Abstract
The nucleotide sequence of the repressor gene C of the temperate phage P2 has been determined. It codes for a nonbasic polypeptide, 99 amino acids long. Twelve repressor-defective mutants have been mapped. All but one are located within the presumed coding part of the gene. There is a strong promoter sequence and an 8-base-pair inverted repeat preceding the gene. The P2 repressor protein shows structural similarity to other DNA-binding proteins. The operator region for the early replication functions was located by sequencing the DNA of three virulent mutants. The sequence indicates that there are two repressor-binding sites. In addition, one of the sites shows sequence homology with part of the operator region of the biotin operon of Escherichia coli.
Full text
PDF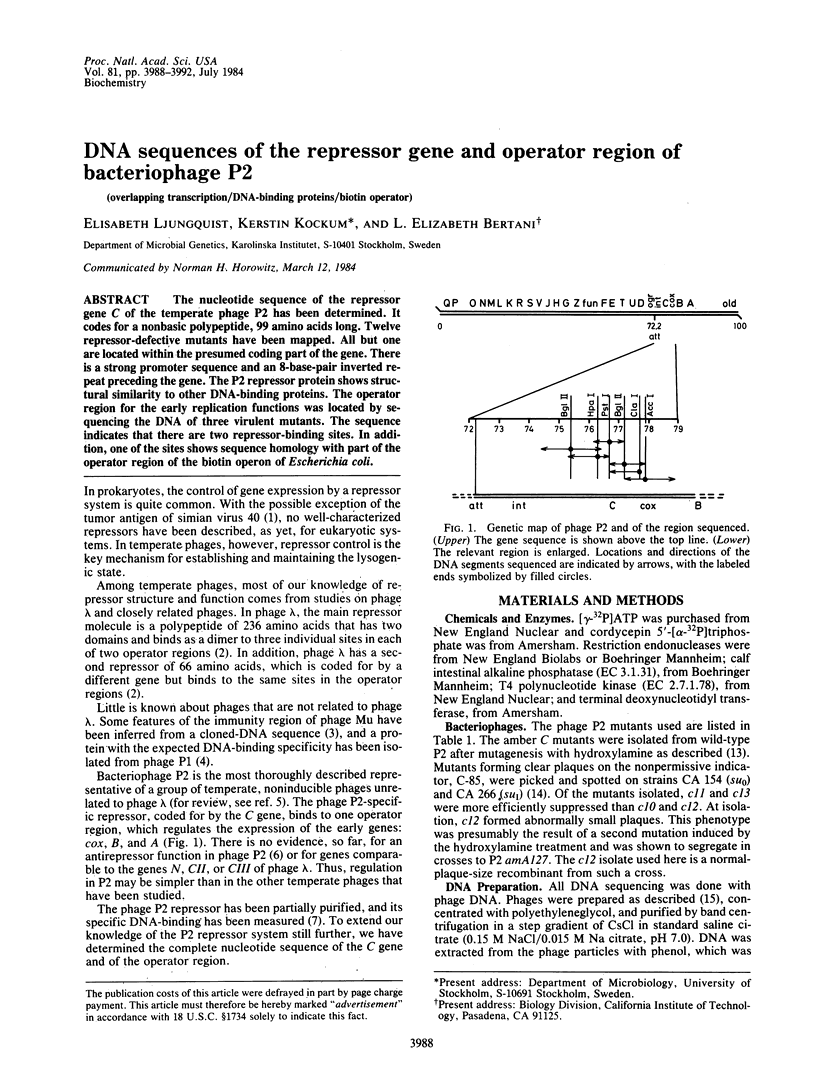




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumstark B. R., Scott J. R. The c1 repressor of bacteriophage P1. I. Isolation of the c1 protein and determination of the P1 DNA region to which it binds. J Mol Biol. 1980 Jul 15;140(4):471–480. doi: 10.1016/0022-2836(80)90266-1. [DOI] [PubMed] [Google Scholar]
- Bertani G. Deletions in bacteriophage P2. Circularity of the genetic map and its orientation relative to the DNA denaturation map. Mol Gen Genet. 1975;136(2):107–137. doi: 10.1007/BF00272034. [DOI] [PubMed] [Google Scholar]
- Bertani L. E. Abortive induction of bacteriophage P2. Virology. 1968 Sep;36(1):87–103. doi: 10.1016/0042-6822(68)90119-0. [DOI] [PubMed] [Google Scholar]
- Bertani L. E., Bertani G. Genetics of P2 and related phages. Adv Genet. 1971;16:199–237. doi: 10.1016/s0065-2660(08)60359-4. [DOI] [PubMed] [Google Scholar]
- Bertani L. E., Bertani G. Preparation and characterization of temperate, non-inducible bacteriophage P2 (host: Escherichia coli). J Gen Virol. 1970 Feb;6(2):201–212. doi: 10.1099/0022-1317-6-2-201. [DOI] [PubMed] [Google Scholar]
- Bertani L. E. Limited multiplication of phages superinfecting lysogenic bacteria and its implication for the mechanism of immunity. Virology. 1965 Dec;27(4):496–511. doi: 10.1016/0042-6822(65)90175-3. [DOI] [PubMed] [Google Scholar]
- Gicquel-Sanzey B., Cossart P. Homologies between different procaryotic DNA-binding regulatory proteins and between their sites of action. EMBO J. 1982;1(5):591–595. doi: 10.1002/j.1460-2075.1982.tb01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Schwarz E. Nucleotide sequence of the cro-cII-oop region of bacteriophage 434 DNA. Nucleic Acids Res. 1979 Mar;6(3):867–881. doi: 10.1093/nar/6.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981 Nov 19;294(5838):217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Lindahl G. On the control of transcription in bacteriophage P2. Virology. 1971 Dec;46(3):620–633. doi: 10.1016/0042-6822(71)90065-1. [DOI] [PubMed] [Google Scholar]
- Lindahl G., Sunshine M. Excision-deficient mutants of bacteriophage P2. Virology. 1972 Jul;49(1):180–187. doi: 10.1016/s0042-6822(72)80019-9. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Bertani L. E. Properties and products of the cloned int gene of bacteriophage P2. Mol Gen Genet. 1983;192(1-2):87–94. doi: 10.1007/BF00327651. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., Takeda Y. Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1428–1432. doi: 10.1073/pnas.79.5.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Otsuka A., Abelson J. The regulatory region of the biotin operon in Escherichia coli. Nature. 1978 Dec 14;276(5689):689–694. doi: 10.1038/276689a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Priess H., Kamp D., Kahmann R., Bräuer B., Delius H. Nucleotide sequence of the immunity region of bacteriophage Mu. Mol Gen Genet. 1982;186(3):315–321. doi: 10.1007/BF00729448. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Shimatake H., Brady C., Rosenberg M. Sequence of Cro gene of bacteriophage lambda. Nature. 1977 Nov 17;270(5634):274–275. doi: 10.1038/270274a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sauer R. T. DNA sequence of the bacteriophage gama cI gene. Nature. 1978 Nov 16;276(5685):301–302. doi: 10.1038/276301a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Pan J., Hopper P., Hehir K., Brown J., Poteete A. R. Primary structure of the phage P22 repressor and its gene c2. Biochemistry. 1981 Jun 9;20(12):3591–3598. doi: 10.1021/bi00515a044. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TESSMAN I., PODDAR R. K., KUMAR S. IDENTIFICATION OF THE ALTERED BASES IN MUTATED SINGLE-STRANDED DNA. I. IN VITRO MUTAGENESIS BY HYDROXYLAMINE, ETHYL METHANESULFONATE AND NITROUS ACID. J Mol Biol. 1964 Aug;9:352–363. doi: 10.1016/s0022-2836(64)80212-6. [DOI] [PubMed] [Google Scholar]
- Tjian R. T antigen binding and the control of SV40 gene expression. Cell. 1981 Oct;26(1 Pt 1):1–2. doi: 10.1016/0092-8674(81)90026-x. [DOI] [PubMed] [Google Scholar]
- Westö A., Ljungquist E. A restriction endonuclease cleavage map of bacteriophage P2. Mol Gen Genet. 1979 Mar 9;171(1):91–102. doi: 10.1007/BF00274019. [DOI] [PubMed] [Google Scholar]
- Westö A., Ljungquist E. Cloning of the immunity repressor determinant of bacteriophage P2 in the pBR322 plasmid. Mol Gen Genet. 1980 Apr;178(1):101–109. doi: 10.1007/BF00267218. [DOI] [PubMed] [Google Scholar]



