Abstract
Background
Previous studies on the association of p53 codon 72 (Arg72Pro) polymorphism with hematological malignancies risk have produced conflicting results. The purpose of this meta-analysis is to define the effect of p53 Arg72Pro polymorphism on hematological malignancies risk.
Methodology/Principal Findings
Through searching PubMed databases (or hand searching) up to April 2012 using the following MeSH terms and keywords: “p53”, “codon 72” “polymorphism” and “leukemia”, or “lymphoma”, or “myeloma”, thirteen were identified as eligible articles in this meta-analysis for p53 Arg72Pro polymorphism (2,731 cases and 7, 356 controls), including nine studies on leukemia (1,266 cases and 4, 474 controls), three studies on lymphoma (1,359 cases and 2,652 controls), and one study on myeloma. The overall results suggested that p53 Arg72Pro polymorphism was not associated with hematological malignancies risk. In stratified analyses, significantly increased non-Hodgkin lymphomas risk was found in p53 Arg72Pro polymorphism heterozygote model (Arg/Pro vs. Arg/Arg: OR = 1.18, 95%CI: 1.02–1.35) and dominant model (Arg/Pro+Pro/Pro vs. Arg/Arg: OR = 1.18, 95%CI: 1.03–1.34), but no significant association was found between leukemia risk and p53 Arg72Pro polymorphism. Further studies showed no association between leukemia risk and p53 Arg72Pro polymorphism when stratified in subtypes of leukemias, ethnicities and sources of controls.
Conclusions/Significance
This meta-analysis indicates that the p53 Arg72Pro polymorphism may contribute to susceptibility to non-Hodgkin lymphomas.
Introduction
Hematological malignancies derived from either of the two major blood cell lineages: myeloid and lymphoid cell lines, include leukemias, lymphomas, myeloma, myelodysplastic syndromes and myeloproliferative diseases. Lymphomas, lymphocytic leukemias, and myelomas are from the lymphoid line, while acute and chronic myelogenous leukemia, myelodyplastic syndromes and myeloproliferative diseases are myeloid in origin. Generally, the overall incidence of hematological malignancies appears to be rising in Western countries but it is very difficult to describe on their epidemiological behavior in a consistent way [1]. In the USA, the number of estimated new cases of hematological malignancies in 2011 was 140,310 and it was predicted to have 53,010 deaths due to hematological malignancies [2]. Hematological malignancies are very heterogeneous diseases with respect to clinical features and acquired genetic alterations. The etiology of hematological malignancies appears to be multifactorial, including the inherited mutations in DNA, and exposure to ionizing radiation, or to chemicals like benzene or cytotoxic therapy. Exposure to these carcinogens may cause DNA damage at the level of hematopoietic progenitors and develop hematological malignancies; however, the majority of cases likely involve genetic variations with a high-risk phenotype [3]. These gene-gene interactions, as well as their interplay with lifestyle-related factors and environmental agents, may be major determinants in hematological malignancy susceptibility [4].
The tumor suppressor p53 plays a pivotal role in response to genotoxic insults from endogenous or environmental agents by orchestrating a diversity of pathways from activation of cell signaling transduction, transcriptional responses, DNA repair to regulation of cell cycle progression and apoptosis [5]. Although p53 mutations are commonly found in different cancers and thought to be associated with carcinogenesis [6], [7], [8], polymorphisms in p53 seem to have a modest effect on cell phenotype, leading to different patterns of cancer susceptibility [9], [10]. The p53 gene locates on chromosome 17p13 and contains 11 exons. The common p53 polymorphisms include p53 codon 72 (c.215C>G; p.R72P; rs1042522), deletion of 16 bp in intron 3 (c.96+41_96+56del16; rs17878362) and IVS6+62A>G (c.672+62A>G; rs1625895) polymorphisms [10]. Among them, p53 codon 72 (Arg72Pro) polymorphism is most widely studied in different cancers [11].
The p53 codon 72 polymorphism is located in exon 4 with CGC to CCC transition, leading to an arginine-to-proline amino-acid substitution in amino-acid position 72 [10]. Laboratory studies have demonstrated the Arg variant is more potent in apoptosis induction whereas the Pro variant is better in inducing cell cycle arrest and DNA damage repair [12], [13], [14]. Recently, many of epidemiological studies have examined the association between p53 Arg72Pro polymorphism and hematological malignancies risk, however, these studies revealed an inconsistent conclusion, probably due to the relatively small size [11], [15], [16], [17]. Therefore, a meta-analysis was performed from all eligible studies to evaluate the association between p53 Arg72Pro polymorphism and hematological malignancies risk in this study.
Materials and Methods
Identification and Eligibility of Relevant Studies
To identify all articles that examined the association of p53 codon 72 polymorphism with hematological malignancies, we conducted a literature search in the PubMed databases up to April 2012 using the following MeSH terms and keywords: “p53”, “codon 72” “polymorphism” and “leukemia”, or “lymphoma”, or “myeloma”. Additional studies were identified by a hand search from references of original studies or review articles on this topic. Eligible studies included in this meta-analysis had to meet the following criteria: (a) an unrelated case-control study, if studies had partly overlapped subjects, only the one with a larger sample size was selected, (b) available genotype frequency, (c) sufficient published data for estimating an odds ratio (OR) with 95% confidence interval (CI) and (d) the genotype frequencies in the control group were consistent with Hardy-Weinberg equilibrium (HWE).
Data Extraction
Two investigators independently extracted data and reached a consensus on all of the items. The following information was extracted from each study: first author, year of publication, country of origin, ethnicity, number of cases and controls, genotype frequency for cases and controls, characteristics for cases, sources of DNA and genotyping methods. Different ethnicity descents were categorized as Asian and Caucasian.
Statistical Analysis
Hardy-Weinberg equilibrium (HWE) was tested by the chi-square test. Crude ORs with 95% CIs were used to assess the strength of association between the p53 Arg72Pro polymorphism and hematological malignancy risk. We first estimated the risks of the Arp/Pro and Pro/Pro genotypes on hematological malignancies, compared with the reference Arg/Arg homozygote, and then evaluated the risks of (Arp/Pro+Pro/Pro vs. Arg/Arg) and (Pro/Pro vs. Arg/Arg + Arp/Pro) on hematological malignancies, assuming dominant and recessive effects of the variant Pro/Pro allele, respectively [11], [18], [19].
Stratified analyses were also performed by types of hematological malignancies, ethnicities and sources of controls. Potential heterogeneity was checked by the χ2-based Q-test. The summary OR estimate of each study was calculated by the random-effects model (the DerSimonian and Laird method).
Publication bias was investigated by funnel plot, and an asymmetric plot suggested possible publication bias. The funnel plot asymmetry was assessed by Egger’s linear regression test. The t test was performed to determine the significance of the asymmetry, and a P value of <0.05 was considered a significant publication bias. All analyses were done with Stata software (version 11.0; StataCorp LP, College Station, TX), using two-sided P values.
Results
Characteristics of Studies
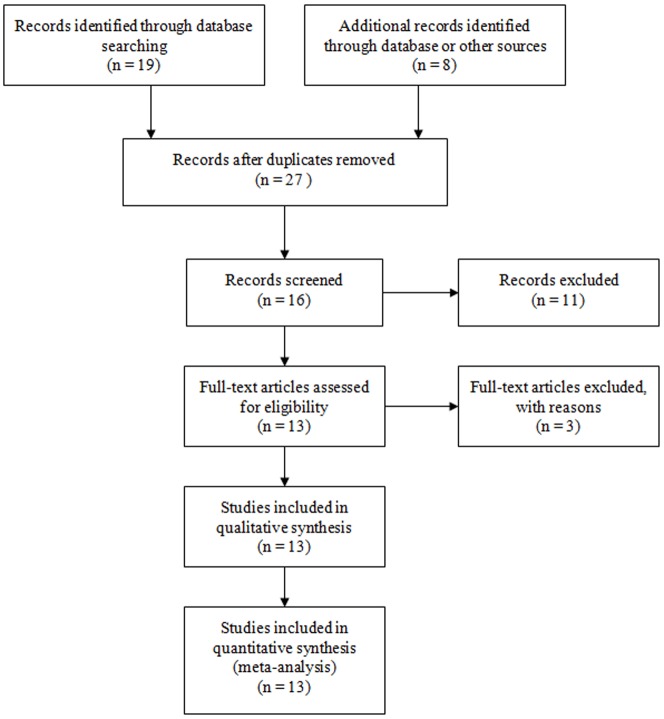
Nineteen abstracts were retrieved through the search “p53”, “codon 72”, “polymorphism” and “leukemia”, and eight studies were identified as eligible studies. Out of the nineteen, seven studies were excluded given that they have not included controls, did not report genotype frequency for controls in their study designs, or reported other diseases [20], [21], [22], [23], [24], [25], [26], one article was review [27], and three studies were in vitro cell biology studies [28], [29], [30]. We also included eligible study with hand searching [31]. By searching “p53”, “polymorphism” and “lymphoma” or “myeloma”, and a hand search from references of original studies or review articles, we included another seven articles [16], [17], [32], [33], [34], [35], [36]. The genotype distributions among the controls of all studies were in agreement with Hardy-Weinberg equilibrium except for three studies [16], [33], [36](Figure 1). As a result, a total of thirteen studies met the inclusion criteria and were identified as eligible articles with 2,711 cases and 7,356 controls, including nine studies of leukemia [15], [31], [37], [38], [39], [40], [41], [42], [43], three studies of lymphoma [17], [32], [34] and one study of myeloma [35]. The selected study characteristics were listed in Table 1. The patients’ demographic characteristics and p53 genotype distribution were listed in Table S1 and S2.
Figure 1. Flow diagram of studies identification.
Table 1. Characteristics of literatures included in the meta-analysis.
| Author | Year | Origin | Ethnicity | Sample size (case/control) | HWE | MAF | Design | Genotype |
| Leukemia | ||||||||
| Nakano Y | 2000 | Japan | Asian | 200/188 | 0.77 | 0.43 | PB | SSCP |
| Bergamaschi G | 2004 | Italy | Caucasian | 96/174 | 0.82 | 0.22 | PB | PCR-RFLP |
| Takeuchi S | 2005 | Japan | Asian | 87/89 | 0.19 | 0.43 | HB | PCR-RFLP |
| Kochethu G | 2006 | UK | Caucasian | 203/97 | 0.50 | 0.34 | PB | PCR-RFLP |
| Phang BH | 2008 | China | Asian | 44/160 | 0.33 | 0.43 | PB | PCR-RFLP |
| Ellis NA | 2008 | USA/UK | Caucasian | 171/3022 | 0.85 | 0.25 | PB | Taqman/PCR-RFLP |
| Xiong X | 2009 | China | Asian | 231/128 | 1.00 | 0.45 | HB | PCR-RFLP |
| Do TN | 2009 | US | Caucasian | 114/414 | 0.90 | 0.25 | PB | Taqman |
| Chauhan PS | 2011 | India | Asian | 120/202 | 0.09 | 0.49 | PB | PCR-RFLP |
| Lymphomas | ||||||||
| Hishida A | 2004 | Japan | Asian | 103/440 | 0.84 | 0.35 | HB | Allele PCR |
| Bittenbring J | 2008 | Germany | Caucasian | 311/512 | 0.81 | 0.25 | HB | Allele PCR |
| Kim HN | 2010 | Korea | Asian | 945/1700 | 0.52 | 0.34 | PB | Taqman |
| Myeloma | ||||||||
| Ortega MM | 2007 | Brazil | Caucasian | 106/230 | 0.09 | 0.37 | HB | PCR-RFLP |
Abbreviations: HB, Hospital based controls; PB, population based controls; PCR, Polymerase chain reaction; PCR-RFLP, Polymerase chain reaction- restriction fragment length polymorphism; SSCP, Single-Strand Conformation Polymorphism; HWE, Hardy-Weinberg equilibrium; MAF, minor allele frequency.
The genotyping for p53 codon 72 polymorphism was performed using polymerase chain reaction (PCR), polymerase chain reaction- restriction fragment length polymorphism (PCR-RFLP), Taqman PCR, or single-strand conformation polymorphism (SSCP) analyses on the genomic DNA from the human blood samples. For ethnic distribution, there were seven studies of Caucasian descent, and six of Asian origin. For the nine studies on leukemia, there were five studies on acute myeloid leukemia (AML) and four studies on others including acute lymphoblastic leukemia (ALL), chronic lymphoblastic leukemia (CLL) and chronic myeloid leukemia (CML). As to ethnic distribution of the leukemia patients, there were five studies of Asians and four studies of Caucasians.
Quantitative Synthesis
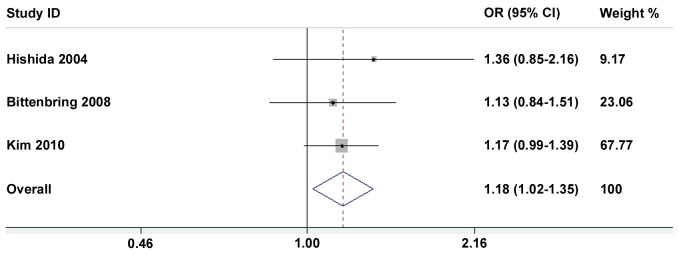
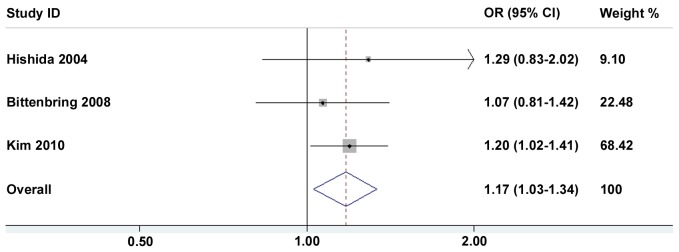
Tables 2 and 3 present in detail the results of the meta-analysis. By pooling all the studies, the p53 Arg72Pro polymorphism was not associated with a hematological malignancies risk, and this negative association maintained in some subgroup analyses such as ethnicities and sources of controls (Table 2). When stratified by hematological malignancies types, no association was found between p53 Arg72Pro polymorphism and leukemia risk (1,266 cases and 4474 controls) in all four models (Table 2). However, p53 Arg72Pro polymorphism heterozygote (Arg72Pro) was significantly correlated increased lymphomas risk (Arg/Pro vs. Arg/Arg: OR = 1.18, 95%CI: 1.02–1.35) (Figure 2), and this association was further confirmed in dominant model (Arg/Pro+Pro/Pro vs. Arg/Arg: OR = 1.18, 95%CI: 1.03–1.34) (Figure 3). Actually, all cases included in these three eligible studies on lymphomas were non-Hodgkin lymphoma patients (NHL) (1,359 cases and 2,652 controls). Thus, our data suggest an association between p53 Arg72Pro polymorphism and NHL risk.
Table 2. Meta-analysis of the p53 codon 72 Arg>Pro polymorphism on hematological malignancy risk.
| Variables | na | Arg/Pro vs. Arg/Arg | Pro/Pro vs. Arg/Arg | Arg/Pro + Pro/Pro vs. Arg/Arg (dominant) | Pro/Pro vs. Arg/Arg + Arg/Pro (recessive) | ||||
| OR (95% CI) | P b | OR (95% CI) | P b | OR (95% CI) | P b | OR (95% CI) | P b | ||
| Total | 13 | 1.08(0.95–1.24) | 0.199 | 1.10(0.83–1.45) | 0.004 | 1.08(0.93–1.26) | 0.033 | 1.05(0.83–1.33) | 0.026 |
| Types | |||||||||
| Leukemia | 9 | 1.03(0.83–1.27) | 0.100 | 1.10(0.71–1.72) | 0.001 | 1.04(0.81–1.34) | 0.010 | 1.07(0.75–1.53) | 0.010 |
| Lymphoma | 3 | 1.18(1.02–1.35) | 0.797 | 1.12(0.80–1.55) | 0.257 | 1.18(1.03–1.34) | 0.719 | 1.02(0.74–1.40) | 0.249 |
| Myeloma | 1 | ||||||||
| Ethnicities | |||||||||
| Asian | 7 | 1.11(0.92–1.34) | 0.235 | 0.99(0.73–1.33) | 0.122 | 1.07(0.86–1.33) | 0.106 | 1.00(0.83–1.21) | 0.389 |
| European | 6 | 1.05(0.85–1.29) | 0.190 | 1.32 (0.74–2.35) | 0.003 | 1.09(0.85–1.40) | 0.037 | 1.31(0.79–2.17) | 0.010 |
| Source of controls | |||||||||
| Hospital based | 5 | 1.00(0.84–1.18) | 0.099 | 0.98(0.73–1.33) | 0.048 | 0.99(0.82–1.19) | 0.027 | 1.00(0.80–1.26) | 0.209 |
| Population based | 8 | 1.02(0.84–1.25) | 0.069 | 1.12(0.73–1.72) | 0.001 | 1.04(0.82–1.31) | 0.006 | 1.11(0.78–1.58) | 0.007 |
Number of comparisons.
P value of Q-test for heterogeneity test.
Table 3. Meta-analysis of the p53 codon 72 Arg>Pro polymorphism on leukemia risk.
| Variables | na | Arg/Pro vs. Arg/Arg | Pro/Pro vs. Arg/Arg | Arg/Pro + Pro/Pro vs. Arg/Arg (dominant) | Pro/Pro vs. Arg/Arg + Arg/Pro (recessive) | ||||
| OR (95% CI) | P b | OR (95% CI) | P b | OR (95% CI) | P b | OR (95% CI) | P b | ||
| Leukemia | 9 | 1.03(0.83–1.27) | 0.100 | 1.10(0.71–1.72) | 0.001 | 1.04(0.81–1.34) | 0.010 | 1.07(0.75–1.53) | 0.010 |
| Ethnicities | |||||||||
| Asian | 5 | 1.01(0.74–1.39) | 0.186 | 0.84(0.57–1.26) | 0.205 | 0.97(0.70–1.34) | 0.119 | 0.85(0.64–1.12) | 0.569 |
| Caucasian | 4 | 1.34(0.73–1.46) | 0.070 | 1.57(0.68–3.61) | 0.003 | 1.12(0.74–1.69) | 0.009 | 1.55(0.77–3.11) | 0.014 |
| Source of controls | |||||||||
| Hospital based | 2 | 1.28(0.85–1.92) | 0.348 | 1.16(0.59–2.28) | 0.185 | 1.22(0.76–1.97) | 0.214 | 1.03(0.67–1.59) | 0.353 |
| Population based | 7 | 0.98(0.76–1.26) | 0.086 | 1.10(0.62–1.94) | 0.001 | 1.00(0.74–1.35) | 0.007 | 1.10(0.69–1.77) | 0.004 |
| Types | |||||||||
| AML | 5 | 1.03(0.78–1.35) | 0.180 | 0.89(0.60–1.30) | 0.197 | 0.99(0.75–1.32) | 0.108 | 0.88(0.67–1.16) | 0.559 |
| Others c | 4 | 1.06(0.67–1.53) | 0.069 | 1.49(0.60–3.71) | 0.002 | 1.09(0.66–1.79) | 0.008 | 1.48(0.69–3.17) | 0.007 |
Number of comparisons.
P value of Q-test for heterogeneity test.
Others include acute lymphoblastic leukemia, chronic lymphocytic Leukemia, chronic myeloid leukemia.
Abbreviations: AML, Acute myeloid leukemia.
Figure 2. Forest plots of heterozygote model (Arg/Pro vs. Arg/Arg) in different subgroups.
The squares and horizontal lines correspond to OR and 95% CI of specific study, and the area of squares reflects study weight (inverse of the variance). The diamond represents the pooled OR and its 95% CI.
Figure 3. Forest plots of dominant model (Arg/Pro + Pro/Pro vs. Arg/Arg) in different subgroups.
The squares and horizontal lines correspond to OR and 95% CI of specific study, and the area of squares reflects study weight (inverse of the variance). The diamond represents the pooled OR and its 95% CI.
We next analyzed the association between p53 Arg72Pro polymorphism and leukemia risk when stratified by the ethnicities, sources of controls, and leukemia types. The results showed that the p53 Arg72Pro polymorphism was not associated with leukemia either in Asians or in Caucasians, and this negative association maintained in other subgroup analyses such as leukemia types and sources of controls (Table 3).
Publication Bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of literatures. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry (Data not shown). Then, the Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still did not show any evidence of publication bias (All P>0.05). The funnel plot can be misleading [44], and Egger’s test may not really show publication bias [45]. To overcome these limitations, we performed the contour-enhanced funnel plot analyses to investigate the potential publication bias [46]. As shown in the Figure S1, no obvious publication bias was observed in the contrasts of Arg/Pro vs. Arg/Arg, Pro/Pro vs. Arg/Arg, dominant and recessive models, respectively.
Discussion
In the present study, we collected all available, published studies and performed a meta-analysis to examine the association between the p53 Arg72Pro polymorphism and susceptibility to hematological malignancies. Thirteen were critically reviewed to clarify controversial results from previous reports. Our meta-analysis showed that significantly increased NHL risks were found in all subjects with p53 Arg72Pro polymorphism heterozygote and dominant model. No significant association was found between p53 Arg72Pro polymorphism and leukemia risk.
Previous meta-analysis showed that p53 Arg72Pro polymorphism was neither associated with hematological malignancies (eight studies), nor associated with leukemia risk (five studies) [11]. When stratified by ethnicities, a protective effect of the p53 codon 72 Pro allele on leukemia was found in Asians even with a small number of studies (331 cases and 437 controls) [11]. With more studies and a larger number of subjects, our meta-analysis study confirmed that p53 Arg72Pro polymorphism was not associated with hematological malignancies or leukemia risk. However, with more than double leukemia cases, we did not found an association between p53 Arg72Pro polymorphism and leukemia risk in Asians (682 cases and 767 controls). We need to point that our meta-analysis study included all the cases from Takeuchi et al. study as leukemia patients, which may introduce bias since there were a few cases of lymphoma patients [42]. However, this did not affect the result when we excluded this study from this meta-analysis (Data not shown). Thus, our study cannot confirm the association between p53 Arg72Pro polymorphism and leukemia risk in Asians and further studies with larger numbers of participants are needed to clarify this association.
On the other hand, the pathogenetic mechanisms of leukemia are different, and stratified analyses are required for different types of Leukemia. Due to the limited number of studies, this meta-analysis only performed subgroup analyses on the association between p53 codon 72 Arg>Pro polymorphism and risk of AML (n = 5), and risk of other types of leukemia (n = 4) (Table 2). Future meta-analysis should analyze the association of genetic variants and different types of leukemia separately by including more emerging studies.
Recently, six studies were conducted to examine the association between p53 Arg72Pro polymorphism and lymphoma risk [16], [17], [32], [33], [34], [36]. In the present meta-analysis, three studies were included [17], [32], [34], and the others were excluded due to the deviation from HWE [16], [33], [36]. Even including these three studies [16], [33], [36], the significant association was still found between p53 Arg72Pro polymorphism and increased risk of all lymphoma (2,845 cases and 4,306 controls) (Arg/Pro vs. Arg/Arg: OR = 1.12, 95%CI: 1.01–1.25), and between p53 Arg72Pro polymorphism and increased risk of NHL (2,547 cases and 4,306 controls) (Arg/Pro vs. Arg/Arg: OR = 1.11, 95%CI: 0.99–1.24) (Data not shown). This was consistent with our data that showed significant association between p53 Arg72Pro polymorphism and increased NHL risk based on limited three studies. However, this meta-analysis has limitation by including indolent and aggressive lymphomas in the same group since pathogenetic mechanisms of different types of lymphomas are different. Therefore, additional well-designed large studies were required to validate the association between p53 Arg72Pro polymorphism and increased risk of lymphomas.
The interaction of different polymorphisms in the same gene, or between different genes, might contribute to hematological malignancies risk. Although the combined effects of different p53 polymorphisms have not been studied, the potential interactions between p53 Arg72Pro polymorphism and other genetic polymorphisms, including those in Murine double minute 2 (MDM2), p73, p21 and Glutathione S-transferase, which are involved in DNA damage repair, apoptosis, cell cycle control, or detoxification of xenobiotic compounds, were found in hematological malignancies [15], [34], [35], [38], [43]. We evaluated the combined effects of these polymorphisms on susceptibility to hematological malignancies, however, due to the limited studies, the data were not sufficient to conduct a meta-analysis.
Heterogeneity for the p53 Arg72Pro polymorphism was observed among these studies. The heterogeneity may be due to various factors, such as diversity in the population characteristics, differences in the number of cases and controls, genotyping methods and study design. Between-study heterogeneity was detected by restricted maximum likelihood-based random-effects meta-regression analysis. Because the number of included studies was limited, we conducted univariate meta-regression model firstly, variables with significant P values ≥0.1 were then entered into the multivariable model. Ethnicity, MAF, source of controls, sample size, publication years and disease types were taken into consideration, and none of these factors showed an evidence of source of heterogeneity (Table S3). To eliminate heterogeneity, we carried out subgroup analysis and used a random-effects model to pool the results whenever significant heterogeneity was present. In addition, some unpublished, eligible publications were not available in the present meta-analysis, which might affect the results.
In conclusion, we found significant associations between the p53 Arg72Propolymorphism and lymphoma (non-Hodgkin lymphoma) risk, but not leukemia risk. However, the number of studies included for our meta-analysis is very limited, and studies based on larger well-designed populations are still needed to clarify the different effects of the p53 Arg72Pro polymorphism in different types of hematological malignancies. Also, studies examining the combined effects of different p53 polymorphisms or different polymorphisms of p53 related genes (e.g., MDM2) should be investigated.
Supporting Information
Contour-enhanced funnel plot for publication bias analysis.
(DOC)
Clinical and demographic characteristics of the patients in each study.
(DOC)
p53 Arg72Pro polymorphism genotype distribution of each study included in the meta-analysis.
(DOC)
Meta-regression analysis.
(DOC)
Acknowledgments
We are grateful to Dr. Klaus Roemer from University of Saarland Medical School, Homburg-Saar, Germany, and Dr. Elena Voropaeva from Novosibirsk State Medical University, Krasniy Prospect, Russia for providing the frequencies of the p53 codon 72 SNP of subjects.
Funding Statement
This study was supported by the grants from Zhejiang Provincial Natural Science Foundation of China (Y12H160154), Health Bureau of Zhejiang Province (2011KYB012), Zhejiang Provincial Administration of Traditional Chinese Medicine (2011ZA010) and the Fundamental Research Funds for the Central Universities (2011QNA7018, 2012QNA7019). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rodriguez-Abreu D, Bordoni A, Zucca E (2007) Epidemiology of hematological malignancies. Ann Oncol 18 Suppl 1i3–i8. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Ward E, Brawley O, Jemal A (2011) Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 61: 212–236. [DOI] [PubMed] [Google Scholar]
- 3. Descatha A, Jenabian A, Conso F, Ameille J (2005) Occupational exposures and haematological malignancies: overview on human recent data. Cancer Causes Control 16: 939–953. [DOI] [PubMed] [Google Scholar]
- 4. Irigaray P, Newby JA, Clapp R, Hardell L, Howard V, et al. (2007) Lifestyle-related factors and environmental agents causing cancer: an overview. Biomed Pharmacother 61: 640–658. [DOI] [PubMed] [Google Scholar]
- 5. Hainaut P, Wiman KG (2009) 30 years and a long way into p53 research. Lancet Oncol 10: 913–919. [DOI] [PubMed] [Google Scholar]
- 6. Goh AM, Coffill CR, Lane DP (2011) The role of mutant p53 in human cancer. J Pathol 223: 116–126. [DOI] [PubMed] [Google Scholar]
- 7. Olivier M, Petitjean A, Marcel V, Petre A, Mounawar M, et al. (2009) Recent advances in p53 research: an interdisciplinary perspective. Cancer Gene Ther 16: 1–12. [DOI] [PubMed] [Google Scholar]
- 8. Oren M, Rotter V (2010) Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2: a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hrstka R, Coates PJ, Vojtesek B (2009) Polymorphisms in p53 and the p53 pathway: roles in cancer susceptibility and response to treatment. J Cell Mol Med 13: 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whibley C, Pharoah PD, Hollstein M (2009) p53 polymorphisms: cancer implications. Nat Rev Cancer 9: 95–107. [DOI] [PubMed] [Google Scholar]
- 11. Francisco G, Menezes PR, Eluf-Neto J, Chammas R (2011) Arg72Pro TP53 polymorphism and cancer susceptibility: a comprehensive meta-analysis of 302 case-control studies. Int J Cancer 129: 920–930. [DOI] [PubMed] [Google Scholar]
- 12. Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M (2003) The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33: 357–365. [DOI] [PubMed] [Google Scholar]
- 13. Pim D, Banks L (2004) p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer 108: 196–199. [DOI] [PubMed] [Google Scholar]
- 14. Siddique M, Sabapathy K (2006) Trp53-dependent DNA-repair is affected by the codon 72 polymorphism. Oncogene 25: 3489–3500. [DOI] [PubMed] [Google Scholar]
- 15. Chauhan PS, Ihsan R, Yadav DS, Mishra AK, Bhushan B, et al. (2011) Association of glutathione S-transferase, EPHX, and p53 codon 72 gene polymorphisms with adult acute myeloid leukemia. DNA Cell Biol 30: 39–46. [DOI] [PubMed] [Google Scholar]
- 16. Havranek O, Spacek M, Hubacek P, Mocikova H, Benesova K, et al. (2011) No association between the TP53 codon 72 polymorphism and risk or prognosis of Hodgkin and non-Hodgkin lymphoma. Leuk Res 35: 1117–1119. [DOI] [PubMed] [Google Scholar]
- 17. Kim HN, Yu L, Kim NY, Lee IK, Kim YK, et al. (2010) Association with TP53 codon 72 polymorphism and the risk of non-Hodgkin lymphoma. Am J Hematol 85: 822–824. [DOI] [PubMed] [Google Scholar]
- 18. He XF, Su J, Zhang Y, Huang X, Liu Y, et al. (2011) Association between the p53 polymorphisms and breast cancer risk: meta-analysis based on case-control study. Breast Cancer Res Treat 130: 517–529. [DOI] [PubMed] [Google Scholar]
- 19. Zhang Z, Wang M, Wu D, Tong N, Tian Y (2010) P53 codon 72 polymorphism contributes to breast cancer risk: a meta-analysis based on 39 case-control studies. Breast Cancer Res Treat 120: 509–517. [DOI] [PubMed] [Google Scholar]
- 20. Feng Z, Zhang C, Kang HJ, Sun Y, Wang H, et al. (2011) Regulation of female reproduction by p53 and its family members. FASEB J 25: 2245–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang HJ, Feng Z, Sun Y, Atwal G, Murphy ME, et al. (2009) Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A 106: 9761–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Majid A, Richards T, Dusanjh P, Kennedy DB, Miall F, et al. (2011) TP53 codon 72 polymorphism in patients with chronic lymphocytic leukaemia: identification of a subgroup with mutated IGHV genes and poor clinical outcome. Br J Haematol 153: 533–535. [DOI] [PubMed] [Google Scholar]
- 23. Phang BH, Chua HW, Li H, Linn YC, Sabapathy K (2011) Characterization of novel and uncharacterized p53 SNPs in the Chinese population–intron 2 SNP co-segregates with the common codon 72 polymorphism. PLoS One 6: e15320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi H, Tan SJ, Zhong H, Hu W, Levine A, et al. (2009) Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am J Hum Genet 84: 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sturm I, Bosanquet AG, Hummel M, Dorken B, Daniel PT (2005) In B-CLL, the codon 72 polymorphic variants of p53 are not related to drug resistance and disease prognosis. BMC Cancer 5: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamaguchi H, Inokuchi K, Sakuma Y, Dan K (2001) Mutation of the p51/p63 gene is associated with blastic crisis in chronic myelogenous leukemia. Leukemia 15: 1729–1734. [DOI] [PubMed] [Google Scholar]
- 27. Hu W, Feng Z, Atwal GS, Levine AJ (2008) p53: a new player in reproduction. Cell Cycle 7: 848–852. [DOI] [PubMed] [Google Scholar]
- 28. Ito A, Morita A, Ohya S, Yamamoto S, Enomoto A, et al. (2011) Cycloheximide suppresses radiation-induced apoptosis in MOLT-4 cells with Arg72 variant of p53 through translational inhibition of p53 accumulation. J Radiat Res (Tokyo) 52: 342–350. [DOI] [PubMed] [Google Scholar]
- 29. Miwa H, Kita K, Saya H, Otsuji A, Masuya M, et al. (1992) Structural alterations of the p53 gene in human leukemias. Leuk Res 16: 1105–1112. [DOI] [PubMed] [Google Scholar]
- 30. Zhang W, Hu G, Deisseroth A (1992) Polymorphism at codon 72 of the p53 gene in human acute myelogenous leukemia. Gene 117: 271–275. [DOI] [PubMed] [Google Scholar]
- 31. Do TN, Ucisik-Akkaya E, Davis CF, Morrison BA, Dorak MT (2009) TP53 R72P and MDM2 SNP309 polymorphisms in modification of childhood acute lymphoblastic leukemia susceptibility. Cancer Genet Cytogenet 195: 31–36. [DOI] [PubMed] [Google Scholar]
- 32. Bittenbring J, Parisot F, Wabo A, Mueller M, Kerschenmeyer L, et al. (2008) MDM2 gene SNP309 T/G and p53 gene SNP72 G/C do not influence diffuse large B-cell non-Hodgkin lymphoma onset or survival in central European Caucasians. BMC Cancer 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hill DA, Wang SS, Cerhan JR, Davis S, Cozen W, et al. (2006) Risk of non-Hodgkin lymphoma (NHL) in relation to germline variation in DNA repair and related genes. Blood 108: 3161–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hishida A, Matsuo K, Tajima K, Ogura M, Kagami Y, et al. (2004) Polymorphisms of p53 Arg72Pro, p73 G4C14-to-A4T14 at exon 2 and p21 Ser31Arg and the risk of non-Hodgkin’s lymphoma in Japanese. Leuk Lymphoma 45: 957–964. [DOI] [PubMed] [Google Scholar]
- 35. Ortega MM, Honma HN, Zambon L, Lorand-Metze I, Costa FF, et al. (2007) GSTM1 and codon 72 P53 polymorphism in multiple myeloma. Ann Hematol 86: 815–819. [DOI] [PubMed] [Google Scholar]
- 36.Pospelova TI, Voropaeva EN, Voevoda MI, Berezina1 OV (2010) p53 polymorphisms as a potential marker for predisposition for non-Hodgkin malignant lymphomas. Haematology and Transfusiology 1.
- 37. Bergamaschi G, Merante S, Orlandi E, Galli A, Bernasconi P, et al. (2004) TP53 codon 72 polymorphism in patients with chronic myeloid leukemia. Haematologica 89: 868–869. [PubMed] [Google Scholar]
- 38. Ellis NA, Huo D, Yildiz O, Worrillow LJ, Banerjee M, et al. (2008) MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood 112: 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kochethu G, Delgado J, Pepper C, Starczynski J, Hooper L, et al. (2006) Two germ line polymorphisms of the tumour suppressor gene p53 may influence the biology of chronic lymphocytic leukaemia. Leuk Res 30: 1113–1118. [DOI] [PubMed] [Google Scholar]
- 40. Nakano Y, Naoe T, Kiyoi H, Kunishima S, Minami S, et al. (2000) Poor clinical significance of p53 gene polymorphism in acute myeloid leukemia. Leuk Res 24: 349–352. [DOI] [PubMed] [Google Scholar]
- 41. Phang BH, Linn YC, Li H, Sabapathy K (2008) MDM2 SNP309 G allele decreases risk but does not affect onset age or survival of Chinese leukaemia patients. Eur J Cancer 44: 760–766. [DOI] [PubMed] [Google Scholar]
- 42. Takeuchi S, Matsushita M, Tsukasaki K, Takeuchi N, Tomonaga M, et al. (2005) P53 codon 72 polymorphism is associated with disease progression in adult T-cell leukaemia/lymphoma. Br J Haematol 131: 552–553. [DOI] [PubMed] [Google Scholar]
- 43. Xiong X, Wang M, Wang L, Liu J, Zhao X, et al. (2009) Risk of MDM2 SNP309 alone or in combination with the p53 codon 72 polymorphism in acute myeloid leukemia. Leuk Res 33: 1454–1458. [DOI] [PubMed] [Google Scholar]
- 44. Lau J, Ioannidis JP, Terrin N, Schmid CH, Olkin I (2006) The case of the misleading funnel plot. BMJ 333: 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zintzaras E, Lau J (2008) Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 61: 634–645. [DOI] [PubMed] [Google Scholar]
- 46. Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L (2008) Contour-enhanced meta-analysis funnel plots help distinguish publication bias from other causes of asymmetry. J Clin Epidemiol 61: 991–996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contour-enhanced funnel plot for publication bias analysis.
(DOC)
Clinical and demographic characteristics of the patients in each study.
(DOC)
p53 Arg72Pro polymorphism genotype distribution of each study included in the meta-analysis.
(DOC)
Meta-regression analysis.
(DOC)