Abstract
Current strategies for determining the structures of membrane proteins in lipid environments by NMR spectroscopy rely on the anisotropy of nuclear spin interactions, which are experimentally accessible through experiments performed on weakly and completely aligned samples. Importantly, the anisotropy of nuclear spin interactions results in a mapping of structure to the resonance frequencies and splittings observed in NMR spectra. Distinctive wheel-like patterns are observed in two-dimensional 1H–15N heteronuclear dipolar/15N chemical shift PISEMA (polarization inversion spin-exchange at the magic angle) spectra of helical membrane proteins in highly aligned lipid bilayer samples. One-dimensional dipolar waves are an extension of two-dimensional PISA (polarity index slant angle) wheels that map protein structures in NMR spectra of both weakly and completely aligned samples. Dipolar waves describe the periodic wave-like variations of the magnitudes of the heteronuclear dipolar couplings as a function of residue number in the absence of chemical shift effects. Since weakly aligned samples of proteins display these same effects, primarily as residual dipolar couplings, in solution NMR spectra, this represents a convergence of solid-state and solution NMR approaches to structure determination.
Keywords: NMR spectroscopy, protein structure, dipolar couplings, membrane proteins, structure determination
Introduction
Among the strongest driving forces in contemporary biochemistry is the availability of the sequences of the genomes of a large and increasing number and variety of organisms, ranging in complexity from viruses to humans. Now, the front page of a daily newspaper is essential reading to keep up with the latest genomes that have been sequenced and their potential economic, medical, and social impact. A major effect of all this sequencing activity, and concern about its consequences, is that the focus of attention is inexorably shifting from the linear arrangement of information in the primary sequences of DNA and polypeptides to the three-dimensional structures of the proteins responsible for expressing the chemical functions that define the life cycles of the organisms. The next ambitious goal in biochemistry is to determine the structures of all of the proteins encoded in the genome of an organism. If all of the expressed proteins were soluble, and adopted stable, folded conformations, then this would be possible, at least in principle, through the methodical application of X-ray crystallography and solution KMR spectroscopy. However, membrane proteins, as well as some other classes of proteins, are not soluble in aqueous solution, at least not in the unaggregated forms required to prepare the samples for the two established experimental methods of structural biology. The development of new approaches to protein structure determination is needed.
“Membrane proteins in health and disease”
The title of the 45th Annual Meeting of the Canadian Society of Biochemistry, Molecular Biology, and Cellular Biology reflects the fact that one third of the proteins encoded in a typical genome are membrane proteins, and they are responsible for many essential biological functions, some of which are unique. When mutations affect their ability to perform these functions, diseases are a consequence. And many membrane proteins are receptors for drugs. Understanding health and ameliorating the effects of diseases requires the determination of the structures of membrane proteins so that the full power of structural biology can be used to decipher the molecular mechanisms of diseases and designing drugs. However, only a few examples of membrane proteins have been amenable to structural analysis using X-ray crystallography and solution NMR spectroscopy, which is hardly surprising given that these methods were developed specifically for soluble, globular proteins. Nonetheless, the structures of the few helical membrane proteins that have been determined provide a tantalizing glimpse of the overall design principles and the subtlety involved in the use of helices as the dominant structural and functional elements of these proteins.
Members of the principal class of membrane proteins are largely helical. Thus, an initial goal of structure determination is to determine the overall topology of the individual helices in terms of their tilt and rotation. In some cases, this results in a description of the protein that is sufficiently detailed to contribute to understanding its function, and in others, it provides an overview of the protein fold and the starting place for the determination of the three-dimensional structure of the protein and its subsequent refinement. NMR spectroscopy is the most powerful and versatile method to describe the structure, dynamics, and interactions of proteins. However, the nature of the samples of membrane proteins in lipid environments is such that further development of a wide range of methods and instruments is essential.
NMR structural studies of proteins
There are three principal spectroscopic considerations for NMR structural studies of proteins: the overall rotational correlation time of the protein, the extent of alignment of the protein in the sample, and the strategy for assignment of the resonances to sites in the protein. Each of these considerations needs to be taken into account in the development of NMR for structural studies of membrane proteins (Opella 1997). For relatively small globular proteins, the sample conditions, instrumentation, experiments, and calculations that lead to structure determination are well established (Cavanagh et al. 1996). The chief requirement for structure determination of globular proteins is that samples can be prepared of isotopically labeled polypeptides that are folded in their native conformation and reorient relatively rapidly in solution. Such samples have been prepared for many hundreds of proteins, and it is likely that this can be done for thousands more of the polypeptide sequences found in genomes (Wuthrich 1998). This is not yet the case for membrane proteins.
Rotational correlation time
The correlation time problem is paramount, since it is not possible to obtain a spectrum of a slowly reorienting or immobile protein without making crucial choices about samples, instrumentation, and experimental methods. It is feasible to apply both solution NMR and solid-state NMR approaches to helical membrane proteins, depending on the choices of lipids and other sample conditions. Figure 1 illustrates the types of lipid assemblies used in NMR structural studies of membrane proteins. Membrane proteins require the presence of lipids to maintain their native conformations and functions (Opella 1997). Membrane proteins can be reconstituted with lipids that self-assemble into micelles, bicelles, or bilayers. The properties of the entire lipid–protein complex determine the effective rotational correlation time of the protein. Micelles and small bicelles can be prepared so that they contain a single polypeptide and reorient rapidly enough for solution NMR spectroscopy. Micelles are spherical aggregates of lipids with the hydrophobic chains on the interior. Samples for solution NMR spectroscopy require the use of relatively high temperatures and high concentration of lipids that form small micelles (McDonnell and Opella 1993), most commonly sodium dodecyl sulfate, dodecyl phosphocholine, or dihexanoyl phosphatidylcholine. Bicelles are bilayer disks and their size can be adjusted through the molar ratios of long-chain and short-chain lipids (Sanders et al. 1993). Bilayers are essentially infinitely large in two dimensions and two molecules thick in the third dimension. Nearly all of the residues in the polypeptide chain are immobile on time scales longer than milliseconds; therefore, the relevant dipolar coupling and chemical shift interactions present in individual backbone sites are not motionally averaged, and as a result, the samples give very broad and poorly resolved spectra when instruments and methods appropriate for proteins in solution are used. However, bilayer samples are suitable for the application of solid-state NMR methods where radiofrequency irradiations replace molecular reorientation as the principal mechanism of averaging the dipolar couplings responsible for the broadening. The solid-state NMR approach that uses stationary, uniaxially aligned samples (Opella et al. 1987) is particularly well suited for membrane proteins in lipid bilayers because the proteins are immobilized by the environment and can be highly aligned (Marassi et al. 1997). The combination of decoupling the heteronuclear dipolar interactions and sample alignment results in narrow, single-line resonances and well-resolved, orientationally dependent spectra that provide the input for structure determination. There is a direct mapping of protein structure onto the solid-state NMR spectra of aligned samples, and the determination of complete three-dimensional structures from the spectra is feasible when multiple orientationally dependent frequencies are measured for nuclei at each residue.
Fig. 1.
Representations of proteins in mice lies (top), bicelles (middle), and bilayers (bottom) (Opella 1997).
Sample alignment
Samples can be prepared with no, weak, or complete alignment of the protein molecules. Complete alignment is possible only with immobile proteins. Rapidly reorienting proteins can have either no or weak alignment The vast majority of NMR studies have been performed on samples without molecular alignment. This is the case for globular proteins, which undergo effectively isotropic reorientation in solution. It is also the case for membrane proteins in micelles and small bicelles. For these samples, structure determination is based on measurements that reflect internal molecular parameters, such as NOEs (nuclear Overhauser enhancements) that give short-range distance measurements and variations in isotropic chemical shifts associated with secondary structure (Almeida and Opella 1997). Unoriented, immobile samples, including membrane proteins in bilayers, can be studied using magic angle sample spinning solid-state NMR spectroscopy (Griffin 1998), where distances and torsion angles are measured through spectroscopic parameters affected by both homo- and heteronuclear dipolar interactions. In the past few years, there has been a great deal of interest in weakly aligned samples of proteins, including soluble, globular proteins in the presence of various types of additives and gel-forming media (Bax et al. 2001). Since the proteins are reorienting rapidly, solution NMR methods are used for the measurement of residual dipolar couplings. It is possible to weakly align membrane proteins in micelles and small bicelles through lanthanide ion (Ma and Opella 2000; Veglia and Opella 2000) and gel methods (Chou et al. 2002). Immobile bilayer samples can be completely aligned to a degree rivaling that observed in single crystals of peptides (Marassi et al. 1997). Membrane proteins in bilayers can be aligned mechanically between glass plates, and membrane proteins in large bicelles can be aligned magnetically and then “flipped” to the desirable parallel orientation through the addition of lanthanide ions (Howard and Opella 1996).
Resonance assignments
The traditional approach to protein structure determination is based on the same overall principles, whether solution NMR or solid-state NMR methods are used and whether the sample is aligned or not. This involves the resolution of resonances through the use of isotopic labels and multidimensional NMR experiments, the measurement of spectral parameters associated with individual resonances, for example, NOEs, J couplings, dipolar couplings, or chemical shift frequencies, the assignment of all resonance to specific sites in the protein, and then the calculation of structures. There are examples of the application of this approach to membrane proteins in micelles (Almeida and Opella 1997) and bilayers (Opella et al. 1999). The availability of orientational information associated with individual resonances means that it is now possible to make effective use of limited amounts of assignment information, for example, some residue-type assignments or a few sequential assignments. It may also be feasible to implement an “assignment-free” approach. The use of either limited or no assignment information prior to calculating structures would greatly speed the process of structure determination by NMR spectroscopy, especially in the case of membrane proteins where assignments are difficult to make in nearly all situations due to overlap of resonances and unfavorable relaxation parameters.
Results and discussion
Dipole–dipole interaction
The local field, which results from the interaction between two nearby nuclei, is a direct source of structural information. Pake’s (1948) seminal paper demonstrated that the dipole–dipole interaction between two spin S = 1/2 nuclei is manifested as a doublet in NMR spectra, with the frequency difference a function of not only the distance between the two nuclei but also the angle between the internuclear vector and the direction of the applied magnetic field. The dipole–dipole interaction provides direct access to geometrical parameters that can be translated into molecular structures. Moreover, it is important for many aspects of solid-state NMR spectroscopy; for example, it is essential to minimize its influence through decoupling to obtain well-resolved spectra. In this regard, it is generally easier to deal with heteronuclear rather than homonuclear dipolar couplings. Heteronuclear dipolar couplings are used extensively to determine the structures of proteins, in particular the 1H–15N interaction at the amide sites in the protein backbone. Uniform labeling with 15N is particularly valuable in proteins because the properties of a “dilute spin” are retained, since the next nearest amide nitrogen is separated by two carbon atoms in the polypeptide backbone (Cross et al. 1982). In addition, each 15N label in an amide site provides three spin interactions for analysis: the 15N chemical shift, the 1H chemical shift, and, of course, the 1H–15N heteronuclear dipolar coupling between the two directly bonded nuclei.
The dipole–dipole interaction is anisotropic; therefore, the value of the splitting varies with molecular orientation. It is maximal for an N–H bond parallel to the field, half-maximal when the bond is perpendicular to the field, and zero when the bond is at the “magic angle”. All of these possibilities are observed in experimental data from aligned proteins. The 1H–15N heteronuclear dipolar interaction has the dual roles of providing a mechanism for resolving among resonances with N–H bonds at different orientations and of providing the input for structure determination in the form of frequency measurements that can be translated into angles between individual bonds and the external axis imposed by the magnetic field. The angular information can then be used in conjunction with the well-established geometry of peptide planes to determine the three-dimensional structure of the polypeptide backbone (Opella et al. 1987). These methods can be extended to additional nitrogen and carbon sites for characterization of side chain conformations.
Separated local field spectroscopy (Waugh 1976) combines several of the elements of high-resolution solid-state NMR spectroscopy to average out the unwanted broadening influences of homonuclear dipolar couplings and double-resonance and multidimensional spectroscopy to average out and separate the heteronuclear dipolar couplings in different parts of the experiment. The chemical shift dimension in two-dimensional separated local field spectra is intrinsically high resolution because it is obtained while decoupling the hydrogens to remove the broadening due to heteronuclear dipolar couplings. Homonuclear dipolar couplings are minimal among the dilute nuclei and generally do not require attention. This enables the dipolar couplings between bonded pairs of 1H and 15N nuclei to be measured for individual 15N sites with different chemical shift frequencies. The original versions of separated local field spectroscopy have more than adequate resolution for studies of peptides or specifically or selectively labeled proteins. However, further improvements in resolution were needed for studies of uniformly 15N labeled proteins.
PISEMA (polarization inversion spin-exchange at the magic angle) (Wu et al. 1994) is a high-resolution version of separated local field spectroscopy. Line widths in the key dipolar frequency dimension are reduced by more than one order of magnitude compared with the conventional separated local field experiment. The combination of narrow lines and favorable scaling factor has such a dramatic effect on the appearance of the spectra that it is now feasible to formulate solid-state NMR experiments where heteronuclear dipolar coupling frequencies complement chemical shifts as a mechanism for spectroscopic resolution as well as the measurement of readily interpretable orientationally dependent frequencies.
PISA (polarity index slant angle) wheels
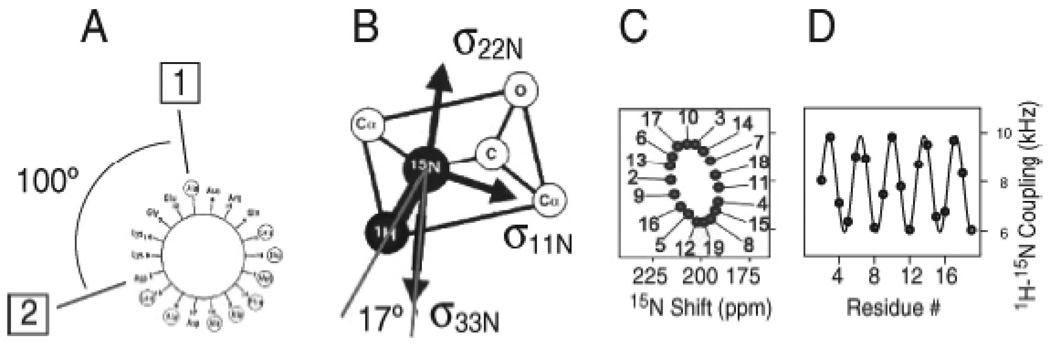
The secondary structure and topology of a membrane protein can be described by the patterns of resonances observed in two-dimensional PISEMA spectra of uniformly 15N labeled polypeptides in aligned bilayers (Marassi and Opella 2000; Wang et al. 2000). The characteristic “wheel-like” patterns observed in these spectra reflect helical wheel projections of residues in both transmembrane and in-plane helices. Therefore, PISA wheels provide direct indices of both secondary structure and topology. The resonance frequencies in both the 1H–15N heteronuclear dipolar and 15N chemical shift dimensions in PISEMA spectra of aligned samples of membrane proteins depend on helix orientation as well as on backbone dihedral angles, the magnitudes and orientations of the principal elements of the amide 15N chemical shift tensor, and the N–H bond length. It is possible to calculate spectra for any protein structure (Bak et al. 2002). The principles involved in the PISA wheel analysis of helices (Marassi and Opella 2000) are illustrated in Fig. 2. In Fig. 2A, the projection down the axis of a helical wheel shows that the 3.6 residues per turn periodicity characteristic of an α-helix results in an arc of 100° between adjacent residues. The drawing of a peptide plane in Fig. 2B shows the orientations of the principal axes of the three operative spin interactions at the 15N-labeled amide site. The 17° difference between the N–H bond axis and the σ33 principal element of the amide 15N chemical shift tensor is of particular importance because of its impact on the spectral appearance of a PISA wheel. The striking wheel-like pattern of resonances calculated from a two-dimensional PISEMA spectrum of an ideal helix is shown in Fig. 2C. A PISA wheel reflects the slant angle (tilt) of the helix, and the assignment of the resonances reflects the polarity index (rotation) of the helix.
Fig. 2.
Principles of PISA wheels (Marassi and Opella 2000). (A) Helical wheel showing the 100° arc between adjacent residues that is a consequence of the periodicity of 3.6 residues per turn in an α-helix; (B) orientations of the principal elements of the spin interaction tensors associated with 15N in a peptide bond; (C) PISA wheel for an ideal α-helix; (D) dipolar wave for an ideal α-helix.
When the helix axis is parallel to the bilayer normal, all of the amide sites have an identical orientation relative to the direction of the applied magnetic field, and therefore, all of the resonances overlap with the same dipolar coupling and chemical shift frequencies. Tilting the helix away from the membrane normal results in variations in the orientations of the amide N–H bond vectors relative to the field. This is seen in the spectra as dispersions of both the heteronuclear dipolar coupling and the chemical shift frequencies. Nearly all transmembrane helices are tilted with respect to the bilayer normal, and it is the combination of the tilt and the 17° difference between the tensor orientations in the molecular frame that makes it possible to resolve many resonances from residues in otherwise uniform helices and is responsible for the wheel-like pattern in PISEMA spectra, such as that illustrated in Fig. 2C.
Dipolar waves
Dipolar waves can serve as maps of protein structure in NMR spectra of both weakly and completely aligned samples (Mesleh et al. 2002). The periodicity inherent in secondary structure elements is key to the use of both PISA wheels and dipolar waves as indices of secondary structure and topology in membrane proteins. Figure 2C displays a classical two-dimensional PISA wheel of an α-helix. Figure 2D illustrates the periodic wave-like variations of the magnitudes of the static heteronuclear dipolar couplings as a function of residue number. Similar patterns are observed in residual dipolar couplings measured for weakly aligned proteins.
Structure determination of the acetylcholine M2 ion-channel peptide
We determined the three-dimensional structure of a functional peptide corresponding to the M2 segment from the α-subunit of the acetylcholine receptor (AchR) by both solution NMR and solid-state NMR methods (Opella et al. 1999). The relatively large quantities of isotopically labeled M2 peptides required for KMR spectroscopy were prepared by expression of recombinant peptides in Escherichia coli. The incorporation of the M2 peptides into lipid bilayers reconstitutes functional, cation-selective channels; the single channel currents recorded from AchR M2 in lipid bilayers, under voltage clamp conditions, have heterogeneous conductances and lifetimes, as expected for monomeric peptides that self-assemble into conductive oligomers of discrete yet variable size. These results indicate that the recombinant and M2 peptide used in the NMR experiments is functional and forms sequence-specific, discrete ion channels in lipid bilayers.
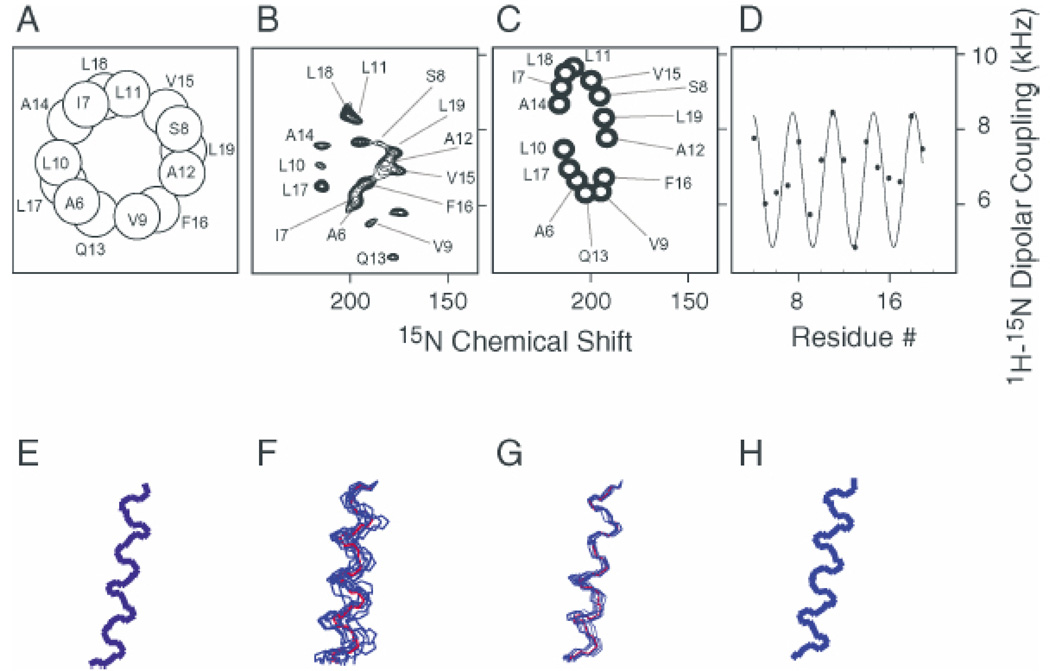
The experimental two-dimensional PISEMA spectrum in Fig. 3A of a sample of the uniformly 15N-labeled M2 peptide in aligned bilayers has excellent resolution, with each amide resonance characterized by 15N chemical shift and 1H–15N dipolar coupling frequencies. The PISA wheel and dipolar wave derived from the experimental data are shown in Figs. 3C and 3D, respectively. The experimental PISEMA spectrum in Fig. 3B is similar to that of the PISA wheel in Fig. 3C, which was calculated for an ideal transmembrane α-helix tilted by 12° with 3.6 residues per turn, as shown in Fig. 3H. Differences between the experimental spectrum and the calculated spectrum are largely due to deviations between the experimentally determined backbone dihedral angles and those of an ideal α-helix and differences among the chemical shift tensors. Without making a single resonance assignment, comparison of the experimental and calculated spectra leads to the conclusion that the AchR M2 helix is transmembrane with a tilt angle of approximately 12°. A similar tilt angle is determined from the dipolar wave fitted to the experimental data in Fig. 3D.
Fig. 3.
Experimental data and PISA wheel and dipolar wave for the AchR. M2 ion channel peptide. (A) Helical wheel; (B) experimental PISEMA spectrum; (C) PISA wheel for an ideal α-helix with a 12° tilt; (D) dipolar wave using the experimental dipolar couplings and resonance assignments in Fig. 3B; (E) structure calculated from the experimental data and resonance assignments in Fig. 3B; (F) superposition of 10 structural fits to the data in Fig. 3B with no assignment information; (G) superposition of 10 structural fits to the data in Fig. 3B with three residue type assignments; (H) ideal α-helix with a 12° tilt corresponding to the PISA wheel in Fig. 3C.
In the experimental PISEMA spectrum of the peptide, the locations of assigned resonances in the wheel-like pattern are nearly identical to those in the helical wheel projection of the peptide also shown in Fig. 3A. Thus, the polarity of the resonances observed in the wheel-like pattern of a PISEMA spectrum provides a direct measure of the angle of the helix rotation about its long axis within the membrane. In principle, one well-resolved two-dimensional PISEMA spectrum of an aligned sample of a uniformly 15N labeled protein provides sufficient information for complete structure determination. The orientationally dependent frequencies associated with each resonance depend on the magnitudes and orientations of the principal elements of the spin interaction tensors in the molecule and on the orientation of the molecular site with respect to the direction of the applied magnetic field. Because the orientation of the bilayer is fixed by the method of sample preparation and the properties of the nuclear spin interaction tensors are generally well characterized, each frequency reflects the orientation of a specific site in the protein with respect to the bilayer. The backbone structure of a protein is defined by the planes formed by the individual rigid peptide bonds and their directly bonded atoms. This is equivalent to the conventional description by vectors representing bonds between nonhydrogen atoms. A standard planar peptide geometry serves as the building block for the structure assembly process. The orientation of a peptide plane consistent with the measured NMR frequencies is defined in terms of polar angles relative to the magnetic field. The input to the computer program consists of the Is MR. frequencies measured from a PISEMA spectrum, the magnitudes and orientations of the principal elements of the amide 15N and 1H chemical shift tensors, and the N–H bond length. Once the orientations of all of the peptide planes in the protein are determined from the experimental data, neighboring planes of fixed orientation are connected through their common α-carbon atom, with the only constraint of a fixed tetrahedral angle of 110°. Another program calculates the ϕ and χ dihedral angles for the two contiguous peptide plane combinations that satisfy tetrahedral angle geometry at the α-carbon. The structure of the AchR peptide determined in this way is shown in Fig. 3E.
The advantage of this direct mathematical analysis is that it is possible to determine the standard deviations in ϕ and μ based on uncertainties in the experimentally determined angles. Another important advantage of the method is that is results in the “piecewise” assembly of structures. Different parts of the protein structure can be determined independently of the rest. This is not possible in methods where distance constraints between different regions of the protein must be established for three-dimensional structure determination. Because the orientation of individual peptide planes is determined relative to a unique external reference, the corresponding errors are not cumulative. However, it does require complete sequential resonance assignments before the structure can be calculated.
Structural fitting
It is also possible to fit structures to the resonance patterns in experimental PISEMA spectra (A. Nevzorov and S.J. Opella, personal communication). This is an alternative approach to structure determination that can be performed where there is either no or only limited amounts of assignment information available. To perform a structural fitting of the NMR data, it is necessary to relate the geometry of a peptide to its spectrum. This starts with the definition of a chain propagator that walks across the spectrum from the resonance of one residue (i) to that of the next residue in the sequence (i + 1). No information from experimental assignments is used. The “assignments” selected by the algorithm vary from those that were made experimentally; nonetheless, the resulting structures are very similar to the “correct” structure shown in Fig. 3E. Ten random shuffles of the order of the resonances resulted in the 10 similar structures that are superimposed in Fig. 3F. The structural fits shown in Fig. 3G also for 10 random shuffles have root mean square deviations (RMSDs) of 1.6 Å (1 Å = 0.1 nm) or less because three residue type assignments are used. In Fig. 3F where no assignment information is used, the RMSDs range from 1.8 to 2.2 Å. The structural fitting algorithm can also be used to determine deviations from an ideal α-helical structure, such as helix bend or twist.
Conclusions and future prospects
NMR structural studies of membrane proteins yield valuable insights into their structure and topology. For example, the structure of the M2 peptide determined by solid-state NMR spectroscopy in lipid bilayers is tilted by 12° and rotated about its helix axis so that the hydrophilic residues face the N-terminal side of the membrane. This has important consequences for ion channel pore geometry and conduction, as it leads to the assembly a symmetric, pentameric, and funnel-like pore with its wide opening at the N-terminal side of the membrane. All of these conclusions about the structure of the peptide in bilayers are immediately apparent from inspection of the assigned PISEMA spectrum prior to complete structure determination. The three-dimensional structures calculated from NMR data have atomic resolution.
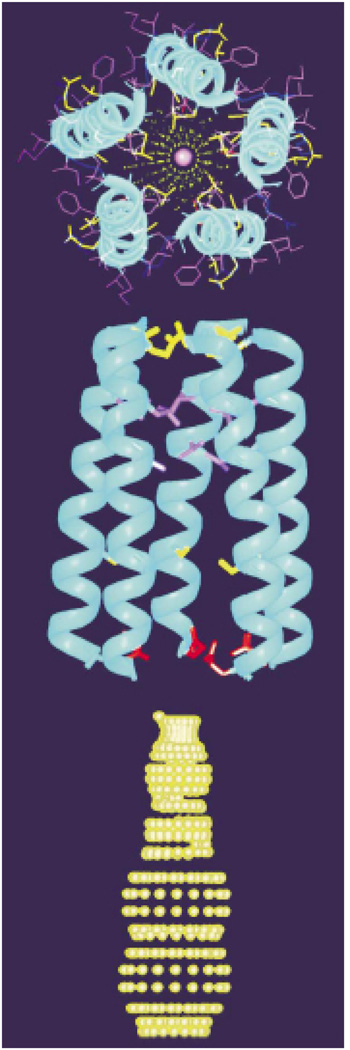
Figure 4 shows a model of the AChR channel pore, constructed from the three-dimensional structure of the AChR M2 helix in lipid bilayers and the known pentameric organization of the channel. The optimized pentameric bundle has a right-handed, interhelical twist with an orientation angle of 12°. A central narrow pore has a diameter ranging from about 3.0 to 8.6 Å. Nonpolar residues are predominantly on the exterior of the bundle, while polar residues line the pore. The residues exposed to the pore lumen are Glu-1, Ser-4, Ser-8, Val-15, Leu-18, and Gln-22, which is in agreement with evidence collected from mutagenesis, affinity labeling, and cysteine accessibility measurements. A side view shows a funnel-shaped bundle, 33 Å in length, with the wide month at the N terminus. A dotted contour depicting the profile of the ion conduction pore calculated from the structure is shown. The channel-lining hydroxyl residues are in agreement with the expectations of a water-filled pore, and the constrictions are compatible with the permeation of both Na and K ions and the model of the pore shown in Fig. 4.
Fig. 4.
Pentameric M2 ion channel in lipid bilayers (Opella et al. 1999)
The structure of the polypeptide shown in Figs. 3 and 4 was determined using solid-state NMR methods that are direct descendents of the initial observations of doublets due to local fields in 1948. The advances required to transform solid-state NMR from a spectroscopic technique to a generally applicable method for determining molecular structures included multiple-pulse sequences, double-resonance methods, and separated local field spectroscopy. They also required improvements in instrumentation, especially the use of high-field magnets, and efficient probes capable of high-power radiofrequency irradiations at high frequencies. The pace of development is accelerating, and the local field is being utilized in an increasing number of ways in spectroscopic investigations of molecular structure and dynamics. Applications to many helical membrane proteins are underway and promise to add to our understanding of membrane proteins in health and disease.
Acknowledgements
This research was supported by grants R37GM24266, PO1GM56538, RO1GM29754, and R01CA82864 from the National Institutes of Health and utilized the Resource for Solid-State NMR of Proteins supported by grant P41RR09731 from the Biomedical Research Technology Program, National Center for Research Resources, National Institutes of Health.
Contributor Information
S.J. Opella, Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0307, U.S.A.
A. Nevzorov, Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0307, U.S.A.
M.F. Mesleh, Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0307, U.S.A.
F.M. Marassi, The Burnham Institute, 10901 North Torrey Pines Road. La Jolla, CA 92037, U.S.A.
References
- Almeida FCL, Opella SJ. fd coat protein structure in membrane environments: structural dynamics of a loop connecting a hydrophobic trans-membrane helix and an amphiapathic helix in a membrane protein. J. Mol. Biol. 1997;270:481–495. doi: 10.1006/jmbi.1997.1114. [DOI] [PubMed] [Google Scholar]
- Bak M, Schultz R, Vosegaard T, Nielsen NC. Specification and visualization of anisotropic interaction tensors in polypeptides and numerical simulations in biological solid-state NMR. J. Magn. Reson. 2002;154:28–45. doi: 10.1006/jmre.2001.2454. [DOI] [PubMed] [Google Scholar]
- Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;330:127–172. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NS. Protein NMR spectroscopy. New York: Academic Press; 1996. [Google Scholar]
- Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J. Am. Chem. Soc. 2002;124:2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- Cross TA, DiVerdi JA, Opella SJ. Strategy for nitrogen NMR of biopolymers. J. Am. Chem. Soc. 1982;104:1759–1761. [Google Scholar]
- Griffin RG. Dipolar recoupling in MAS spectra of biological solids. Nat. Struct. Biol. NMR Suppl. 1998;II:508–512. doi: 10.1038/749. [DOI] [PubMed] [Google Scholar]
- Howard KP, Opella SJ. High resolution solid-state NMR spectra of integral membrane proteins reconstituted into magnetically oriented phospholic bilayers. J. Magn. Reson. 1996;112:91–94. doi: 10.1006/jmrb.1996.0116. [DOI] [PubMed] [Google Scholar]
- Ma C, Opella SJ. Lanthanide ions bind specifically to an added “EF-hand” and orient a membrane protein in micelles for solution NMR spectroscopy. J. Magn. Reson. 2000;146:381–384. doi: 10.1006/jmre.2000.2172. [DOI] [PubMed] [Google Scholar]
- Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J. Magn. Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marassi FM, Ramamoorthy A, Opella SJ. Complete resolution of the solid-state NMR spectrum of a uniformly 15N-labeled membrane protein in phospholipid bilayers. Proc. Natl. Acad. Sci. U.S. A. 1997;94:8551–8556. doi: 10.1073/pnas.94.16.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell PA, Opella SJ. Effect of detergent concentration on multidimensional solution NMR spectra of membrane proteins in micelles. J. Magn. Reson. 1993;B102:120–125. [Google Scholar]
- Mesleh MF, Veglia G, DeSilva TM, Marassi FM, Opella SJ. Dipolar waves as NMR maps of protein structure. J. Am. Chem. Soc. 2002;124:4206–4207. doi: 10.1021/ja0178665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opella SJ. NMR and membrane proteins. Nat. Struct. Biol. NMR Suppl. 1997;I:845–848. [PubMed] [Google Scholar]
- Opella SJ, Stewart PL, Valentine KG. Structural analysis of solid-state NMR measurement of peptides and proteins. Q. Rev. Biophys. 1987;19:7–49. doi: 10.1017/s0033583500004017. [DOI] [PubMed] [Google Scholar]
- Opella SJ, Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M. Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nat. Struct. Biol. 1999;6:374–379. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pake GE. Nuclear resonance absorption in hydrated crystals: fine structure of proton line. J. Chem. Phys. 1948;16:327–336. [Google Scholar]
- Sanders CR, Hare BJ, Howard K, Prestegard JH. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Prog. NMR Spectrosc. 1993;26:421–444. [Google Scholar]
- Veglia G, Opella SJ. Lanthanide ion binding to adventitious sites aligns membrane proteins in micelles for solution NMR spectroscopy. J. Am. Chem. Soc. 2000;122:11 733–11 734. [Google Scholar]
- Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. Imaging membrane protein helical wheels. J. Magn. Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- Waugh JS. Uncoupling of local field spectra in nuclear magnetic resonance: determination of atomic positions in solids. Proc. Natl. Acad. Sci. U.S. A. 1976;78:1894–1897. doi: 10.1073/pnas.73.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Ramamoorthy A, Opella SJ. High-resolution heteronuclear dipolae solid-state NMR spectroscopy. J. Magn. Reson. 1994;A109:270–272. [Google Scholar]
- Wuthrich K. The second decade-into the third millennium. Nat. Struct. Biol. NMR. Suppl. 1998;II:492–495. doi: 10.1038/728. [DOI] [PubMed] [Google Scholar]