eid1112.050898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola P, Raoult D. Ticks and tickborne bacterial disease in humans: an emerging infection threat. Clin Infect Dis. 2001;32:897–928. doi: 10.1086/319347. [DOI] [PubMed] [Google Scholar]
- Petrovec M, Bidovec A, Sumner JW, Nicholson WL, Childs JE, Avsic-Zupanc T. Infection with Anaplasma phagocytophila in cervids from Slovenia: evidence of two genotypic lineages. Wien Klin Wochenschr. 2002;114:641–647. [PubMed] [Google Scholar]
- Lotric-Furlan S, Rojko T, Petrovec M, Avsic-Zupanc T, Strle F. Epidemiological, clinical and laboratory characteristics of patients with human granulocytic anaplasmosis in Slovenia. Wien Klin Wochenschr. 2006;118:708–713. doi: 10.1007/s00508-006-0700-4. [DOI] [PubMed] [Google Scholar]
- Barandika JF, Hurtado A, García-Esteban C, Gil H, Escudero R, Barral M, Jado I, Juste RA, Anda P, García-Pérez AL. Tick-borne zoonotic bacteria in wild and domestic small mammals in northern Spain. Appl Environ Microbiol. 2007;73:6166–6171. doi: 10.1128/AEM.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuen S. Anaplasma phagocytophilum - the most widespread tick-borne infection in animals in Europe. Vet Res Commun. 2007;1:79–84. doi: 10.1007/s11259-007-0071-y. [DOI] [PubMed] [Google Scholar]
- Torina A, Vicente J, Alongi A, Scimeca S, Turlá R, Nicosia S, Di Marco V, Caracappa S, de la Fuente J. Observed prevalence of tick-borne pathogens in domestic animals in Sicily, Italy during 2003–2005. Zoonoses Public Health. 2007;54:8–15. doi: 10.1111/j.1863-2378.2007.00989.x. [DOI] [PubMed] [Google Scholar]
- Sixl W, Petrovec M, Marth E, Wüst G, Stünzner D, Schweiger R, Avsic-Zupanc T. Investigation of Anaplasma phagocytophila infections in Ixodes ricinus and Dermacentor reticulatus ticks in Austria. Ann NY Acad Sci. 2003;990:94–97. doi: 10.1111/j.1749-6632.2003.tb07343.x. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Naranjo V, Ruiz-Fons F, Vicente J, Estrada-Peña A, Almazán C, Kocan KM, Martín MP, Gortázar C. Prevalence of tick-borne pathogens in ixodid ticks (Acari: Ixodidae) collected from European wild boar (Sus scrofa) and Iberian red deer (Cervus elaphus hispanicus) in central Spain. Eur J Wild Res. 2004;50:187–196. doi: 10.1007/s10344-004-0060-1. [DOI] [Google Scholar]
- de la Fuente J, Naranjo V, Ruiz-Fons F, Höfle U, de Mera IG F, Villanúa D, Almazán C, Torina A, Caracappa S, Kocan KM, Gortázar C. Potential vertebrate reservoir hosts and invertebrate vectors of Anaplasma marginale and A phagocytophilum in central Spain. Vector Borne Dis. 2005;5:390–401. doi: 10.1089/vbz.2005.5.390. [DOI] [PubMed] [Google Scholar]
- Hulinska D, Langrova K, Pejcoch M, Pavlasek I. Detection of Anaplasma phagocytophilum in animals by real-time polymerase chain reaction. APMIS. 2004;112:239–247. doi: 10.1111/j.1600-0463.2004.apm11204-0503.x. [DOI] [PubMed] [Google Scholar]
- Strasek Smrdel K, Tozon N, Duh D, Petrovec M, Avsic Zupanc T. Diversity of groESL sequences of Anaplasma phagocytophilum among dogs in Slovenia. Clin Microbiol Infec. 2009;15(Suppl 2):79–80. doi: 10.1111/j.1469-0691.2008.02201.x. [DOI] [PubMed] [Google Scholar]
- Michalik J, Stańczak J, Cieniuch S, Racewicz M, Sikora B, Dabert M. Wild boars as hosts of human-pathogenic Anaplasma phagocytophilum variants. Emerg Infect Dis. 2012;18:998–1001. doi: 10.3201/eid1806.110997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torina A, Alongi A, Naranjo V, Scimeca S, Nicosia S, Di Marco V, Caracappa S, Kocan KM, de la Fuente J. Characterization of Anaplasma infections in Sicily, Italy. Ann NY Acad Sci. 2008;1149:90–93. doi: 10.1196/annals.1428.065. [DOI] [PubMed] [Google Scholar]
- Ruiz-Fons F, de Mera IG F, Acevedo P, Höfle U, Vicente J, de la Fuente J, Gortazar C. Ticks (Acari: Ixodidae) parasitizing Iberian red deer (Cervus elaphus hispanicus) and European wild boar (Sus scrofa) from Spain: geographical and temporal distribution. Vet Parasitol. 2006;140:133–142. doi: 10.1016/j.vetpar.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Estrada-Peña A, Acevedo P, Ruiz-Fons F, Gortázar C, de la Fuente J. Evidence of the Importance of Host Habitat Use in Predicting the Dilution Effect of Wild Boar for Deer Exposure to Anaplasma spp. PLoS One. 2008;3:e2999. doi: 10.1371/journal.pone.0002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo A, Pérez-Martínez L, Santibáñez S, Santibáñez P, Palomar AM, Oteo JA. Anaplasma spp. in wild mammals and Ixodes ricinus from the North of Spain. Vector Borne Zoonotic Dis. 2011;11:3–8. doi: 10.1089/vbz.2009.0214. [DOI] [PubMed] [Google Scholar]
- Masuzawa T, Uchishima Y, Fukui T, Okamoto Y, Muto M, Koizumi N, Yamada A. Detection of Anaplasma phagocytophilum from wild boars and deer in Japan. Jpn J Infect Dis. 2011;64:333–336. [PubMed] [Google Scholar]
- Castellaw AH, Chenney EF, Varela-Stokes AS. Tick-borne disease agents in various wildlife from Mississippi. Vector Borne Zoonotic Dis. 2011;11:439–442. doi: 10.1089/vbz.2009.0221. [DOI] [PubMed] [Google Scholar]
- Carlyon JA, Chan WT, Galán J, Roos D, Fikrig E. Repression of rac2 mRNA expression by Anaplasma phagocytophila is essential to the inhibition of superoxide production and bacterial proliferation. J Immunol. 2002;169:7009–7018. doi: 10.4049/jimmunol.169.12.7009. [DOI] [PubMed] [Google Scholar]
- Borjesson DL, Kobayashi SD, Whitney AR, Voyich JM, Argue CM, Deleo FR. Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J Immunol. 2005;174:6364–6372. doi: 10.4049/jimmunol.174.10.6364. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Ayoubi P, Blouin EF, Almazán C, Naranjo V, Kocan KM. Gene expression profiling of human promyelocytic cells in response to infection with Anaplasma phagocytophilum. Cell Microbiol. 2005;7:549–559. doi: 10.1111/j.1462-5822.2004.00485.x. [DOI] [PubMed] [Google Scholar]
- Pedra JH, Sukumaran B, Carlyon JA, Berliner N, Fikrig E. Modulation of NB4 promyelocytic leukemic cell machinery by Anaplasma phagocytophilum. Genomics. 2005;86:365–377. doi: 10.1016/j.ygeno.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Sukumaran B, Carlyon JA, Cai JL, Berliner N, Fikrig E. Early transcriptional response of human neutrophils to Anaplasma phagocytophilum infection. Infect Immun. 2005;73:8089–8099. doi: 10.1128/IAI.73.12.8089-8099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Kioi M, Han J, Puri RK, Goodman JL. Anaplasma phagocytophilum-induced gene expression in both human neutrophils and HL-60 cells. Genomics. 2008;92:144–151. doi: 10.1016/j.ygeno.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Galindo RC, Ayoubi P, García-Pérez AL, Naranjo V, Kocan KM, Gortazar C, de la Fuente J. Differential expression of inflammatory and immune response genes in sheep infected with Anaplasma phagocytophilum. Vet Immunol Immunopath. 2008;126:27–34. doi: 10.1016/j.vetimm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Huber W, Irizarry R. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer New York, Dudoit S; 2005. [Google Scholar]
- Benjamini Y, Hockberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Asanovich KM, Bakken JS, Madigan JE, Aguero-Rosenfeld M, Wormser GP, Dumler JS. Antigenic diversity of granulocytic Ehrlichia isolates from humans in Wisconsin and New York and a horse in California. J Infect Dis. 1997;176:1029–1034. doi: 10.1086/516529. [DOI] [PubMed] [Google Scholar]
- Munderloh UG, Tate CM, Lynch MJ, Howerth EW, Kurtti TJ, Davidson WR. Isolation of an Anaplasma sp. organism from white-tailed deer by tick cell culture. J Clin Microbiol. 2003;41:4328–4335. doi: 10.1128/JCM.41.9.4328-4335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo FR, Melo VS, Ramos CA, Madruga CR, Soares CO, Kessler RH, Almeida NF, Araújo GS, Alves LC, Torres Júnior RA, Fragoso SP, Arauco PR, Bacanelli G, Oliveira MB, Santos LR. Development of enzyme-linked immunosorbent assays based on recombinant MSP1a and MSP2 of Anaplasma marginale. Mem Inst Oswaldo Cruz. 2005;100:765–769. doi: 10.1590/S0074-02762005000700016. [DOI] [PubMed] [Google Scholar]
- Ririe KM, Rasmussen RP, Wittwer C. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta CT) Method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Massung RF, Owens JH, Ross D, Reed KD, Petrovec M, Bjoersdorff A, Coughlin RT, Beltz GA, Murphy CI. Sequence analysis of the ank gene of granulocytic Ehrlichiae. J Clin Microbiol. 2000;38:2917–2922. doi: 10.1128/jcm.38.8.2917-2922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrdel K, Serdt M, Duh D, Knap N, Županc T. Anaplasma phagocytophilum in ticks in Slovenia. Parasites & Vectors. 2010;3:102. doi: 10.1186/1756-3305-3-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Ezekowitz RA. Phagocytosis and comparative innate immunity: learning on the fly. Nat Rev Immunol. 2008;8:131–141. doi: 10.1038/nri2240. [DOI] [PubMed] [Google Scholar]
- Garyu JW, Dumler JS. Anaplasma phagocytophilum infection reduces expression of phagocytosis-related receptors on neutrophils. Ann NY Acad Sci. 2005;1063:416–419. doi: 10.1196/annals.1355.075. [DOI] [PubMed] [Google Scholar]
- Niu H, Yamaguchi M, Rikihisa Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell Microbiol. 2008;10:593–605. doi: 10.1111/j.1462-5822.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- Severo MS, Stephens KD, Kotsyfakis M, Pedra JH. Anaplasma phagocytophilum: deceptively simple or simply deceptive? Future Microbiol. 2012;7:719–731. doi: 10.2217/fmb.12.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Naranjo V, Höfle U, Vicente J, Martín MP, Ruiz-Fons F, Gortazar C, Kocan KM, de la Fuente J. Genes differentially expressed in oropharyngeal tonsils and mandibular lymph nodes of tuberculous and non-tuberculous European wild boars naturally exposed to Mycobacterium bovis. FEMS Immunol Med Microbiol. 2006;46:298–312. doi: 10.1111/j.1574-695X.2005.00035.x. [DOI] [PubMed] [Google Scholar]
- de Mera IG F, de la Lastra JM P, Ayoubi P, Naranjo V, Kocan KM, Gortazar C, de la Fuente J. Differential expression of inflammatory and immune response genes in mesenteric lymph nodes of Iberian red deer (Cervus elaphus hispanicus) naturally infected with Mycobacterium bovis. Dev Com Immunol. 2008;32:85–91. doi: 10.1016/j.dci.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Galindo RC, de la Fuente J. Transcriptomics data integration reveals Jak-STAT pathway as a common pathway affected by pathogenic intracellular bacteria in natural reservoir hosts. J Proteomics Bioinform. 2012;5:108–115. [Google Scholar]
- Takaki H, Minoda Y, Koga K, Takaesu G, Yoshimura A, Kobayashi T. TGF-beta1 suppresses IFN-gamma-induced NO production in macrophages by suppressing STAT1 activation and accelerating iNOS protein degradation. Genes Cells. 2006;11:871–882. doi: 10.1111/j.1365-2443.2006.00988.x. [DOI] [PubMed] [Google Scholar]
- Galindo RC, Muñoz PM, de Miguel MJ, Marin CM, Blasco JM, Gortazar C, Kocan KM, de la Fuente J. Differential expression of inflammatory and immune response genes in rams experimentally infected with a rough virulent strain of Brucella ovis. Vet Immunol Immunopath. 2009;127:295–303. doi: 10.1016/j.vetimm.2008.10.326. [DOI] [PubMed] [Google Scholar]
- Galindo RC, Ayoubi P, Naranjo V, Gortazar C, de la Fuente J. Gene expression profiles of European wild boar naturally infected with Mycobacterium bovis. Vet Immunol Immunopath. 2009;129:119–125. doi: 10.1016/j.vetimm.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Carlyon JA, Fikrig E. Invasion and survival strategies of Anaplasma phagocytophilum. Cell Microbiol. 2003;11:743–754. doi: 10.1046/j.1462-5822.2003.00323.x. [DOI] [PubMed] [Google Scholar]
- Woldehiwet Z. Immune evasion and immunosuppression by Anaplasma phagocytophilum, the causative agent of tick-borne fever of ruminants and human granulocytic anaplasmosis. Vet J. 2008;175:37–44. doi: 10.1016/j.tvjl.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Choi KS, Dumler JS. Mitogenic component in polar lipid-enriched Anaplasma phagocytophilum membranes. Clin Vaccine Immunol. 2007;14:1260–1265. doi: 10.1128/CVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Loewenich FD, Scorpio DG, Reischl U, Dumler JS, Bogdan C. Frontline: control of Anaplasma phagocytophilum, an obligate intracellular pathogen, in the absence of inducible nitric oxide synthase, phagocyte NADPH oxidase, tumor necrosis factor, Toll-like receptor (TLR)2 and TLR4, or the TLR adaptor molecule MyD88. Eur J Immunol. 2004;34:1789–1797. doi: 10.1002/eji.200425029. [DOI] [PubMed] [Google Scholar]
- Ehlers S. Commentary: adaptive immunity in the absence of innate immune responses? The un-Tolled truth of the silent invaders. Eur J Immunol. 2004;34:1783–1788. doi: 10.1002/eji.200425250. [DOI] [PubMed] [Google Scholar]
- Kim HY, Mott J, Zhi N, Tajima T, Rikihisa Y. Cytokine gene expression by peripheral blood leukocytes in horses experimentally infected with Anaplasma phagocytophila. Clin Diagn Lab Immunol. 2002;9:1079–1084. doi: 10.1128/CDLI.9.5.1079-1084.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RJ, Birtles RJ, Radford AD, Woldehiwet AD. Recurrent bacteraemia in sheep infected persistently with Anaplasma phagocytophilum. J Comp Path. 2012. in press. http://dx.doi.org/10.1016/j.jcpa.2012.02.005. [DOI] [PubMed]
- Wesche H, Korherr C, Kracht M, Falk W, Resch K, Martin MU. The interleukin-1 receptor accessory protein (IL-1RAcP) is essential for IL-1-induced activation of interleukin-1 receptor-associated kinase (IRAK) and stress-activated protein kinases (SAP kinases) J Biol Chem. 1997;272:7727–7731. doi: 10.1074/jbc.272.12.7727. [DOI] [PubMed] [Google Scholar]
- van de Sande WW, Fahal A, Verbrugh H, van Belkum A. Polymorphisms in genes involved in innate immunity predispose toward mycetoma susceptibility. J Immunol. 2007;179:3065–3074. doi: 10.4049/jimmunol.179.5.3065. [DOI] [PubMed] [Google Scholar]
- Beetz S, Wesch D, Marischen L, Welte S, Oberg HH, Kabelitz D. Innate immune functions of human gammadelta T cells. Immunobiol. 2008;213:173–182. doi: 10.1016/j.imbio.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Rikihisa Y. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin Microbiol Rev. 2011;24:469–489. doi: 10.1128/CMR.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altstaedt J, Kirchner H, Rink L. Cytokine production of neutrophils is limited to interleukin-8. Immunol. 1996;89:563–568. doi: 10.1046/j.1365-2567.1996.d01-784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand RJ, Dai S, Gribar SS, Richardson W, Kohler JW, Hoffman RA, Branca MF, Li J, Shi XH, Sodhi CP, Hackam DJ. A role for connexin 43 in macrophage phagocytosis and host survival after bacterial peritoneal infection. J Immunol. 2008;181:8534–8543. doi: 10.4049/jimmunol.181.12.8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocan KM, Busby AT, Allison RW, Breshears MA, Coburn L, Galindo RC, Ayllón N, Blouin EF, de la Fuente J. Sheep experimentally-infected with a human isolate of Anaplasma phagocytophilum serve as a host for infection of Ixodes scapularis. Ticks Tick Borne Dis. 2012. in press. http://dx.doi.org/10.1016/j.ttbdis.2012.01.004. [DOI] [PubMed]
- Reichard MV, Manzano Roman R, Kocan KM, Blouin EF, de la Fuente J, Snider TA, Heinz RE, West MD, Little SE, Massung RF. Inoculation of white-tailed deer (Odocoileus virginianus) with Ap-V1 or NY-18 strains of Anaplasma phagocytophilum and microscopic demonstration of Ap-V1 in Ixodes scapularis adults that acquired infection from deer as nymphs. Vector-Borne Zoon Dis. 2009;9:565–568. doi: 10.1089/vbz.2008.0106. [DOI] [PubMed] [Google Scholar]
- Blas-Machado U, de la Fuente J, Blouin EF, Almazán C, Kocan KM, Mysore JV. Experimental infection of C3H/HeJ mice with the NY18 isolate of Anaplasma phagocytophilum. Vet Pathol. 2007;44:64–73. doi: 10.1354/vp.44-1-64. [DOI] [PubMed] [Google Scholar]
- Franzén P, Aspan A, Egenvall A, Gunnarsson A, Karlstam E, Pringle J. Molecular evidence for persistence of Anaplasma phagocytophilum in the absence of clinical abnormalities in horses after recovery from acute experimental infection. J Vet Intern Med. 2009;23:636–642. doi: 10.1111/j.1939-1676.2009.0317.x. [DOI] [PubMed] [Google Scholar]