The phosphoglycolipid antibiotic moenomycin directly blocks bacterial cell wall biosynthesis by inhibiting peptidoglycan glycosytransferases.[1] The enzyme MoeO5, encoded by the moe gene cluster 1 in Streptomyces ghanaensis, catalyzes the initial step of moenomycin production in which the C15-hydrocarbon moiety of farnesyl pyrophosphate (FPP) is transferred to the 2-hydroxyl group of 3-phosphoglycerate (3PG), forming an ether bond (Figure 1).[2] The reaction is similar to that of geranylgeranylglyceryl phosphate synthase (GGGPS) for synthesizing Archaea-type phospholipids.[3] However, unlike GGGPS whose product retains the all-trans configuration of the isoprenyl chain, the farnesyl group transferred by MoeO5 undergoes a trans-to-cis isomerization at the C2-C3 double bond.[4] The crystal structures of GGGPS and the bacterial homologue PcrB reveal a triose phosphate isomerase (TIM)-barrel fold, which had not been observed previously in prenyltransferases (PTs).[3, 5] The sequences of GGGPS and PcrB share 35% amino acid identity, but MoeO5 shows only 10% identity to both enzymes (Figure S1). Here we report the X-ray crystallographic structures of MoeO5 bound to the product 2-Z,E-farnesyl-3-phosphoglycerate (FPG), a substrate analogue farnesyl thiopyrophosphate (FsPP), magnesium ion (Mg2+), and pyrophosphate ion (PPi) that, together with additional biochemical and bioinformatics results, shed light on the possible mechanisms of action of this unusual enzyme.
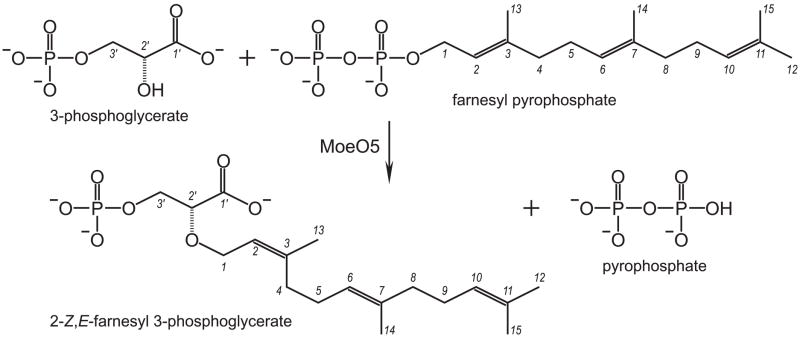
Figure 1.
The first step in moenomycin biosynthesis. The reaction is catalyzed by MoeO5, which transfers the C15 farnesyl group from FPP to 3PG. The carbon atoms are numbered 1 – 15 in FPP and 1′ – 3′ in 3PG. Importantly, the original all-trans configuration is converted to 2-cis,6-trans (Z,E) upon the farnesyl transfer, forming FPG and PPi.
Molecular-replacement (MR) approaches to determine the MoeO5 structure by using GGGPS and PcrB as search models were not successful, reflecting perhaps the significant variations in the protein sequences. Because MoeO5 contains no Cys residue, to solve the structure by using multiple isomorphous replacement (MIR), we produced the mutant H97C for efficient preparation of mercury-based MIR derivatives (Table S1). The other structures were solved by MR (Figure S2 and Table 1). See the Supporting Information (SI) for details. MoeO5 crystallizes as a homodimer (Figure S3 and Table S2). The dimer interface buries 1200 Å2, or more than 10% surface area, on each monomer and mainly involves hydrophobic residues in helices α4 and α5. The cis-peptide of Pro141 binds to a Mg2+ ion at the molecular dyad (Figure S3). These helices also mediate dimerization in GGGPS and PcrB, but here one of the TIM barrels is rotated by 180° (Figure S4). More description of the protein structure as well as the bound ligands can be found in the SI. Despite its Cα deviation of about 2.0 Å from the GGGPS and PcrB monomer (Figure S5), the similar protein fold with a connecting loop (denoted λ3) between strands β3 and β4 clearly places MoeO5 among this new class of TIM-barrel PTs.
Table 1.
Data collection and refinement statistics for the MoeO5 crystals. All positive reflections were used in the refinement. Values in parentheses are for the outermost resolution shells.
| Native | FsPP (3 hours) | FsPP (overnight) | PPi | 3PG | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P21 | P1 | P1 | P1 | P1 |
| Unit-cell | |||||
| a (Å) | 59.0 | 46.6 | 46.7 | 46.7 | 46.8 |
| b (Å) | 84.5 | 58.9 | 58.6 | 59.7 | 58.7 |
| c (Å) | 59.6 | 58.8 | 58.9 | 59.1 | 59.2 |
| α (°) | 90.0 | 97.6 | 97.7 | 66.7 | 97.8 |
| β(°) | 112.3 | 108.4 | 112.2 | 71.1 | 112.6 |
| γ(°) | 90.0 | 112.5 | 108.6 | 66.0 | 108.2 |
| Resolution (Å) | 25 – 1.39 (1.44 – 1.39) | 25 – 1.57 (1.63 – 1.57) | 25 – 1.8 (1.86 – 1.80) | 25 – 1.66 (1.72 – 1.66) | 25 – 1.66 (1.72 – 1.66) |
| Unique reflections | 105467 (10291) | 70651 (6964) | 46862 (4623) | 59540 (5880) | 59984 (5912) |
| Redundancy | 5.1 (5.0) | 4.0 (4.0) | 4.0 (3.9) | 4.0 (4.0) | 4.0 (4.0) |
| Completeness (%) | 97.5 (95.5) | 96.8 (95.1) | 96.6 (95.2) | 96.1 (94.8) | 96.2 (95.0) |
| Average I/s(I) | 24.8 (3.4) | 42.4 (6.4) | 37.3 (9.0) | 37.1 (9.4) | 39.9 (11.4) |
| Rmerge (%) | 6.9 (55.6) | 3.3 (26.6) | 3.2 (9.9) | 3.1 (9.8) | 3.5 (8.6) |
| Refinement | |||||
| No. of reflections | 99738 (8573) | 69049 (6216) | 46545 (4495) | 59066 (5627) | 59609 (5666) |
| Rwork (95% of data) | 0.168 (0.222) | 0.163 (0.203) | 0.161 (0.179) | 0.161 (0.184) | 0.158 (0.179) |
| Rfree (5% of data) | 0.189 (0.250) | 0.191 (0.243) | 0.192 (0.209) | 0.186 (0.227) | 0.187 (0.220) |
| R.m.s.d. bonds (Å) | 0.020 | 0.019 | 0.020 | 0.020 | 0.020 |
| R.m.s.d. angles (°) | 1.9 | 1.9 | 2.0 | 1.9 | 2.0 |
| Dihedral angles | |||||
| Most favored (%) | 96.2 | 95.8 | 95.8 | 96.2 | 96.0 |
| Allowed (%) | 2.6 | 3.0 | 3.2 | 2.8 | 2.8 |
| Disallowed (%) | 1.2 | 1.2 | 1.0 | 1.0 | 1.2 |
| No. of non-H atoms | |||||
| Protein | 3927 | 3927 | 3943 | 3932 | 3927 |
| Water | 590 | 467 | 405 | 432 | 470 |
| Ligand | 53 | 53 | 72 | 62 | 59 |
| Average B (Å2) | |||||
| Protein | 13.1 | 18.6 | 18.0 | 13.9 | 14.1 |
| Water | 26.9 | 31.6 | 28.6 | 26.1 | 26.4 |
| Ligand | 12.6 | 21.2 | 25.3 | 17.6 | 15.8 |
| PDB ID code | 3VK5 | 3VKA | 3VKB | 3VKC | 3VKD |
The wild-type MoeO5 co-crystallized with a bound product FPG, of which the C15 tail moiety makes a U-turn mainly at the C10-C11 bond (Figure 2), rather than extending straight into a nonpolar groove along helix α4 as supposed in GGGPS and PcrB.[3,5] The equivalent groove in MoeO5 is obstructed by the λ3 loop (Figure S6), which contains His97 and is four-residues longer than in GGGPS and PcrB (Figure S1).[4] This loop may act as a swinging door for binding and enclosure of the prenyl substrate in GGGPS,[3] but the door is virtually locked up in MoeO5 and the resulting small volume seems barely able to accommodate a C10 group. However, substitution of a Tyr residue of GGGPS and PcrB by Ala157 in strand β5 makes a “nook” for the bent C15 group in MoeO5 (Figure S6). Neither the C10 GPP nor the C20 GGPP is a substrate of MoeO5, and soaking with the thio-analogue GGsPP has no effect on the bound FPG. Clearly the cavity is specific for FPP, and the bent conformation is likely to be important in precisely positioning the C15 group for the trans-to-cis conversion.
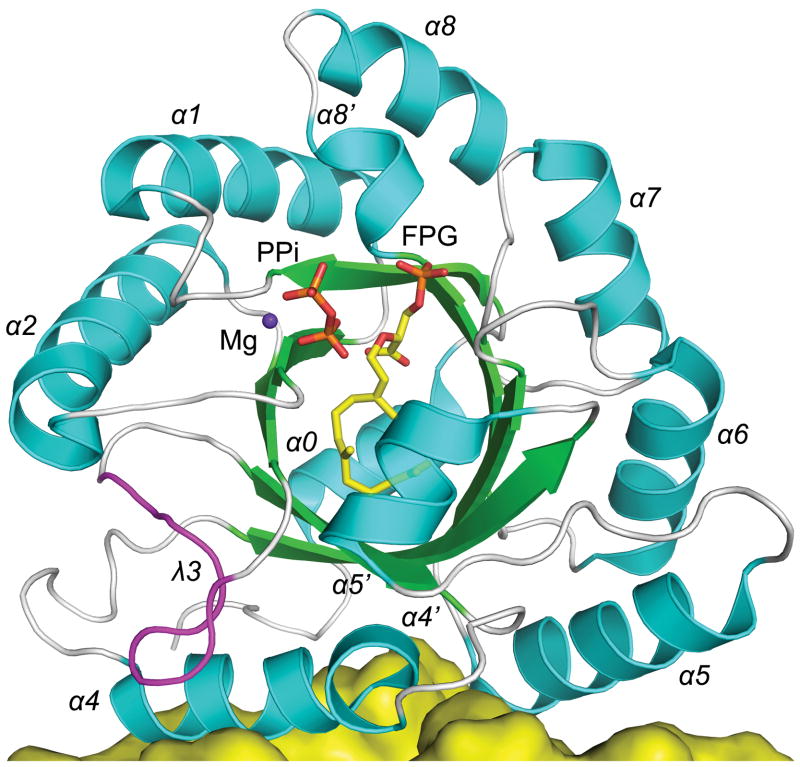
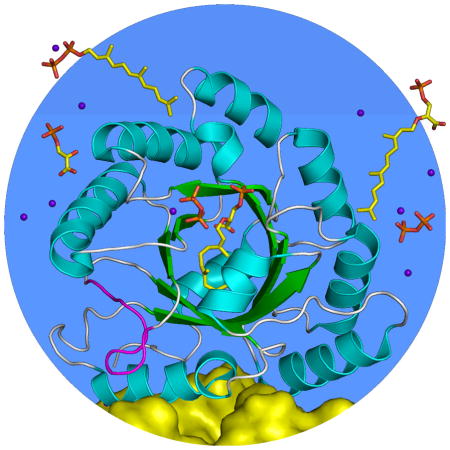
Figure 2.
Structure of the MoeO5 monomer. The central eight-stranded β-barrel and the surrounding α-helices (numbered 0 – 8) in the ribbon diagram are colored green and cyan. The unusual loop of λ3 is highlighted in magenta. The bound FPG and Mg-PPi complex (from the FsPP-soak crystal) are depicted as ball-and-stick models. A fraction of the other monomer in the MoeO5 dimer is also shown, as a yellow surface presentation.
On the other hand, the 3PG head group binding site is equivalent to that for glycerol 1-phosphate (G1P) in GGGPS (Figure S7).[3] The 3′-phosphate, sandwiched between loop β7-α7 and helix α8′, is fastened by three backbone NH groups (Figure 3a). The 1′-carboxyl group, together with His97, also binds to a water molecule. When the crystals were soaked with FsPP, partial substitution of FPG occurred in three hours, and it was mostly replaced overnight. The β-phosphate group of FsPP occupies the same position as does the 3′-phosphate of FPG, but the α-phosphate is loosely bound (Figure 3b). Each active site also shows a Mg-PPi complex adjacent to the FsPP, which may represent an alternative disposition of the PPi moiety of FsPP (Figure S8). Here, the Mg2+ ion binds to Asp41, which is highly conserved in MoeO5, GGGPS and PcrB (Table S3). A similar site containing Mg2+ and PO43− was found with a 3PG soak, but PPi alone did not bind to this site (detailed in the SI). Like most other PTs, MoeO5 requires Mg2+ for activity and the network of interactions with Mg2+ is expected to facilitate FPP ionization, the first step in catalysis.
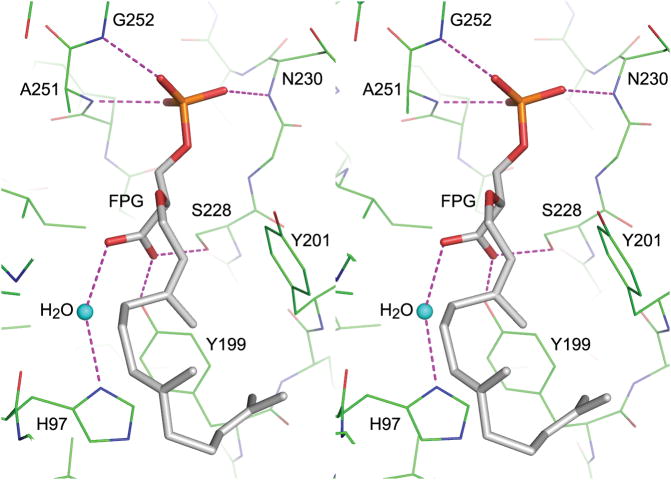
Figure 3.
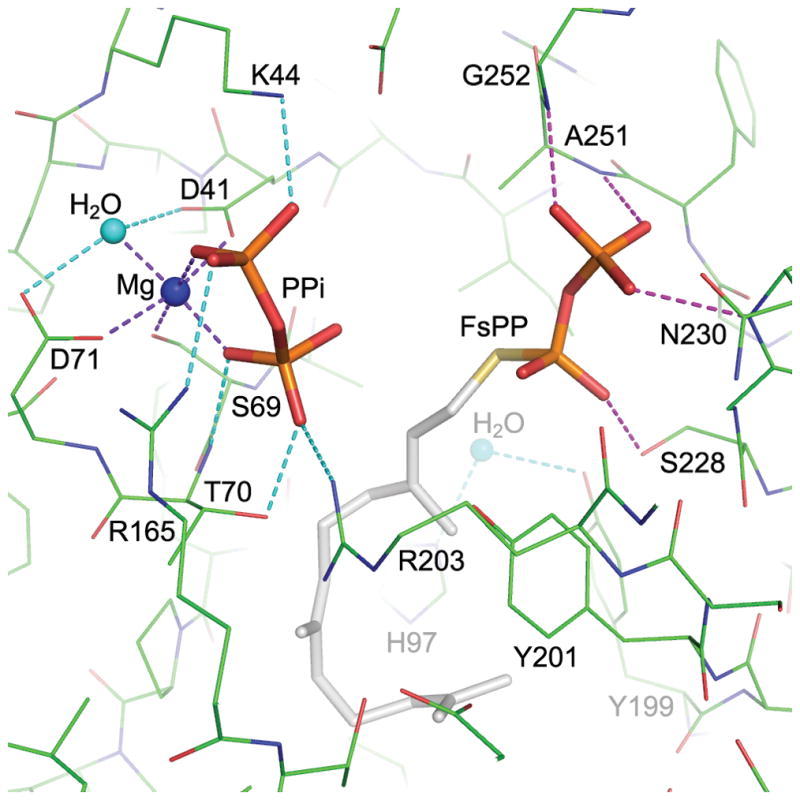
MoeO5 interactions with the bound ligands. a) In a stereo view, the FPG is depicted by thick stick model with gray carbon atoms, and the surrounding amino acids are shown as thin sticks with green carbons. Hydrogen bonds between FPG and the protein are shown as dashed lines. The water molecule bound to His97 is consistently observed in all wild-type crystals. b) The bound FsPP and Mg-PPi along with the active-site amino acids are shown in a similar way as in a). The coordinate bonds to the Mg2+ ion and other hydrogen bonds to the ligands are shown as dashed lines. Note that PPi is closer to C1 than C3 of the FsPP.
In GGGPS and PcrB the prenyl carbocation is directly attached to the C3′-hydroxyl of G1P, and the trans-configuration is conserved (Figure 4a). However, in MoeO5 Doud et al. recently proposed catalytic mechanisms in which either nerolidyl pyrophosphate (NPP) was formed after ionization, facilitating bond rotation and the trans-to-cis conversion (Figure 4b), or that Z,E-FPP was an intermediate.[4] The observation that trans-FPP and NPP react at the same rate while cis-FPP reacts ~5x faster (Figure S9) suggests that all three species may be involved. Another possibility is that the His97-bound water molecule, which is 3.6 Å from the C3 atom, can be activated and associate with the farnesyl cation, transiently forming nerolidol (NOH), facilitating bond rotation (Figure 4c), and we do find that H97C is an inactive mutant, supporting a vital role of His97, although R,S-trans-NOH (+ 3PG + PPi + Mg2+) is not a substrate.
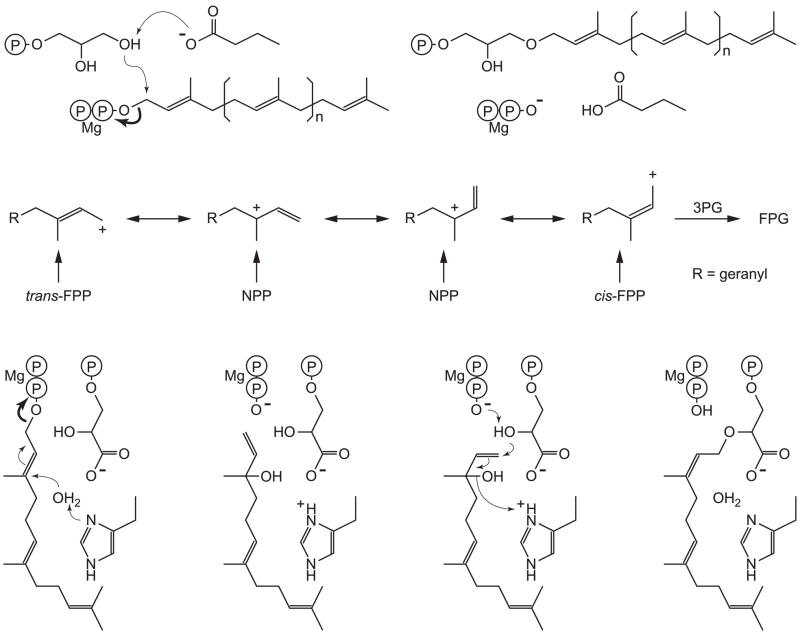
Figure 4.
Proposed mechanisms for the TIM-barrel PTs. The reactions start by ionization (bold arrow) of the prenyl pyrophosphate, which requires a bound Mg2+. a) In catalysis by GGGPS and PcrB, the 3′-OH of G1P is deprotonated by Glu167 (Glu160 in PcrB) and associates directly with the carbocation, forming an ether bond. The prenyl chain length varies from n = 3 in GGGPS to n = 5 in PcrB. b) In MoeO5, trans-FPP, Z,E-FPP and NPP all react to form FPG, consistent with an ionization/isomerization mechanism, but shorter and longer chain species do not. c) His97 is essential for catalysis and may play a role in isomerization, possibly via NOH.
In summary, MoeO5 forms a TIM barrel structure and binds FPG in a curved pocket, mainly as a result of its long λ3 loop. An FPP ionization site containing Asp41 and Mg2+ is located nearby, and the results obtained here are consistent with formation of a Z,E-FPP intermediate. We also find that His97 in the λ3 loop is essential for activity. Except for using Asp41 to bind Mg-PPi, the cis-bond formation catalyzed by MoeO5 is distinct from those of other cis-PTs.[6] To further study this novel mechanism by mutagenesis, kinetic analysis and other physicochemical measurements, the various ligand-complex structures presented here provide a good starting point.
Supplementary Material
Footnotes
This work was supported by National Basic Research Program of China (grant 2011CB710800 to RTG), Tianjin Municipal Science and Technology Commission (10ZCKFSY06000 to RTG), and the National Institutes of Health (AI074233 to EO). We thank the National Synchrotron Radiation Research Center of Taiwan for beam-time allocation and data-collection assistance.
Contributor Information
Feifei Ren, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China.
Dr. Tzu-Ping Ko, Wang Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan.
Xinxin Feng, Department of Chemistry, University of Illinois, Urbana, IL 61801, USA.
Dr. Chun-Hsiang Huang, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
Dr. Hsiu-Chien Chan, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
Dr. Yumei Hu, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
Dr. Ke Wang, Department of Chemistry, University of Illinois, Urbana, IL 61801, USA
Prof. Dr. Yanhe Ma, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
Prof. Dr. Po-Huang Liang, Wang Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan
Prof. Dr. Andrew H.-J. Wang, Wang Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan
Prof. Dr. Eric Oldfield, Email: eo@chad.scs.uiuc.edu, Department of Chemistry, University of Illinois, Urbana, IL 61801, USA
Prof. Dr. Rey-Ting Guo, Email: guo_rt@tib.cas.cn, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
References
- 1.Ostash B, Walker S. Nat Prod Rep. 2010;27:1594–1617. doi: 10.1039/c001461n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Ostash B, Saghatelian A, Walker S. Chem Biol. 2007;14:257–267. doi: 10.1016/j.chembiol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ostash B, Doud EH, Lin C, Ostash I, Perlstein DL, Fuse S, Wolpert M, Kahne D. Biochemistry. 2009;48:8830–8841. doi: 10.1021/bi901018q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payandeh J, Fujihashi M, Gillon W, Pai EF. J Biol Chem. 2006;281:6070–6078. doi: 10.1074/jbc.M509377200. [DOI] [PubMed] [Google Scholar]
- 4.Doud EH, Perlstein DL, Wolpert M, Cane DE, Walker S. J Am Chem Soc. 2011;133:1270–1273. doi: 10.1021/ja109578b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Guldan H, Matysik FM, Bocola M, Sterner R, Babinger P. Angew Chem Int Ed. 2011;50:8188–8191. doi: 10.1002/anie.201101832. [DOI] [PubMed] [Google Scholar]; b) Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, et al. Proteins. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- 6.a) Guo RT, Ko TP, Chen AP, Kuo CJ, Wang AH, Liang PH. J Biol Chem. 2005;280:20762–20774. doi: 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]; b) Liang PH. Biochemistry. 2009;48:6562–6570. doi: 10.1021/bi900371p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.