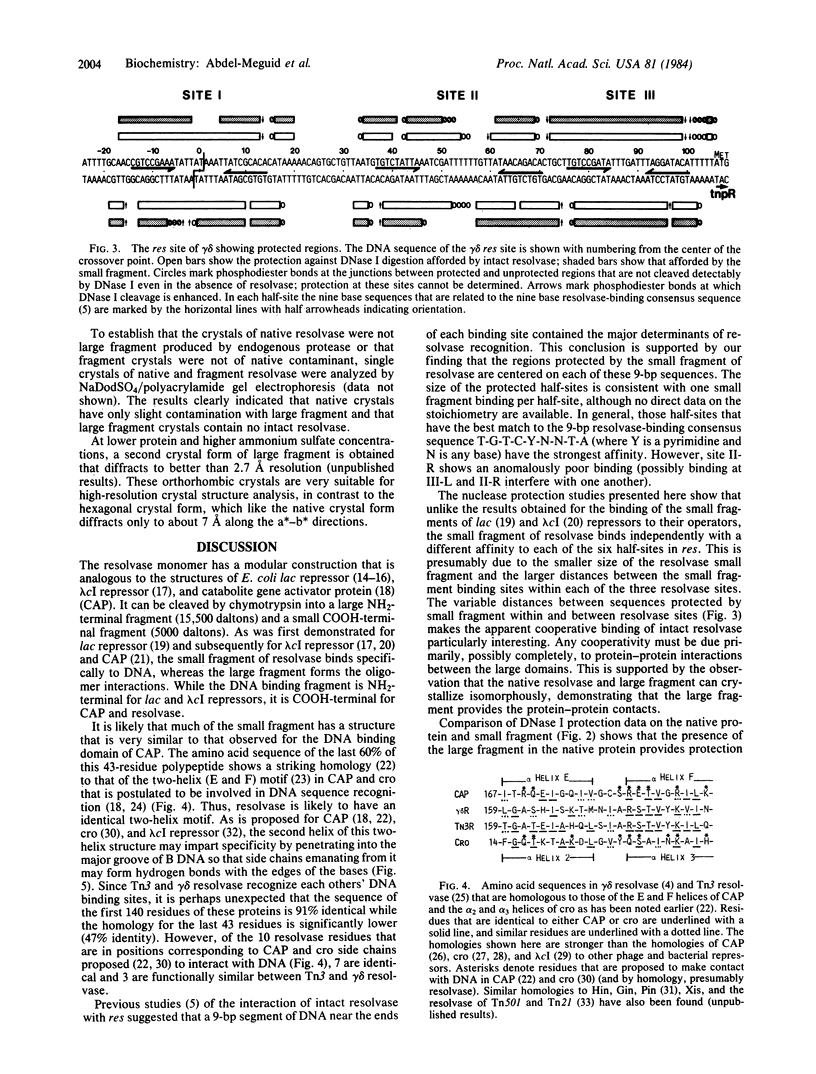
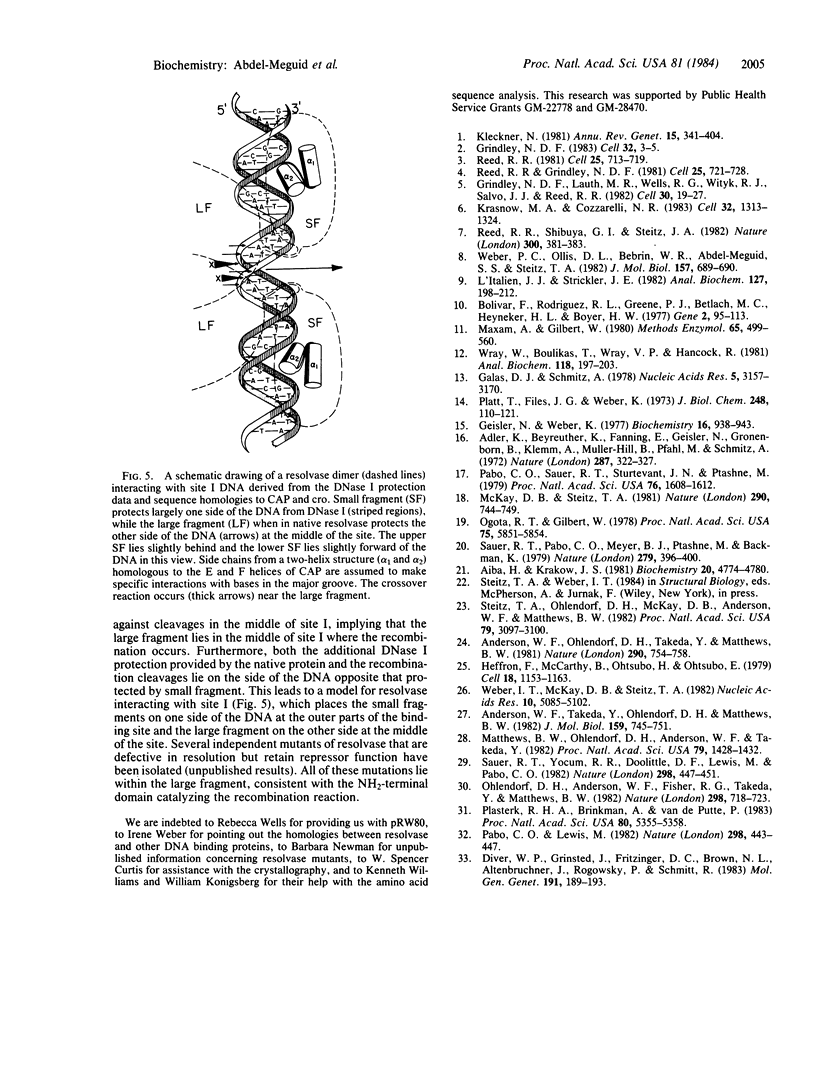
Abstract
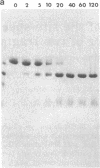
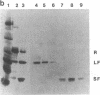
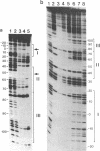
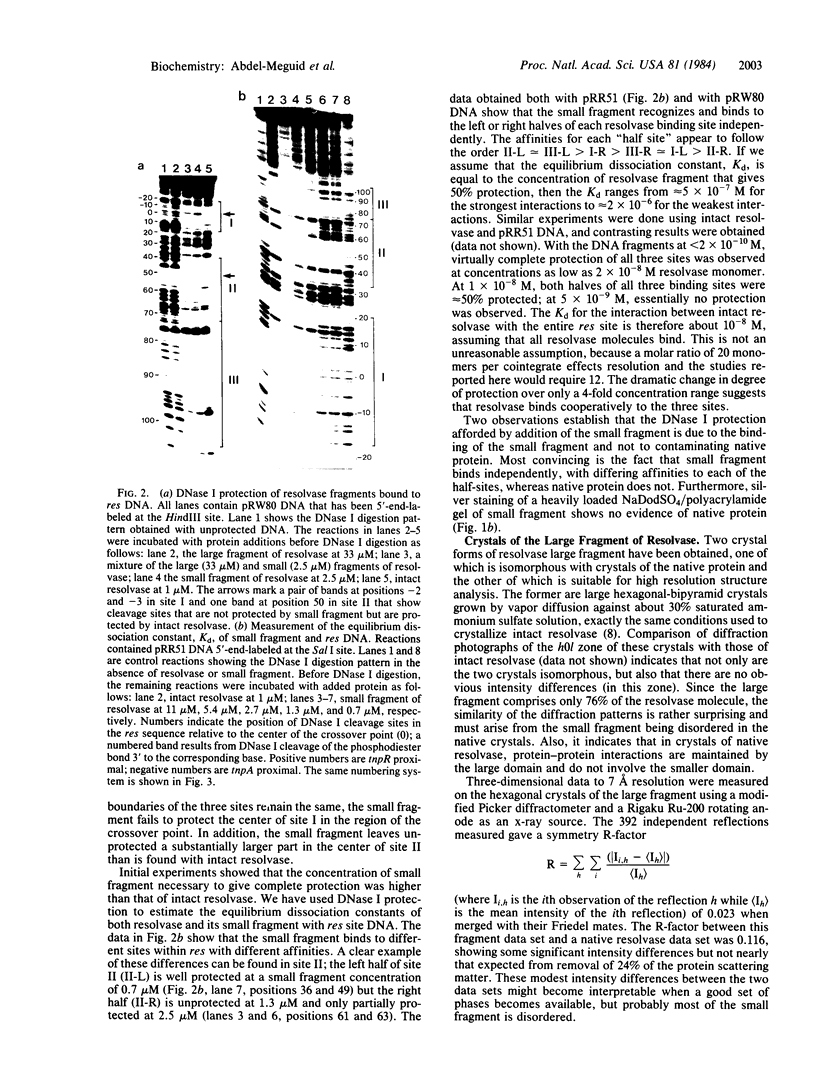
The 20,500-dalton gamma delta resolvase monomer can be cleaved by chymotrypsin into a 5000-dalton COOH-terminal fragment and a 15,500-dalton NH2-terminal fragment that have been purified. Two crystal forms of the large fragment have been obtained, one of which is isomorphous with crystals of the native protein, showing that the large fragment makes the protein-protein contacts in the crystal and that the small fragment is segmentally disordered relative to the large fragment. Nuclease protection demonstrates that the small fragment binds specifically to all three DNA binding sites protected by resolvase. However, unlike native resolvase, which binds to all three complete sites with equal affinity, the small fragment binds to each of the six half sites with a different affinity. It has not been possible to demonstrate specific DNA binding of the larger fragment. Thus, resolvase has a modular construction analogous to that found for some repressors and activators; its COOH-terminal domain recognizes specific sequences in the DNA and its NH2-terminal domain mediates protein-protein interactions and probably has the enzymatic activity.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler K., Beyreuther K., Fanning E., Geisler N., Gronenborn B., Klemm A., Müller-Hill B., Pfahl M., Schmitz A. How lac repressor binds to DNA. Nature. 1972 Jun 9;237(5354):322–327. doi: 10.1038/237322a0. [DOI] [PubMed] [Google Scholar]
- Aiba H., Krakow J. S. Isolation and characterization of the amino and carboxyl proximal fragments of the adenosine cyclic 3' ,5'-phosphate receptor protein of Escherichia coli. Biochemistry. 1981 Aug 4;20(16):4774–4780. doi: 10.1021/bi00519a038. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Anderson W. F. Proposed alpha-helical super-secondary structure associated with protein-dna recognition. J Mol Biol. 1982 Aug 25;159(4):745–751. doi: 10.1016/0022-2836(82)90111-5. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N., Weber K. Isolation of amino-terminal fragment of lactose repressor necessary for DNA binding. Biochemistry. 1977 Mar 8;16(5):938–943. doi: 10.1021/bi00624a020. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Lauth M. R., Wells R. G., Wityk R. J., Salvo J. J., Reed R. R. Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of gamma delta and Tn3. Cell. 1982 Aug;30(1):19–27. doi: 10.1016/0092-8674(82)90007-1. [DOI] [PubMed] [Google Scholar]
- Grindley N. D. Transposition of Tn3 and related transposons. Cell. 1983 Jan;32(1):3–5. doi: 10.1016/0092-8674(83)90490-7. [DOI] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- L'Italien J. J., Strickler J. E. Application of high-performance liquid chromatographic peptide purification to protein microsequencing by solid-phase Edman degradation. Anal Biochem. 1982 Nov 15;127(1):198–212. doi: 10.1016/0003-2697(82)90165-8. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., Takeda Y. Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1428–1432. doi: 10.1073/pnas.79.5.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T., Sturtevant J. M., Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1608–1612. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Brinkman A., van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T., Files J. G., Weber K. Lac repressor. Specific proteolytic destruction of the NH 2 -terminal region and loss of the deoxyribonucleic acid-binding activity. J Biol Chem. 1973 Jan 10;248(1):110–121. [PubMed] [Google Scholar]
- Reed R. R., Grindley N. D. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell. 1981 Sep;25(3):721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Shibuya G. I., Steitz J. A. Nucleotide sequence of gamma delta resolvase gene and demonstration that its gene product acts as a repressor of transcription. Nature. 1982 Nov 25;300(5890):381–383. doi: 10.1038/300381a0. [DOI] [PubMed] [Google Scholar]
- Reed R. R. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981 Sep;25(3):713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Pabo C. O., Meyer B. J., Ptashne M., Backman K. C. Regulatory functions of the lambda repressor reside in the amino-terminal domain. Nature. 1979 May 31;279(5712):396–400. doi: 10.1038/279396a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T., McKay D. B., Steitz T. A. Two helix DNA binding motif of CAP found in lac repressor and gal repressor. Nucleic Acids Res. 1982 Aug 25;10(16):5085–5102. doi: 10.1093/nar/10.16.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Ollis D. L., Bebrin W. R., Abdel-Meguid S. S., Steitz T. A. Crystallization of resolvase, a repressor that also catalyzes site-specific DNA recombination. J Mol Biol. 1982 Jun 5;157(4):689–690. doi: 10.1016/0022-2836(82)90508-3. [DOI] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]