Abstract
Emerging questions in cell biology necessitate nanometer-scale imaging in live cells. Here we present scanning angle interference microscopy, capable of localizing fluorescent objects with nanometer-scale precision along the optical axis in motile cellular structures. We use this approach to resolve nano-topographical features of the cell membrane and cytoskeleton, as well as the temporal evolution, three-dimensional architecture, and nano-scale dynamics of focal adhesion complexes.
Despite recent advances in super-resolution methodologies, real-time imaging with nano-scale precision remains challenging, especially along the optical axis. Current techniques (reviewed in1) have revealed the architecture of molecular complexes such as focal adhesions with unprecedented detail in fixed cells, yet the dynamic behavior of these structures remains largely unknown2. A need still exists for imaging methodologies capable of recording nano-scale structural details and movements in living cells as occurs during processes such as motility, where nano-scale dynamics of molecules are presumably coordinated across large spatial regions of the cell3-5.
The leading approach for measuring the topography of lipid membranes with nanometer-scale z-resolution is fluorescence interference contrast microscopy (FLIC)6. In FLIC, axially varying structured illumination is used to determine the vertical position of nanometer-sized objects. To eliminate periodic replication artifacts and obtain unambiguous positions, the pattern of structured illumination is manipulated by varying the angle of incidence of excitation light to provide additional spatial information7. Although FLIC has the potential to visualize the rapid movements and spatial organization of proteins, interference contrast methodologies have not been widely adopted in biology, likely due to the technical challenges of implementation, including the assembly of custom microscopy suites and/or the generation of micro-fabricated substrates, and the assumptions that must be made regarding fluorophore molecular orientation.
Here we modify FLIC to develop a new interference contrast method with nanometer precision, which we term scanning angle interference microscopy. This approach can be implemented on commercial total internal reflection (TIRF) microscopes with no hardware modifications (Supplementary Note), and it overcomes many of the limitations of FLIC. We determine unambiguous positions of fluorescent structures over a wide axial range (approaching a micron or more) by actively scanning the incidence angle of the excitation. Furthermore, by manipulating the polarization state of light, we have eliminated the requirement for knowledge of the orientation of fluorescence dipoles in the labeled structure. This approach enables imaging with several nanometer axial precision and temporal sampling rates on the order of one second.
Samples for scanning angle interference microscopy are prepared on reflective silicon wafers with a thin layer of silicon oxide that functions as a sample substrate (Fig. 1a). Axially varying patterns of excitation light intensity are generated through the interference of incident and reflected light, and the structure of the pattern is manipulated by controlling the incidence angle of the excitation light. To quantify performance, we adsorbed nano-sized fluorescent beads on silicon substrates with oxide spacer layers of varying thickness and scanned them with excitation light of varying incidence angle. The recorded fluorescence intensity profiles across varying incidence angles were then fit to an optical model to obtain the axial position of the bead center from the silicon surface with nanometer-scale precision. The use of excitation light linearly polarized perpendicular to the plane of incidence (s-polarized) improved interference intensity contrast by as much as five-fold over non-polarized or circularly polarized light (Supplementary Fig. 1), eliminated the dependence of the optical model on fluorophore dipole orientation (See Optical theory, Online Methods), and significantly improved the accuracy of object localization (Supplementary Fig. 1).
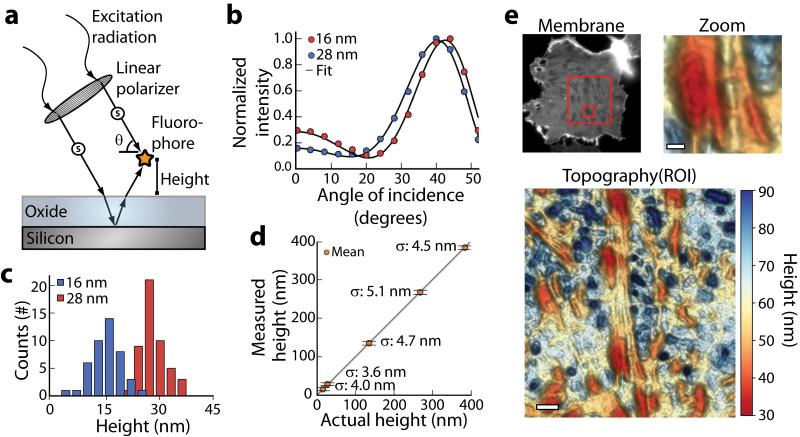
Figure 1. Scanning Angle Interference Microscopy.
(a) Axially varying excitation intensity arises from interference between direct and reflected light off a silicon substrate. The structure of the illumination pattern is controlled by varying the polarization and angle of incidence of the excitation light, and is manipulated to probe the height of a fluorophore above a reflective substrate. A thin layer of oxide is used to space the sample above the silicon surface. (b) Average emission intensity and fits to the optical model for beads at the indicated axial positions, and (c) the corresponding histograms of axial position measured for individual beads. (d) Measured axial position of fluorescent beads adsorbed on silicon oxide layers of varying thickness plotted against their expected position. Data are plotted as the mean, horizontal bars show standard deviation σ (n = 45 - 50). Height is reported as the axial distance of the bead center above the reference point z = 500 nm. (e) Epifluorescence image (Membrane) and three-dimensional scanning angle interference reconstruction (Topography and Zoom) of an epithelial cell ventral membrane labeled with DiO. Boxes in the epifluorescence image indicate regions shown in ROI (larger box) and Zoom (smaller box). Height is reported as the absolute distance of the membrane above the silicon oxide surface. Scale bars, 2 μm (ROI), 750 nm (Zoom).
Measured bead heights above the silicon were on average within a few nanometers of expected heights based on thin film measurements of the silicon oxide (Fig. 1b-d and Supplementary Fig. 2) with the standard deviation ranging from 3.6 to 5.1 nm (Fig. 1d). Beads varying in axial position by as little as 13 nm had distinct interference contrast profiles, allowing their height differences to be resolved (Fig. 1b-c). Similarly, analysis of mCherry and mEmerald conjugates of the focal adhesion protein paxillin in cells positioned at four heights above the silicon substrate revealed a standard deviation of just 1.8 nm from the mean height across eight independent measurements (Supplementary Fig. 3). We found that a signal to noise ratio of approximately two or better yielded the most accurate results in cellular samples (Supplementary Fig. 4). The error of measurement in our approach due to natural refractive index variations in cells was theoretically predicted to be ~3.8 nm for structures positioned 0 to 350 nm above the substrate (Supplementary Fig. 5). These predictions were experimentally borne out with a nearly identical mean height and standard deviation obtained for beads absorbed on the silicon substrates when they were imaged through either buffer or cells (Supplementary Fig. 6). Lateral resolution of the technique in cells was approximately diffraction limited (full width at half maximum ~300 nm; Supplementary Fig. 7).
We used scanning angle interference microscopy to image the vertical positions of three types of cellular structures: the plasma membrane (Fig. 1e), the microtubule cytoskeletal network (Fig. 2a), and molecules within the focal adhesion complex (Fig. 2b). Visual reconstructions of dye-labeled, fixed ventral cell membranes revealed fine axial detail, displaying distinct topographical features corresponding to the sites of focal adhesion complexes, the cortical actin network proximal to the plasma membrane, and the variations in glycocalyx thicknesses ranging from 40 to 85 nm (Fig. 1e and Supplementary Fig. 8). In fixed epithelial cells with fluorescently-labeled microtubules, the scanning angle interference images showed a downward bending of the microtubule network in the lamella (Fig. 2a). Notably, the measured microtubule heights ranged from approximately 70 to 350 nanometers, which is beyond the working range of other techniques that offer high axial precision, including variable incidence angle TIRF, interference-based photo-activation localization microscopy, and standard implementations of FLIC.
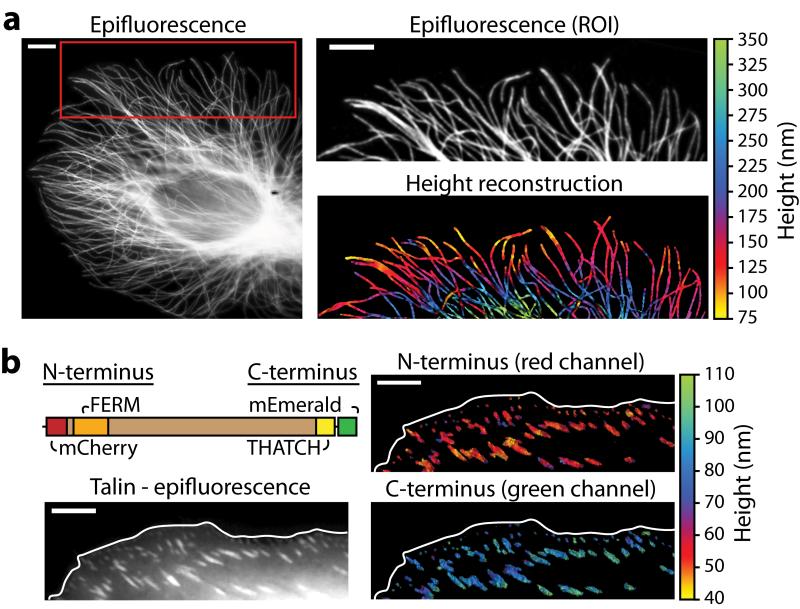
Figure 2. Scanning angle interference imaging of microtubules.
(a) Epifluorescence image (left panel) of microtubules in a fixed epithelial cell. The red box marks the region of interest (ROI) shown in the right panels; immunofluorescence (top) and the corresponding scanning angle interference image (bottom) are shown. (b) The schematic shows dual-labeled talin, which binds to integrin and actin through its FERM and THATCH domains, respectively. The images show the difference in height of the integrin-binding N-terminus of talin and the actin-binding C-terminus. Scale bars – 5 μm.
We next applied scanning angle interference microscopy as a molecular ruler in live cells. We generated a construct for expressing talin, a focal adhesion molecule with a rod-like structure 60 nm in length, as a fusion with mCherry at its N-terminus and mEmerald at its C-terminus. We found that the C-terminus was situated on average 37 nm higher than the N-terminus, corresponding to a molecular orientation of 51 degrees relative to the vertical axis. The precision of measurement with our approach was comparable to advanced single molecule localization microscopy methods implemented on two-objective and interference based microscopes (Fig. 2b), however in contrast to these approaches, our method permits the acquisition of these measurements with 1-10 second sampling rates in living cells8-9. Scanning angle interference microscopy does not currently incorporate vertical sectioning and is therefore ideally suited for the measurement of discrete structures with nano-scale thickness, < 150 nm, such as plasma membranes and associated proteins, cytoskeletal filaments, microtubules, and vesicles (Supplementary Fig. 9). For fluorophores distributed vertically in a diffraction-limited spot, it provides an average height of the molecules, as observed with a talin construct double-labeled with mCherry on its C- and N-termini (Supplementary Fig. 9).
The axial precision and ability to image multiple color channels, coupled with the dynamic capability of scanning angle interference microscopy, makes the method useful for exploring the molecular mechanisms and topological arrangements underlying dynamic mechanical processes. We simultaneously measured the heights of paxillin-mEmerald and vinculin-mCherry conjugates, two structural molecules localized to focal adhesions. We observed qualitatively that paxillin increased in height as compared to vinculin in the adhesions that formed adjacent to sites of cell-cell contact following the collision of two motile cells (Fig. 3a and Supplementary Movie). In more quantitative measurements, we observed that paxillin and vinculin were vertically stratified in motile cells; paxillin was at an average height of 61±6.5 nm and vinculin at an average height of 99±7.2 nm (P < 0.001; n > 20 cells; Supplementary Fig. 3). Finally, we imaged paxillin and vinculin during cycles of cell retraction in the maturing adhesions of these motile cells (Fig 3b and Supplementary Movie). Imaging randomly selected adhesions in these motile cells revealed that the average height of paxillin in maturing adhesions was significantly lower than the average height of paxillin in stable adhesions in non-retracting regions of the cells (Fig. 3c), as well as its height in small, newly formed adhesions in protruding regions (n = 5 cells; P < 0.001; Supplementary Fig. 11). We found that paxillin was typically lowered in maturing adhesions within 5 – 10 minutes of the start of cell retraction cycles. By contrast, axial movement of vinculin in maturing adhesions was more random (Fig. 3c and Supplementary Fig. 10).
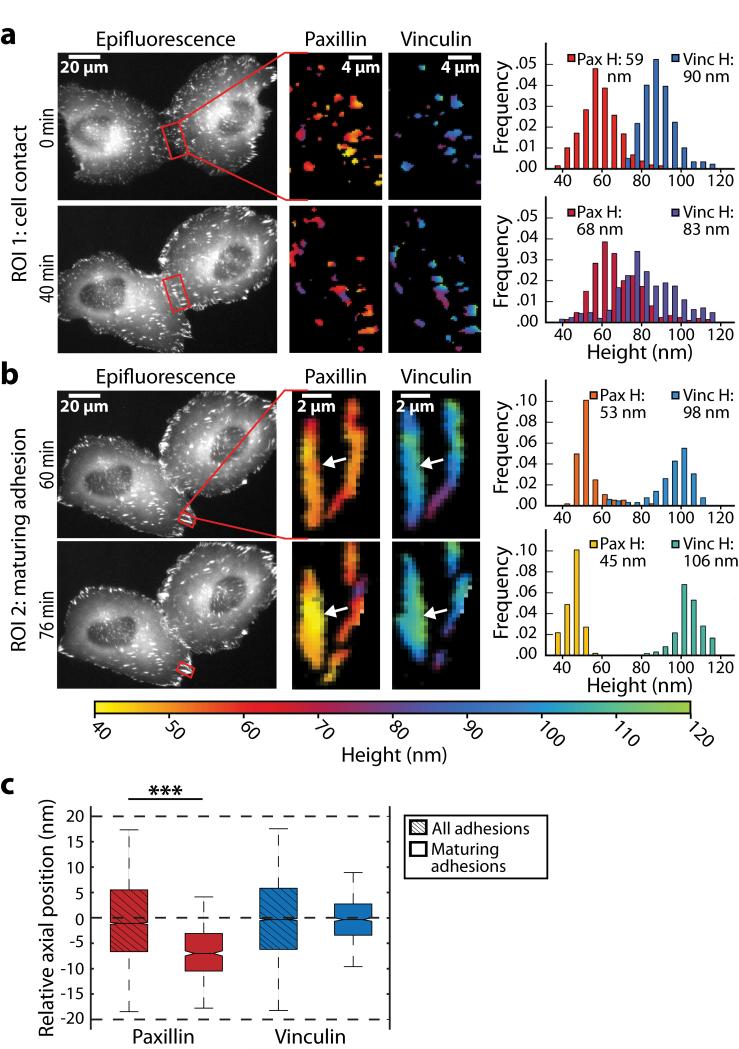
Figure 3. Nano-scale dynamics of adhesion proteins in migrating cells.
(a, b) Live-cell time-lapse multicolor scanning angle interference imaging of epithelial cells expressing paxillin-mEmerald and mCherry-vinculin following cell-cell contact (a) or in an assembling adhesion during cell retraction (b). The left panels show paxillin epifluorescence images at the indicated time point, the region of interest (ROI) is boxed in red. The middle and right panels show heights of paxillin and vinculin in the ROI. The plots show distributions of paxillin and vinculin heights in ROI, mean height (H) above the oxide surface is calculated over 35 to 120nm. In (a) the histogram shows the measured heights of paxillin and vinculin at each pixel corresponding to site of adhesion. In (b) the histogram displays the heights of the molecules at the subset of ROI pixels within the boundary of the left adhesion (indicated by white arrow). (c) The plot shows the relative heights of paxillin and vinculin in maturing adhesions during cell retraction. Axial position is reported relative to the average height of molecules in all adhesions in the cell (P < 0.001; n=3 cells, >= 9 adhesions).
Although the mechanism of downward paxillin movement during cell migration remains unclear, one possibility is that these vertical shifts correspond to increased mechanical engagement of the cytoskeleton with the adhesion complex, which is known to stimulate adhesion maturation. Consistent with this notion, we did not observe any downward displacement of paxillin in cells treated with the Rho kinase inhibitor Y-27632 (data not shown). Nevertheless, the findings raise the possibility that adhesion complexes could transduce mechanical information through the spatial reorganization of their molecular components. The experimental data also highlight the ability of this imaging approach to visualize the dynamics of cellular structures in response to force and to provide the experimentalist with quantitative insight. We therefore propose that scanning angle interference microscopy may be a useful new tool for unraveling cellular mechanisms of mechanotransduction.
In summary, we have demonstrated that scanning angle interference microscopy is capable of localizing biological structures with nanometer-scale precision along the optical axis in living cells on time scales suitable for observing dynamic processes. The combination of high axial and temporal resolution, extended depth of field, ease of implementation, and the ability to image live cells in multiple color channels distinguishes this approach from existing super-resolution techniques. Scanning angle interference microscopy will enable the investigation of the many dynamic processes that occur proximal to the cell membrane – for instance, signaling linked to receptor activation and mechanotransduction, endocytosis, exocytosis, and extracellular-matrix remodeling - and should thus be broadly applicable to emerging questions in membrane transport, signal transduction, and mechanobiology.
ONLINE METHODS
Preparation of reflective substrates for cell culture
N-type [100]-orientation silicon wafers with 500 nm or 1.9 μm of silicon oxide were purchased from Addison Engineering. Wafers with 500 nm or greater silicon oxide were selected to maximize interference contrast (Supplementary Fig. 12). Wafers were cut into approximately 1 cm × 1 cm chips by scoring with a diamond-tip pen. They were cleaned by sonicating in acetone for 20 minutes, rinsing with water, sonicating for 20 minutes in 1 M potassium hydroxide, and rinsing with water. The wafers were then chemically activated to enable conjugation of extracellular matrix (ECM) proteins for cell adhesion. The wafers were incubated for one hour in 0.5% (3-aminopropyl)trimethoxysilane (APS) in water, sonicated five times in water for five minutes to remove excess APS, incubated for one hour in 0.5% gluteraldeyde in phosphate buffered saline (PBS; pH = 7.4), sonicated five times in water for five minutes, and dried under nitrogen gas. The wafers were sterilized under a UV lamp and incubated overnight at 4 °C in 10 μg ml−1 human plasma fibronectin (Millipore).
Preparation of reflective substrates with adsorbed nanobeads
To validate the setup, silicon wafers were prepared with defined layers of silicon oxide that functioned as spacers between the silicon surface and adsorbed fluorescent nanobeads on the oxide surface. The SiO2 layers were grown on bare P-type [100]-orientation silicon wafers (Addison Engineering) using silane chemistry (5% SiH4 / 95% N2) of plasma enhanced chemical vapor deposition (PECVD process). The plasma was generated by mixing 170 sccm silane (first precursor) and 710 sccm NO2 (second precursor) in an Oxford PlasmaLab 100 system. The NO2 was used as the process oxidant and its flow rate was enough to obtain high quality SiO2 films with no excess Si and no Si-H bonding. The chamber pressure was maintained at 1000 mT. The radio-frequency (RF) power to generate the plasma of ionized gasses was 20 W. The wafer stage temperature was set at 300 °C and the chiller control was also set at 70 °C to promote the development of nonporous films. Under these conditions, the silicon oxide deposition rate was 78.6 nm min−1. The thickness of each oxide layer was checked using an n & k 1500 system (n & k Technology, Inc.). 20-nm carboxylate-modified yellow-green fluorescent spheres (Invitrogen) were diluted to a final concentration of 5×108 beads ml−1 in PBS + 100 mM NaCl and sonicated for five minutes to disperse aggregates. Silicon wafers were cut and cleaned as above and incubated in the bead solution for five minutes at room temperature to allow beads to adsorb through hydrophobic interaction. The wafers were then washed once with PBS + 100 mM NaCl, dried in air at 37 °C, and stored at room temperature.
Preparation of fluorescent reference slides
Laser illumination in widefield epi-fluorescence is subject to variations in intensity owing to interference fringing. To quantify the variation in intensity, reference slides with a monolayer of fluorescent dye were prepared using silane conjugates of fluorescein and rhodamine B. The conjugates were synthesized by reacting 1 mg of fluorescein isocyanate or 1.37 mg of rhodamine B isocyanate with 9.25 mg of APS in 1.25 mL anhydrous ethanol. Reactions were conducted under nitrogen gas at room temperature for two hours with constant stirring. Immediately following the reaction, the conjugates were centrifuged at 20,000 g for 10 minutes to pellet unwanted aggregates and then diluted ten-fold in absolute ethanol. Clean glass bottom dishes (#1.5 glass) were incubated with the diluted silane conjugates for 30 minutes at room temperature, rinsed three times with water, and stored in water until imaging.
Lentiviral and expression constructs
To obtain transcriptionally inducible paxillin-mCherry and paxillin-mEmerald, the fusions were cloned into the 2nd generation lentiviral vector pLV Hygro Tet. pLV Hygro Tet was prepared from pLV tTRKRAB (Addgene #12249; a generous gift of Didier Trono) by inserting in place of the EEF1α promoter and tTRKRAB transgene, a hygromycin resistance gene expression cassette consisting of an SV40 promoter and bovine growth hormone polyadenylation sequence in reverse orientation for selection in transduced cells back to back with the regulatable hybrid hepatamerized Tet operator minimal CMV promoter10 followed by a central polypurine tract and the multiple cloning site (MCS) from pcDNA3.1+ (Invitrogen). The paxillin-mCherry gene was excised from a N1 clonetech-style vector with the restriction enzymes Nhe-I (made blunt) and Not-I and inserted into the PME-I and Not-I sites of the pLV Hygro Tet MCS to make pLV paxillin-mCherry Hygro Tet. Similarly, paxillin-mEmerald was cloned to make pLV paxillin-mEmerald Hygro Tet. A second lentiviral construct, pLV rtTAs-M2 IRES Neo, was also prepared from pLV tTRKRAB to provide EEF1α promoter mediated constitutive expression of the synthetic tetracycline inducible reverse transcriptional transactivator rtTAs-M2 (generous gift of Wolfgang Hillen and Hermann Bujard) from a bicistronic mRNA coexpressing neomycin phosphotransferase downstream of an internal ribosome entry site for selection of transduced cells with G418. The structure and sequence of pLV Hygro Tet, pLV paxillin-mCherry Hygro Tet, pLV paxillin-mEmerald Hygro Tet, and pLV rtTAs-M2 IRES Neo are available for download at www.lablife.org (Weaver lab).
A validated shRNAi (TRCN0000299022) in the lentivector pLKO.1 Puro and targeting the human talin1 3′UTR was obtained from Sigma. Murine talin1 tagged with mCherry at the N-terminus and mEmerald at the C-terminus was prepared from mCherry N-terminally and mEmerald C-terminally tagged murine Talin1 constructs previously described8 ligating a SalI-NotI fragment from the mEmerald construct containing the C-terminal end of talin1 fused to mEmerald with a AgeI-SalI fragment from the mCherry construct containing mCherry fused to the N-terminal end of talin1 with the NotI-AgeI vector fragment from pmCherry-N1 (Clontech). Murine talin1 tagged at N and C termini with mCherry was prepared from this latter construct by substituting an AgeI-NotI mCherry fragment from pmCherry-N1 for mEmerald. The construct for expression of mCherry-vinculin (C-terminal fusion) was described previously11.
Cell culture and imaging sample preparation
MCF10A human mammary epithelial cells were grown at 37 °C and 5% CO2 in DMEM F12 (Invitrogen) supplemented with 5% donor horse serum (Invitrogen), 20 ng ml−1 epidermal growth factor (Peprotech), 10 μg ml−1 insulin (Sigma), 0.5 μg ml−1 hydrocortisone (Sigma), 0.1 μg ml−1 cholera toxin (Sigma), and 100 units ml−1 penicillin/streptomycin. For stable expression of paxillin-mEmerald or paxillin-mCherry, recombinant lentivirus was prepared by transient transfection into HEK-293T cells and used to transduce MCF10A cells as previously described12. Stable cell lines were selected in 200 ng ml−1 G418 and 380 ng ml−1 hygromycin and sorted on a FACSAria II cell sorter (BD Biosciences) for paxillin-mEmerald or paxillin-mCherry expression. Expression of paxillin-mEmerald or paxillin-mCherry was induced with 200 ng ml−1 doxycycline in growth media for one day prior to experimentation. Transient transfection was with Lonza Nucleofector Kit V (program T024). Talin fluorescent protein conjugates were co-transfected with shRNA targeting endogenous MCF10A talin1.
For all experiments, cells were replated on fibronectin-conjugated silicon wafers and cultured overnight before live-cell imaging, fixation, or cell roofing. Except for microtubule imaging, cells were plated on wafers with 500-nm silicon oxide and fixation was performed in 2% paraformaldehyde for 20 minutes. For cell membrane imaging, the dorsal cell membrane was removed through cell roofing according to previously described protocols13. Briefly, cells were swelled in ice-cold hypotonic buffer (15 mM KCl, 15 mM Hepes, 1 mM CaCl2, 1 mM MgCl2, pH 7.4) for 4 minutes. Swollen cells were then sonicated with a probe-type sonicator (Microson XL2000; Misonix) using a short pulse (<1 s, Power Setting 8) with the sonicator probe (1/8” diameter) approximately 3 mm above the cells. Following sonication, the remaining ventral plasma membranes were rinsed once in PBS and fixed. Membranes were labeled with fluorescent dye post-fixation by incubating in 1 μM Vybrant DiO (Invitrogen) in PBS for one hour at room temperature. Rho kinase inhibition was with 10 μM of the inhibitor Y-27632 (Cayman Chemical) in growth media. For the microtubule imaging, cells were plated on wafers with 1.9 μm silicon oxide overnight and fixed by incubating in 0.25% glutaraldehyde for 30 seconds and then transferring them to a solution of 0.25% glutaraldehyde with 0.1% Triton X-100 in PBS. Cells were then rinsed with PBS and covered with a freshly prepared drop of 0.2% sodium borohydride in PBS, which was exchanged three times over 30 minutes. This was followed by two rinses with PBS and a 10 minute incubation with a solution of PBS, 2% BSA, and 0.1% Triton X-100. Microtubules were labeled with rat monoclonal anti-tubulin antibody (AbD Serotec) and Alexa488-conjugated anti-rat secondary antibody (Invitrogen).
Image acquisition
Imaging was performed on a motorized objective-type TIRF inverted microscope system (Ti-E Perfect Focus System; Nikon) equipped with 488 nm and 561 nm lasers, electronic shutters, an electron-multiplying charged-coupled device camera (QuantEM 512; Photometrics) and controlled by NIS-Elements software (Nikon). A linear glass polarizing filter (Edmunds Optics) was placed in the excitation laser path to polarize the light perpendicular to the plane of incidence along which the excitation laser light emanates from the objective. On the Nikon microscope, the polarizer was placed in the filter cube housing the dichroic mirror, where the excitation filter would normally sit. The relationship between motor position of the TIRF illuminator and actual angle of laser light out of the objective was calibrated by varying motor position and measuring the position of the focused laser spot along the ceiling of the room housing the microscope or with a protractor. The corresponding angles in aqueous samples were determined from Snell’s Law. Motor units corresponding to desired laser incidence angles for image acquisition were programmed into NIS-Elements to facilitate automated data acquisition. Samples were imaged with a 60x / NA 1.2 water-immersion objective (CFI Plan Apo VC; Nikon) or a 100x / NA 1.45 oil-immersion objective (CFI Plan Apo Lambda; Nikon) with additional magnification using a 1.5x optovar (0.18 μm/pixel for 60x, 0.11 μm/pixel for 100x). Silicon substrates were placed sample-side down in 35-mm #1.5 glass bottom dishes in PBS or phenol red-free growth media with 10 mM Hepes (pH 7.4) to maintain pH for live-cell imaging. A buoyancy adjusted weight of approximately 1.75 g was placed on the wafer to keep the wafer sample surface flat against the glass bottom. Live-cells were maintained at 37 °C using a blower-style heater (ASI 400; Nevtek). For imaging microtubules on silicon substrates with 1.9 μm of silicon oxide, a sequence of 33 epi-fluorescence images of each sample was obtained by scanning through laser incidence angles below the critical angle from −34 degrees to +34 degrees in two degree increments along the plane of incidence (Supplementary Fig. 12). For all other samples, a sequence of 27 epi-fluorescence images was obtained from −52 to +52 degrees in four degree increments. Typical camera exposure times were on the order of 10-100 ms (with an EM gain of 200) resulting in acquisition rates on the order of 3 - 60 s per image sequence. A sequence of images of the reference slides was obtained similarly for each color channel.
Optical theory
The electromagnetic theory describing silicon surface-generated standing waves of excitation radiation has previously been reported14. The layered silicon oxide/silicon structure is treated as a single virtual layer using transfer matrix methods that account for internal reflections and refractions. Reflection and refraction at known sample interfaces, including the cell membrane and sample buffer or the cell membrane and cell cytoplasm, were determined to have a negligible effect on the structure of the electric field and were therefore excluded from the optical theory (Supplementary Fig. 13). For incident light polarized perpendicular to the plane of incidence, the relative local electric field at a position H in the sample above the silicon oxide is given by:
| (1) |
where rTE is the transverse electric (TE) component, perpendicular to the plane of incidence, of the Fresnel coefficient of reflection between the sample interface and the virtual silicon oxide/silicon layer and φ is the phase difference of the direct and reflected light at axial position H given by:
| (2) |
The effective Fresnel coefficient is obtained from the characteristic transfer matrix MTE according to:
| (3) |
| (4) |
| (5) |
| (6) |
where ki is the wavenumber in material i; nSi, nox, nb are the refractive index of the silicon, silicon oxide, and sample, respectively; θSi, θox, θb are the angles of incidence in the silicon, silicon oxide, and sample, respectively; and dox is the thickness of the silicon oxide layer. The angles of incidence in silicon oxide and silicon were calculated according to Snell’s Law.
Image analysis
For height reconstructions, intensity profiles from acquired image sequences were fit to the optical theory on a pixel-by-pixel basis. To correct for uneven laser illumination across the image, the intensity of each pixel, and for live cell imaging an all-pixel average, in each sample image was adjusted according to the corresponding intensity in the reference slide image sequence:
| (7) |
Si,n is the intensity of pixel i in sample image n, Ri,n is the intensity of pixel i in reference image n, and C is the average background intensity of the camera device obtained by snapping an image on the microscope with the shutters closed and averaging the intensity over all image pixels. Emission by fluorophores on the microscope is proportional to the excitation radiation intensity, which is proportional to the square of Equation 1. The detected pattern of intensity variation at a given pixel in the sample image sequence was thus fit according to:
| (8) |
Here the scaling parameter A accounts for variation in detected intensity due to factors including mean excitation laser intensity, fluorophore density, fluorophore photo-physics, self-interference of emitted photons, and efficiency of emitted photon detection. The offset parameter B accounts for background fluorescence in the sample images. Parameters A, B, and H were fit through constrained nonlinear least-squares optimization using Trust Region algorithms (Math Kernel Library; Intel). Fitted data whose scaling parameter was below a defined minimum (e.g. dim structures) were excluded from further analysis.
Software
A custom software package for image analysis and height reconstruction of scanning angle interference image sequences was written in C++ and compiled for the Linux operating system. Additional programs for data analysis and 3D visualization were written in Matlab and Python using libraries distributed by Enthought, Inc.
Supplementary Material
ACKNOWLEDGEMENTS
We graciously thank J. Lakins for guidance on lentiviral vector design and the construction of talin fusion constructs and T. Onuta for assistance with the fabrication of silicon wafers at the Maryland Nanocenter FabLab. We also thank M. Krummel and S. Peck for helpful discussions and preliminary hardware development. Image acquisition for this study was performed at the Nikon Imaging Center at the UCSF. This work was supported by BCRP DoD Era of Hope grant W81XWH-05-1-0330, NIH/NCI TMEN grant 1U54CA163155-01, NCI grant U54CA143836-01, and NIH/NCI R01 CA138818-01A1.
Footnotes
AUTHOR CONTRIBUTIONS
M.J.P. and V.M.W conceived and initiated the project. M.J.P. and K.S.T. designed the instrumentation. M.J.P., C.C.D. and M.G.R designed and performed experiments. M.W.D. designed fluorescent protein constructs. M.J.P. and M.G.R. wrote the analysis software. V.M.W. and J.T.L. supervised the project. All authors wrote the paper.
REFERENCES
- 1.Huang B, Babcock H, Zhuang XW. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shtengel G, et al. P. Natl. Acad. Sci. USA. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DuFort CC, Paszek MJ, Weaver VM. Nat. Rev. Mol. Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore SW, Roca-Cusachs P, Sheetz MP. Dev. Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons JT, Horwitz AR, Schwartz MA. Nat. Rev. Mol. Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambacher A, Fromherz P. J. Opt. Soc. Am. B. 2002;19:1435–1453. [Google Scholar]
- 7.Ajo-Franklin CM, Ganesan PV, Boxer SG. Biophys. J. 2005;89:2759–2769. doi: 10.1529/biophysj.105.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanchanawong P, et al. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu K, Babcock HP, Zhuang X. Nat. Methods. 2012;9:185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gossen M, Bujard H. Proc. Natl. Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnette DT, et al. Nature Cell Biol. 2011;13:371–382. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barde I, Salmon P, Trono D. Current Protocols in Neuroscience. John Wiley & Sons, Inc.; 2001. [DOI] [PubMed] [Google Scholar]
- 13.Drees F, Reilein A, Nelson WJ. Methods Mol. Biol. 2004;294:303–320. doi: 10.1385/1-59259-860-9:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambacher A, Fromherz P. Appl. Phys. A-Mater. 1996;63:207–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.