Abstract
Background
A number of case-control studies were conducted to investigate the association of common type 2 diabetes (T2D) risk gene polymorphisms with gestational diabetes mellitus (GDM). However, these studies have yielded contradictory results. We therefore performed a meta-analysis to derive a more precise estimation of the association between these polymorphisms and GDM, hence achieve a better understanding to the relationship between T2D and GDM.
Methods
PubMed, EMBASE, ISI web of science and the Chinese National Knowledge Infrastructure databases were systematically searched to identify relevant studies. Data were abstracted independently by two reviewers. A meta-analysis was performed to examine the association between 9 polymorphisms from 8 genes and susceptibility to GDM. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated. Heterogeneity among articles and their publication bias were also tested.
Results
We identified 22 eligible studies including a total of 10,336 GDM cases and 17,445 controls. We found 8 genetic polymorphisms were significantly associated with GDM in a random-effects meta-analysis. These polymorphisms were in or near the following genes: TCF7L2 (rs7903146), MTNR1B (rs10830963), IGF2BP2 (rs4402960), KCNJ11 (rs5219), CDKAL1 (rs7754840), KCNQ1 (rs2237892 and rs2237895) and GCK (rs4607517); while no association was found for PPARG with GDM risk. Similar results were also observed under dominant genetic model for these polymorphisms.
Conclusions
This meta-analysis found 8 genetic variants associated with GDM. The relative contribution and relevance of the identified genes in the pathogenesis of GDM should be the focus of future studies.
Introduction
Gestational diabetes mellitus (GDM) is defined as glucose intolerance that is first detected during pregnancy [1]. It is characterized by impaired insulin secretion and action [2], [3]. Gestational diabetes complicates about 1–3% of all pregnancies in the western world [4], whereas 5–10% among of Asian women [5]. Although its exact etiology is unknown, accumulating evidence recognizes GDM as a quintessential multifactorial disease in which environmental triggers interact with genetic variants [6]. Given the fact that women with a history of GDM are at an increased risk of developing type 2 diabetes (T2D) later in their lives [7] and women with a family history of diabetes may be predisposed to an increased risk of GDM [8], it is plausible to hypothesize that GMD may share the same risk factors and genetic susceptibilities with T2D. However, knowledge regarding the genetics of GDM is very limited so far [9].
Recently, spectacular advance was made in identifying susceptible genes involved in T2D through genome-wide association strategy (GWAS) [10], [11]. Consequently, a number of novel genetic variants (PPARG, KCNJ11, IGF2BP2, KCNQ1, TCF7L2, CDKAL1, and MTNR1B) were shown to increase the risk of T2D in reproducible studies. Therefore, several studies have examined the association of these newly identified loci using a candidate gene approach for GDM. It has been reported that the pathophysiological changes of GDM are similar to those observed in T2D, which is characterized by peripheral insulin resistance accompanied by an insulin secretory defect [12], [13]. Functional studies showed that these new diabetogenic genes took part in many steps of the process, for instance, impaired β-cell function (CDKAL1, IGF2BP2, KCNQ1, KCNJ11, MTNR1B), insulin resistance (PPARG, TCF7L2), and abnormal utilization of glucose (GCK) [14]–[23].
Genetic association studies can be problematic to reproduce due to inadequate statistical power, multiple hypothesis testing, population stratification, publication bias, and phenotypic heterogeneity. Considering the lack of sufficient evidence about the effect of candidate genes of T2D on GDM and the conflicting results reported, we therefore performed a meta-analysis to assess the association between the most commonly studied polymorphisms in the PPARG, CDKAL1, KCNQ1, IGF2BP2, TCF7L2, KCNJ11, MTNR1B, GCK genes and GDM risk.
Materials and Methods
Literature search strategy
Genetic association studies polymorphism and GDM risk published up to April, 2012 were identified through systematic searches in PubMed, EMBASE, ISI Web of Science and the Chinese National Knowledge Infrastructure (CNKI) databases. No language restrictions were applied. The search strategy consisted of multiple queries combining: ‘gestational diabetes mellitus’ and ‘variations’ or ‘polymorphisms’. In addition, the names of specific genes and polymorphisms were combined with the topic ‘gestational diabetes mellitus’. All reference lists from the main reports and relevant reviews were hand searched for additional eligible studies.
Eligible studies and data extraction
Eligible studies had to meet all the following criteria: (1) original papers containing independent data, (2) identification of gestational diabetes mellitus cases was confirmed pathologically, (3) case–control or cohort studies and (4) genotype distribution information in cases and controls or odds ratio (OR) with its 95% confidence interval (CI) and P-value. The major reasons for exclusion of studies were (1) overlapping data; (2) case-only studies, family based studies, and review articles.
Data extraction was performed independently by two reviewers. Review reports from the two were than compared to identify any inconsistency, and differences were resolved by further discussion among all authors. For each included study, the following information was extracted from each report according to a fixed protocol: first author's surname, publication year, definition and numbers of cases and controls, frequency of genotypes, Hardy–Weinberg equilibrium status, source of controls, mean age of cases and controls, body mass index (BMI), ethnicity, and genotyping method.
Statistical methods
The strength of association between the genetic polymorphism and GDM was accessed by calculating odds ratio (OR) with 95% confidence interval (CI). For single nucleotide polymorphisms (SNPs), the frequency of the risk allele was compared between diabetic cases and non-diabetic controls. Additional pooled estimates were also given with corresponding results under dominant genetic model.
Cochran's chi-square-based Q statistic test was performed in order to assess possible heterogeneity between the individual studies and thus to ensure that each group of studies was suitable for meta-analysis [24]. ORs were pooled according to the method of DerSimonian and Laird that takes into account the variation between studies, and 95% CI were constructed using Woolf's method [25], [26]. The Z test was used to determine the significance of the pooled OR. Pre-specified stratified analyses were performed to explain heterogeneity or investigate whether the reported association was present in a subgroup. Stratified analysis was performed for ethnicity (Caucasian vs East Asian origin).
Funnel plots was used to provide diagnosis of the potential publication bias. Egger's regression test was also conducted to identify small study effects [27]. Chi-square test was used to check if there was significant deviation from Hardy–Weinberg equilibrium (HWE) among the control subjects in each study. All statistical analyses were carried out with the Stata software version 10.0 (Stata Corporation, College Station, TX). The type I error rate was set at 0.05. All the p-values were for two-sided analysis.
Results
Characteristics of studies
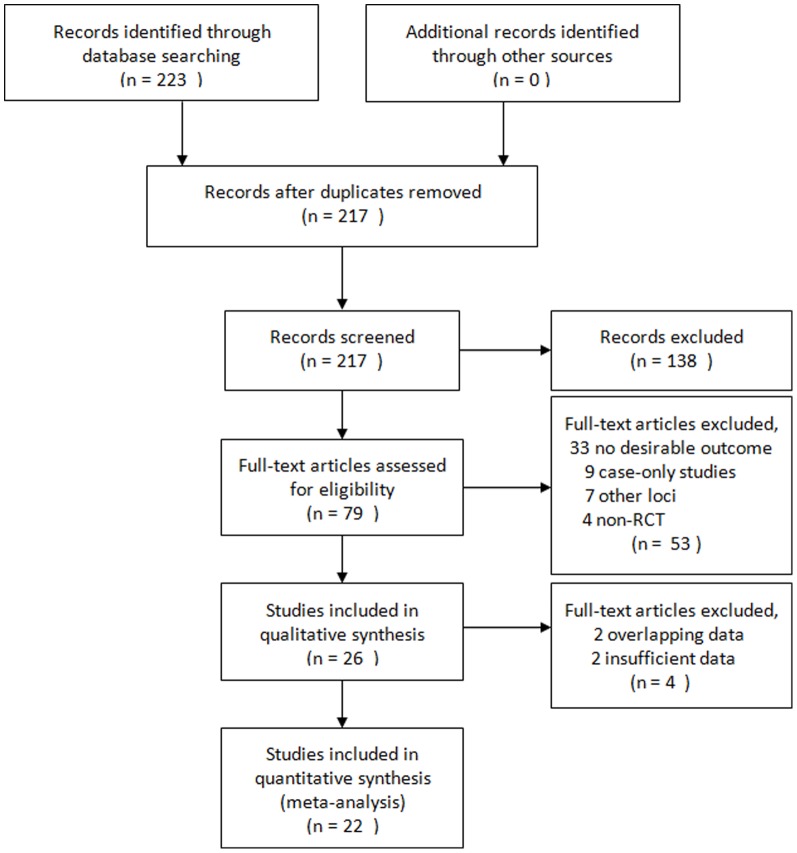
In all, we included 22 studies in this meta-analysis, with a total of 10,336 cases and 17,445 controls concerning 9 genetic variants in or near 8 genes. The detailed characteristics of the studies included were shown in Table 1. The study selection process is shown in Figure 1. These polymorphisms were found to occur in frequencies consistent with Hardy–Weinberg equilibrium in the control populations of the vast majority of the published studies. Details of analyses of all assessed genetic variants are provided in Table 2.
Table 1. Characteristics of the studies included in the meta-analysis.
| Study | Year | Ethnicity | Case | Control | No. of case/control | Genotyping method | Age of case/control | BMI of case/control |
| Kwak [28] | 2012 | Korean | OGTT confirmed | Normal fasting glucose | 1399/2025 | Affymetrix array, TaqMan | 32.1/62.5 | 24.2/24.4 |
| Kim [29] | 2011 | Korean | OGTT confirmed | Normal glucose tolerant | 908/966 | TaqMan | 33.2/32.2 | 23.3/21.4 |
| Deng [30] | 2011 | Chinese | GDM patients | Non-diabetic participants | 87/91 | Sequencing | 31.8/29.7 | 23.6/21.5 |
| Heude [31] | 2011 | French | OGTT confirmed | Normal glucose tolerant | 109/1571 | TaqMan | NA/NA | NA/NA |
| Wang [32] | 2011 | Chinese | OGTT confirmed | Normal glucose tolerant | 705/1029 | TaqMan | 32.0/30.0 | 21.7/21.5 |
| Pappa [33] | 2011 | Greek | GDM per IGDW criteria | Normal glucose tolerant | 148/107 | RFLP | 32.5/26.7 | 26.0/24.3 |
| Papadopoulou [34] | 2011 | Swede | OGTT confirmed | Normal glucose tolerant | 803/1110 | TaqMan | NA/NA | NA/NA |
| Shin [35] | 2010 | Korean | OGTT confirmed | Healthy | 930/628 | TaqMan | 33.2/NA | 23.3/NA |
| Cheng [36] | 2010 | Chinese | OGTT confirmed | Normal glucose tolerant | 55/173 | RFLP | 27.0/29.6 | NA/NA |
| Kwak [37] | 2010 | Korean | OGTT confirmed | Normal fasting glucose | 869/632 | TaqMan | 27.9/64.7 | 23.1/23.3 |
| Santos [38] | 2010 | Euro-Brazilian | GDM per ADA criteria | Healthy | 150/600 | RFLP | 31.7/24.9 | 33.5/24.8 |
| Zhou [39] | 2009 | Chinese | OGTT confirmed | Normal glucose tolerant | 520/916 | RFLP | 32.5/30.7 | 21.4/21.4 |
| Cho [40] | 2009 | Korean | OGTT confirmed | Normal fasting glucose | 868/632 | TaqMan | 32.0/64.7 | 23.1/23.3 |
| Zhu [41] | 2009 | Chinese | GDM patients | Non-diabetic participants | 179/180 | RFLP | 28.1/27.4 | 24.6/23.4 |
| Lauenborg [14] | 2008 | Dane | OGTT confirmed | Normal glucose tolerant | 276/2383 | TaqMan | 43.1/45.2 | 28.9/25.0 |
| Shaat [42] | 2007 | Swede | OGTT confirmed | Non-diabetic participants | 637/1232 | TaqMan | 32.3/30.5 | NA/NA |
| Tok [43] | 2006 | Turkish | OGTT confirmed | Normal glucose tolerant | 100/62 | RFLP | 33.5/31.6 | 26.6/23.0 |
| Shaat [44] | 2006 | Swede | OGTT confirmed | Non-diabetic participants | 641/1229 | RFLP | NA/NA | NA/NA |
| Shaat [45] | 2005 | Swede | OGTT confirmed | Normal glucose tolerant | 588/1180 | TaqMan | 32.2/30.5 | NA/NA |
| Shaat [46] | 2004 | Swede | OGTT confirmed | Normal glucose tolerant | 500/550 | RFLP | 32.3/NA | 30.1/NA |
| Chiu [47] | 1994 | American | OGTT confirmed | Non-diabetic participants | 174/99 | SSCP | 28.3/22.1 | 33.9/22.5 |
| Unpublished data | / | Chinese | OGTT confirmed | Normal glucose tolerant | 50/50 | RFLP | 30.5/28.8 | 31.0/27.9 |
OGTT: Oral Glucose Tolerance Test, IGDW: International Gestational Diabetes Workshop, ADA: American Diabetes Association, NA: Not Available.
Figure 1. The flow chart of the included studies.
Table 2. Results of the pooled data analyses for the 9 studied polymorphisms and gestational diabetes mellitus risk.
| Variants per gene | Risk allele | Total/subgroup | No. data sets | No. of case/control | Risk allele | Dominant model | ||||
| OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | |||||
| PPARG rs1801282 | C | Total | 11 | 2908/6940 | 1.01 (0.96–1.06) | 0.80 | 1.00 | 1.14 (0.68–1.91) | 0.63 | 0.45 |
| Caucasian | 5 | 1559/5721 | 1.00 (0.94–1.06) | 0.98 | 0.99 | 1.01 (0.58–1.76) | 0.98 | 0.44 | ||
| East Asian | 4 | 1149/1035 | 1.02 (0.93–1.11) | 0.70 | 0.99 | 2.43 (0.38–15.43) | 0.35 | 0.27 | ||
| TCF7L2 rs7903146 | T | Total | 6 | 3148/6550 | 1.51 (1.39–1.65) | <10−5 | 0.77 | 1.69 (1.51–1.89) | <10−5 | 0.51 |
| Caucasian | 4 | 1812/4681 | 1.51 (1.38–1.65) | <10−5 | 0.48 | 1.71 (1.49–1.96) | <10−5 | 0.28 | ||
| East Asian | 2 | 1336/1869 | 1.55 (1.16–2.09) | 0.004 | 0.90 | 1.56 (1.24–2.22) | 0.001 | 0.75 | ||
| MTNR1B rs10830963 | G | Total | 5 | 3094/4111 | 1.34 (1.18–1.52) | <10−5 | 0.02 | 1.46 (1.25–1.72) | <10−5 | 0.11 |
| IGF2BP2 rs4402960 | T | Total | 4 | 2304/5228 | 1.21 (1.08–1.36) | 0.001 | 0.09 | 1.25 (1.07–1.49) | 0.003 | 0.06 |
| East Asian | 3 | 2030/2894 | 1.24 (1.07–1.44) | 0.004 | 0.07 | 1.27 (1.12–1.43) | 0.0002 | 0.81 | ||
| KCNJ11 rs5219 | T | Total | 5 | 2305/5569 | 1.15 (1.06–1.24) | 0.0004 | 0.99 | 1.25 (1.10–1.42) | 0.001 | 0.88 |
| Caucasian | 3 | 991/3698 | 1.17 (1.05–1.30) | 0.005 | 0.98 | 1.25 (1.07–1.46) | 0.006 | 0.72 | ||
| East Asian | 2 | 1314/1871 | 1.13 (1.02–1.26) | 0.03 | 0.93 | 1.11 (1.03–1.20) | 0.02 | 0.29 | ||
| CDKAL1 rs7754840 | C | Total | 4 | 2959/3675 | 1.43 (1.20–1.71) | <10−4 | 0.0003 | 1.51 (1.33–1.82) | <10−4 | 0.008 |
| KCNQ1 rs2237892 | C | Total | 3 | 2285/2168 | 1.20 (1.09–1.31) | <10−4 | 0.70 | 1.42 (1.18–1.71) | 0.0002 | 0.97 |
| KCNQ1 rs2237895 | C | Total | 3 | 2286/2168 | 1.20 (1.09–1.31) | 0.0001 | 0.75 | 1.31 (1.16–1.48) | <10−4 | 0.54 |
| GCK rs4607517 | A | Total | 5 | 2135/4193 | 1.12 (1.02–1.23) | 0.01 | 0.41 | 1.15 (1.01–1.30) | 0.04 | 0.43 |
Genetic variants involved in β-cell function
Six genetic variants in five genes thought to be related to β-cell function were reproducibly associated with GDM. rs4402960 of IGF2BP2 was associated with GDM (OR = 1.21, 95% CI = 1.08–1.36, P = 0.001; Supplementary figure 1) in four studies (n = 8,732), also in the subgroup among East Asian diabetes mellitus patients (OR = 1.24, 95% CI = 1.07–1.44, P = 0.004). The rs10830963 in MTNR1B were studied in five studies concerning diabetic patients form East Asian population. The overall OR of the G allele for GDM was 1.34 (95% CI = 1.18–1.52, P<10−5; Supplementary figure 2). CDKAL1 rs7754840 polymorphism was studied in four studies and was associated with GDM in the meta-analysis (OR = 1.43, 95% CI = 1.20–1.71, P<10−4; Supplementary figure 3). All four studies contained diabetic patients of East Asian descent. For KCNJ11 rs5219, our meta-analysis gave an overall OR of 1.15 (95% CI = 1.06–1.24, P = 0.0004; Supplementary figure 4) without statistically significant between-study heterogeneity. Significantly increased GDM risks were also found for Caucasian and East Asian populations when stratified by ethnicity. Significantly increased GDM risks were found for rs2237892 and rs2237895 of KCNQ1 with per-allele OR of 1.20 (95% CI = 1.09–1.31, P<10−4; Supplementary figure 5) and 1.20 (95% CI = 1.09–1.31, P = 0.0001; Supplementary figure 6) respectively. In addition, statistically significant results were also observed for these polymorphisms under dominant genetic model. After adjusting for multiple testing using Bonferroni correction, all significant associations for these polymorphisms under the allelic comparison and dominant genetic model remained.
Genetic variants involved in insulin resistance
A variant in PPARG, rs1801282, was the most studied polymorphism in GDM, with 11 data sets resulting in a pooled odds ratio of 1.01 (95% CI = 0.96–1.06, P = 0.80, Supplementary figure 7). Subsidiary analyses of ethnicity yielded a per-allele OR for East Asians of 1.02 (95% CI: 0.93–1.11, P = 0.70) and for Caucasians of 1.00 (95% CI: 0.94–1.06, P = 0.98). Similar results were also detected under dominant genetic model.
rs7903146 of TCF7L2, which is an important component in Wnt signaling pathway involved in development of the pancreas and islets, was associated with GDM (C allele: OR = 1.51, 95% CI = 1.39–1.65, P<10−5, Supplementary figure 8; dominant model: OR = 1.69, 95% CI = 1.51–1.89, P<10−5). Stratification by ethnicity indicated that the polymorphism was significantly associated with GDM for East Asians and Caucasians in all genetic models. After Bonferroni correction, significant associations still maintained for the polymorphism.
Genetic variants involved in glucose utilization
rs4607517 of GCK, which is first rate-limiting step in the glycolysis pathway, was significantly associated with GDM in the meta-analysis (OR = 1.12, 95% CI = 1.02–1.23, P = 0.01, Supplementary figure 9). Similar results were also found using dominant genetic model with OR of 1.15 (95% CI = 1.01–1.30, P = 0.04). However, for GCK rs4607517 association was no longer statistically significant using dominant genetic model after Bonferroni correction.
Publication bias
Begger's funnel plot and Egger's test were used to identify the potential publication biases of the literature, the shapes of the funnel plots appeared to be symmetrical (Supplementary figure 10–18) for polymorphisms in PPARG, TCF7L2, MTNR1B, IGF2BP2, KCNJ11, CDKAL1, KCNQ1 and GCK, suggesting that there was no obvious publication bias. Egger's test was used to provide further statistical evidence; similarly, the results showed no significant publication bias in this meta-analysis for these polymorphisms (P>0.05 for all polymorphisms).
Discussion
Large sample and unbiased epidemiological studies of predisposition genes polymorphisms could provide insight into the in vivo relationship between candidate genes and complex diseases. In this meta-analysis, 8 genetic variants were found to be associated with increased GDM susceptibility. Genetic studies of several T2D associated variants in relation to GDM has been performed previously, but this is the first complete overview assessing for these genetic variants that are reproducibly associated with the presence of GDM. This information could lead to improved insight into underlying pathogenetic mechanisms and the relationship between GDM and T2D. These results support a role for the following in the pathogenesis of GDM: impaired β-cell function, insulin resistance and abnormal utilization of glucose. During pregnancy, women are faced with increased adiposity and increased insulin resistance. The insulin resistance that develops during pregnancy is explained in part by the increased production of human placental lactogen, estrogen, and prolactin [48]–[50]. Those who have limited β-cell capacity for the compensation of insulin resistance are likely to develop GDM [9]. Women with GDM are assumed to have decreased β-cell insulin secretory function similar to T2D [9]. After parturition, nearly one-half of these women progress to T2D within 5 years [51]–[53]. Therefore, GDM is often regarded as a herald of type 2 diabetes in later life. Functional studies remain to be performed to establish the precise roles of these variants and pathways.
The identification of GDM susceptibility variants can lead to novel biological insights and improved measures of individual etiological processes, as indicated previously [54]. Individual etiological processes could allow preventive and therapeutic interventions in complex disease to be tailored to individuals on the basis of their genetic profiles. From prediction studies with genetic variants for T2D, it has been shown that 20 established genetic variants in T2D have an AUC of 0.54 (0.5 means no predictive value, 1.0 is perfect prediction), in contrast to the Framingham offspring and Cambridge risk scores (AUC of 0.78 and 0.72, respectively). Interestingly, addition of genetic information to phenotype-based risk models did not improve prediction [55]. It is also possible that for GDM the genotypic risk does not exceed the risk contributed by conventional risk factors (e.g. BMI, age, term of pregnancies), which means that the predictive value of risk variants for GDM would be limited [56]. Although genetic prediction and use of personalized medicine in GDM remains a new undertaking, prediction is likely to improve as additional disease variants are detected and replicated [57].
Novel biological insights may lead to development of new therapeutic targets, biomarkers and opportunities for disease prevention. Hypothesis-free approaches, such as GWAS, are most promising in this respect. At present, it seems wise to focus on assessing the relevance of previously detected genetic variants. As common SNPs associated with GDM and detected by GWAS may represent rare genetic variants with large effects, sequencing the regions surrounding highly significant and replicated genomic regions to detect rare variants appears to be reasonable. Follow-up in vitro and in vivo studies could then assess the functional relevance of these variants in GDM.
In interpreting the results, some limitations of this meta-analysis should be addressed. Publication bias is a concern in all meta-analyses even though the use of a statistical test did not show it. Negative studies are less likely to be published, potentially leading to an overestimation of effects. Moreover, non-significant genetic associations might have been underreported in published articles. Therefore, the effect estimates of the present study should be interpreted with caution, especially in cases where associations were based on small numbers of studies and/or small sample numbers. Second, in the subgroup analysis by ethnicity, the number of studies and subjects analyzed was small, and the statistical power was so low that caution should be taken in interpreting these results. Finally, the overall outcomes were based on individual unadjusted ORs, while a more precise evaluation should be adjusted by other potentially suspected factors including age, BMI, and environmental factors.
To the best of our knowledge, this study was the first comprehensive meta-analysis to assess the relationship between the T2D related gene polymorphisms and GDM susceptibility. Our meta-analysis identified 8 genetic variants associated with GDM. As studies among other populations are currently limited, further studies including a wider spectrum of subjects should be carried to investigate the role of those variants in other populations, which should lead to better, comprehensive understanding of the association between the genetic polymorphism and GDM. For future studies, gene–gene and gene–environment interactions should also be considered.
Supporting Information
Meta-analysis of the association between IGF2BP2 rs4402960 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between MTNR1B rs10830963 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between CDKAL1 rs7754840 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between KCNJ11 rs5219 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between KCNQ1 rs2237892 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between KCNQ1 rs2237895 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between PPARG rs1801282 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between TCF7L2 rs7903146 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between GCK rs4607517 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Begg's funnel plot of PPARG rs1801282 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.15).
(TIFF)
Begg's funnel plot of TCF7L2 rs7903146 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.18).
(TIFF)
Begg's funnel plot of MTNR1B rs10830963 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.69).
(TIFF)
Begg's funnel plot of IGF2BP2 rs4402960 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.70).
(TIFF)
Begg's funnel plot of KCNJ11 rs5219 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.76).
(TIFF)
Begg's funnel plot of CDKAL1 rs7754840 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.25).
(TIFF)
Begg's funnel plot of KCNQ1 rs2237892 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.34).
(TIFF)
Begg's funnel plot of KCNQ1 rs2237895 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.77).
(TIFF)
Begg's funnel plot of GCK rs4607517 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.65).
(TIFF)
PRISMA 2009.
(DOC)
Acknowledgments
We thank everyone who helped with this study.
Funding Statement
No current external funding sources for this study.
References
- 1. Metzger BE (1991) Summary and recommendations of the Third International Workshop-Conference on Gestational Diabetes Mellitus. Diabetes Suppl 2 197–201. [DOI] [PubMed] [Google Scholar]
- 2. Buchanan TA, Metzger BE, Freinkel N, Bergman RN (1990) Insulin sensitivity and B-cell responsiveness to glucose during late pregnancy in lean and moderately obese women with normal glucose tolerance or mild gestational diabetes. Am J Obstet Gynecol 162: 1008–1014. [DOI] [PubMed] [Google Scholar]
- 3. Ryan EA, O'Sullivan MJ, Skyler JS (1985) Insulin action during pregnancy. Studies with the euglycemic clamp technique. Diabetes 34: 380–389. [DOI] [PubMed] [Google Scholar]
- 4. Hadden DR (1985) Geographic, ethnic, and racial variations in the incidence of gestational diabetes mellitus. Diabetes 34 (Suppl 2) 8–12. [DOI] [PubMed] [Google Scholar]
- 5. Shaat N, Groop L (2007) Genetics of gestational diabetes mellitus. Curr Med Chem 14: 569–83. [DOI] [PubMed] [Google Scholar]
- 6. Martin AO, Simpson JL, Ober C, Frienkel N (1985) Frequency of diabetes mellitus in mothers of probands with gestational diabetes mellitus: possible maternal influence on the predisposition to gestational diabetes. Am J Obstet Gynecol 151: 471–5. [DOI] [PubMed] [Google Scholar]
- 7. Bellamy L, Casas JP, Hingorani AD, Williams D (2009) Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet 373: 1773–9. [DOI] [PubMed] [Google Scholar]
- 8. Williams MA, Qiu C, Dempsey JC, Luthy DA (2003) Familial aggregation of type 2 diabetes and chronic hypertension in women with gestational diabetes mellitus. J Reprod Med 48: 955–962. [PubMed] [Google Scholar]
- 9. Buchanan TA, Xiang AH (2005) Gestational diabetes mellitus. J Clin Invest 115: 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, et al. (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, et al. (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885. [DOI] [PubMed] [Google Scholar]
- 12. Buchanan TA (2001) Pancreatic B-cell defects in gestational diabetes: implications for the pathogenesis and prevention of type 2 diabetes. J Clin Endocrinol Metab 86: 989–993. [DOI] [PubMed] [Google Scholar]
- 13. Kautzky-Willer A, Prager R, Waldhausl W, Pacini G, Thomaseth K, et al. (1997) Pronounced insulin resistance and inadequate beta-cell secretion characterize lean gestational diabetes during and after pregnancy. Diabetes Care 20: 1717–1723. [DOI] [PubMed] [Google Scholar]
- 14. Lauenborg J, Grarup N, Damm P, Borch-Johnsen K, Jørgensen T, et al. (2009) Common Type 2 Diabetes Risk Gene Variants Associate with Gestational Diabetes. J Clin Endocrinol Metab 94 (1) 145–150. [DOI] [PubMed] [Google Scholar]
- 15. Groenewoud MJ, Dekker JM, Fritsche A, Reiling E, Nijpels G, et al. (2008) Variants of cdkal1 and igf2b 2 affect first-phase insulin secretion during hyperglycaemic clamps. Diabetologia 51: 1659–1563. [DOI] [PubMed] [Google Scholar]
- 16. Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, et al. (2007) A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 39: 770–775. [DOI] [PubMed] [Google Scholar]
- 17. Pascoe L, Frayling TM, Weedon MN, Mari A, Tura A, et al. (2008) Beta Cell glucose sensitivity is decreased by 39% in non-diabetic individuals carrying multiple diabetes-risk alleles compared with those with no risk alleles. Diabetologia 51: 1989–1992. [DOI] [PubMed] [Google Scholar]
- 18. Grarup N, Rose CS, Andersson EA, Andersen G, Nielsen AL, et al. (2007) Studies of association of variants near the HHEX, CDKN2A/B and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects validation and extension of genome-wide association studies. Diabetes 56: 3105–3111. [DOI] [PubMed] [Google Scholar]
- 19. Tam CHT, Ho JSK, Wang Y, Lee HM, Lam VKL, et al. (2010) Common Polymorphisms in MTNR1B, G6PC2 and GCK Are Associated with Increased Fasting Plasma Glucose and Impaired Beta-Cell Function in Chinese Subjects. Plos One 5: e11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lango H, Palmer CN, Morris AD, Zeggini E, Hattersley AT, et al. (2008) Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes 57: 3129–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, et al. (2007) Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 117: 2155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei FY, Nagashima K, Ohshima T, Saheki Y, Lu YF, et al. (2005) Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat Med 11: 1104–1108. [DOI] [PubMed] [Google Scholar]
- 23. Ubeda M, Rukstalis JM, Habener JF (2006) Inhibition of cyclindependent kinase 5 activity protects pancreatic beta cells from glucotoxicity. J Biol Chem 281: 28858–28864. [DOI] [PubMed] [Google Scholar]
- 24. Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10: 101–129. [Google Scholar]
- 25. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 26. Woolf B (1955) On estimating the relation between blood group and disease. Ann Hum Genet 19: 251–253. [DOI] [PubMed] [Google Scholar]
- 27. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwak SH, Kim SH, Cho YM, Go MJ, Cho YS, et al. (2012) A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 61: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim JY, Cheong HS, Park BL, Baik SH, Park S, et al. (2011) Melatonin receptor 1 B polymorphisms associated with the risk of gestational diabetes mellitus. BMC Med Genet 12: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heude B, Pelloux V, Forhan A, Bedel JF, Lacorte JM, et al. (2011) Association of the Pro12Ala and C1431T variants of PPARgamma and their haplotypes with susceptibility to gestational diabetes. J Clin Endocrinol Metab 96: E1656–1660. [DOI] [PubMed] [Google Scholar]
- 31. Deng Z, Shu QQ, Chen YH, Xiang MH, Li X, et al. (2011) [Association of genetic variant rs10830963 of melatonin receptor 1B gene in women with gestational diabetes mellitus]. Zhonghua Wei Chan Yi Xue Za Zhi 14: 666–669. [Google Scholar]
- 32. Wang Y, Nie M, Li W, Ping F, Hu Y, et al. (2011) Association of six single nucleotide polymorphisms with gestational diabetes mellitus in a Chinese population. PLoS One 6: e26953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pappa KI, Gazouli M, Economou K, Daskalakis G, Anastasiou E, et al. (2011) Gestational diabetes mellitus shares polymorphisms of genes associated with insulin resistance and type 2 diabetes in the Greek population. Gynecol Endocrinol 27: 267–272. [DOI] [PubMed] [Google Scholar]
- 34. Papadopoulou A, Lynch KF, Shaat N, Hakansson R, Ivarsson SA, et al. (2011) Gestational diabetes mellitus is associated with TCF7L2 gene polymorphisms independent of HLA-DQB1*0602 genotypes and islet cell autoantibodies. Diabet Med 28: 1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shin HD, Park BL, Shin HJ, Kim JY, Park S, et al. (2010) Association of KCNQ1 polymorphisms with the gestational diabetes mellitus in Korean women. J Clin Endocrinol Metab 95: 445–449. [DOI] [PubMed] [Google Scholar]
- 36. Cheng Y, Ma Y, Peng T, Wang J, Lin R, et al. (2010) [Genotype discrepancy between maternal and fetal Pro12Ala polymorphism of PPARG2 gene and its association with gestational diabetes mellitus]. Zhonghua Fu Chan Ke Za Zhi 45: 170–173. [PubMed] [Google Scholar]
- 37. Kwak SH, Kim TH, Cho YM, Choi SH, Jang HC, et al. (2010) Polymorphisms in KCNQ1 are associated with gestational diabetes in a Korean population. Horm Res Paediatr 74: 333–338. [DOI] [PubMed] [Google Scholar]
- 38. Santos IC, Frigeri HR, Rea RR, Almeida AC, Souza EM, et al. (2010) The glucokinase gene promoter polymorphism −30G>A (rs1799884) is associated with fasting glucose in healthy pregnant women but not with gestational diabetes. Clin Chim Acta 411: 892–893. [DOI] [PubMed] [Google Scholar]
- 39. Zhou Q, Zhang K, Li W, Liu JT, Hong J, et al. (2009) Association of KCNQ1 gene polymorphism with gestational diabetes mellitus in a Chinese population. Diabetologia 52: 2466–2468. [DOI] [PubMed] [Google Scholar]
- 40. Cho YM, Kim TH, Lim S, Choi SH, Shin HD, et al. (2009) Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia 52: 253–261. [DOI] [PubMed] [Google Scholar]
- 41. Zhu Y, Wu Y (2009) [Relationship between Prol2Ala polymorphism in peroxisome proliferators-activated receptor gamma 2 gene and gestational diabetes mellitus]. Shi Yong Yi Xue Za Zhi 25: 1963–1965. [Google Scholar]
- 42. Shaat N, Lernmark A, Karlsson E, Ivarsson S, Parikh H, et al. (2007) A variant in the transcription factor 7-like 2 (TCF7L2) gene is associated with an increased risk of gestational diabetes mellitus. Diabetologia 50: 972–979. [DOI] [PubMed] [Google Scholar]
- 43. Tok EC, Ertunc D, Bilgin O, Erdal EM, Kaplanoglu M, et al. (2006) PPAR-gamma2 Pro12Ala polymorphism is associated with weight gain in women with gestational diabetes mellitus. Eur J Obstet Gynecol Reprod Biol 129: 25–30. [DOI] [PubMed] [Google Scholar]
- 44. Shaat N, Karlsson E, Lernmark A, Ivarsson S, Lynch K, et al. (2006) Common variants in MODY genes increase the risk of gestational diabetes mellitus. Diabetologia 49: 1545–1551. [DOI] [PubMed] [Google Scholar]
- 45. Shaat N, Ekelund M, Lernmark A, Ivarsson S, Almgren P, et al. (2005) Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia 48: 2544–2551. [DOI] [PubMed] [Google Scholar]
- 46. Shaat N, Ekelund M, Lernmark A, Ivarsson S, Nilsson A, et al. (2004) Genotypic and phenotypic differences between Arabian and Scandinavian women with gestational diabetes mellitus. Diabetologia 47: 878–884. [DOI] [PubMed] [Google Scholar]
- 47. Chiu KC, Go RC, Aoki M, Riggs AC, Tanizawa Y, et al. (1994) Glucokinase gene in gestational diabetes mellitus: population association study and molecular scanning. Diabetologia 37: 104–110. [DOI] [PubMed] [Google Scholar]
- 48. Kim H, Toyofuku Y, Lynn FC, Chak E, Uchida T, et al. (2010) Serotonin regulates pancreatic beta cell mass during pregnancy. Nat Med 16: 804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Di Cianni G, Miccoli R, Volpe L, Lencioni C, Del Prato S (2003) Intermediate metabolism in normal pregnancy and in gestational diabetes. Diabetes Metab Res Rev 19: 259–270. [DOI] [PubMed] [Google Scholar]
- 50. Karnik SK, Chen H, McLean GW, Heit JJ, Gu X, et al. (2007) Menin controls growth of pancreatic beta-cells in pregnant mice and promotes gestational diabetes mellitus. Science 318: 806–809. [DOI] [PubMed] [Google Scholar]
- 51. Kim C, Newton KM, Knopp RH (2002) Gestational diabetes and the incidence of type 2 diabetes: a systematic review. Diabetes Care 25: 1862–1868. [DOI] [PubMed] [Google Scholar]
- 52. Lee H, Jang HC, Park HK, Metzger BE, Cho NH (2008) Prevalence of type 2 diabetes among women with a previous history of gestational diabetes mellitus. Diabetes Res Clin Pract 81: 124–129. [DOI] [PubMed] [Google Scholar]
- 53. Metzger BE, Cho NH, Roston SM, Radvany R (1993) Prepregnancy weight and antepartum insulin secretion predict glucose tolerance five years after gestational diabetes mellitus. Diabetes Care 16: 1598–1605. [DOI] [PubMed] [Google Scholar]
- 54. McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369. [DOI] [PubMed] [Google Scholar]
- 55. Talmud PJ, Hingorani AD, Cooper JA, Marmot MG, Brunner EJ, et al. (2010) Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ 340: b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wald NJ, Hackshaw AK, Frost CD (1999) When can a risk factor be used as a worthwhile screening test? BMJ 319: 1562–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kraft P, Hunter DJ (2009) Genetic risk prediction – are we there yet? N Engl J Med 360: 1701–1703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Meta-analysis of the association between IGF2BP2 rs4402960 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between MTNR1B rs10830963 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between CDKAL1 rs7754840 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between KCNJ11 rs5219 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between KCNQ1 rs2237892 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between KCNQ1 rs2237895 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between PPARG rs1801282 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between TCF7L2 rs7903146 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Meta-analysis of the association between GCK rs4607517 polymorphism and the risk for gestational diabetes mellitus.
(TIF)
Begg's funnel plot of PPARG rs1801282 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.15).
(TIFF)
Begg's funnel plot of TCF7L2 rs7903146 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.18).
(TIFF)
Begg's funnel plot of MTNR1B rs10830963 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.69).
(TIFF)
Begg's funnel plot of IGF2BP2 rs4402960 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.70).
(TIFF)
Begg's funnel plot of KCNJ11 rs5219 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.76).
(TIFF)
Begg's funnel plot of CDKAL1 rs7754840 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.25).
(TIFF)
Begg's funnel plot of KCNQ1 rs2237892 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.34).
(TIFF)
Begg's funnel plot of KCNQ1 rs2237895 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.77).
(TIFF)
Begg's funnel plot of GCK rs4607517 polymorphism and gestational diabetes mellitus risk (Egger test, P = 0.65).
(TIFF)
PRISMA 2009.
(DOC)