SUMMARY
Current models of HIV-1 morphogenesis hold that newly synthesized viral Gag polyproteins traffic to and assemble at the cell membrane into spherical protein shells. The resulting late-budding structure is thought to be released by the cellular ESCRT machinery severing the membrane tether connecting it to the producer cell. Using electron tomography and scanning transmission electron microscopy, we find that virions have a morphology and composition distinct from late-budding sites. Gag is arranged as a continuous but incomplete sphere in the released virion. In contrast, late-budding sites lacking functional ESCRT exhibited a nearly closed Gag sphere. The results lead us to propose that budding is initiated by Gag assembly, but is completed in an ESCRT-dependent manner before the Gag sphere is complete. This suggests that ESCRT functions early in HIV-1 release—akin to its role in vesicle formation—and is not restricted to severing the thin membrane tether.
INTRODUCTION
HIV-1 assembly and budding are directed by viral Gag polyproteins, which are thought to assemble at the cell membrane into a spherical protein shell, coincident with the induction of membrane curvature (Demirov and Freed, 2004; Morita and Sundquist, 2004). Once the spherical Gag shell is complete, the resulting late-budding structure (appearing like a lollipop in electron microscopy [EM] sections) is thought to be released by the cellular ESCRT machinery severing the thin membrane tether that connects it to the producer cell. ESCRT is recruited by a PTAP motif in the HIV-1 Gag polyprotein and consists of several large multiprotein assemblies (ESCRT-I, -II, and -III). Gag binds the ESCRT-I component TSG101, while the actual release is thought to be mediated by ESCRT-III, which is disassembled by the VPS4 ATPase during this process (Hurley, 2008; Morita and Sundquist, 2004; Williams and Urbe, 2007).
HIV-1 is initially released as an immature, noninfectious particle comprised of a layer of uncleaved Gag polyproteins underneath the virion membrane. Subsequently, it is converted into the mature, infectious virion by the viral protease cleaving Gag in five distinct positions (Adamson and Freed, 2007). Based on the hexagonal symmetry and measured center-to-center spacing of 8 nm for the immature lattice, it was suggested that an immature HIV-1 particle with an average radius of 73 nm consists of 4900 Gag polyproteins arranged in a spherical layer underneath the virion membrane (Briggs et al., 2004). Since formation of the mature capsid requires only 1200–1500 CA proteins (derived in stoichiometric amounts by proteolytic cleavage of Gag), not all of the Gag molecules appear to be needed for the internal structure of the infectious virion. The presence of surplus CA in mature HIV-1 was supported by hydrogen-deuterium exchange experiments revealing two roughly equal CA populations with different exchange rates (Lanman et al., 2004).
Recently, Wright et al. studied immature HIV-1 particles using cryo-electron tomography (cryo-ET) (Wright et al., 2007). This method permits analysis of the entire three-dimensional structure by reconstructing a three-dimensional volume (tomogram) from a series of tilted projection images of native, frozen particles (Lucic et al., 2005). The cryo-ET study confirmed the overall arrangement and lattice of the Gag layer, but the authors reported this layer to be incomplete, covering roughly 40% of the inner membrane surface. Completeness of the Gag lattice had not been determined in previous ultrastructural studies because of the inherent limitations of sectioned preparations and projection images. Wright et al. also reported that ordered Gag patches in the virion were arranged in an irregular pattern, separated by large regions without ordered density. This observation suggested that HIV-1 budding may occur by the concerted action of multiple Gag patches rather than a continuous Gag lattice, while such structures have not been detected in previous ultrastructural studies. However,notomographic analysisof HIV-1 budding sites has been reported to date, and accordingly, no complete three-dimensional reconstruction of viral budding sites is available.
Here, we present three-dimensional structural data on HIV-1 budding sites and released immature virions, obtained by electron tomography along with scanning transmission electron microscopy (STEM) mass measurements of immature and mature virions. The results lead us to propose that HIV-1 budding is initiated by assembly of a continuous Gag layer obtaining its radius of curvature early in assembly, but is completed in an ESCRT-dependent manner well before the Gag shell is complete. Accordingly, extracellular particles contain an incomplete but continuous Gag layer covering approximately two-thirds of the virion membrane. These findings suggest that ESCRT acts much earlier in HIV-1 release—akin to its role in vesicle formation, where it is involved at all stages—and its function is not restricted to severing the thin membrane tether of the completed late bud. The observed late-budding structures may thus represent assembly complexes lacking a functional ESCRT machinery where Gag can reach its equilibrium assemblage, but do not appear to be normal precursors of extracellular HIV-1.
RESULTS
The Gag Layer in Immature HIV-1 Particles
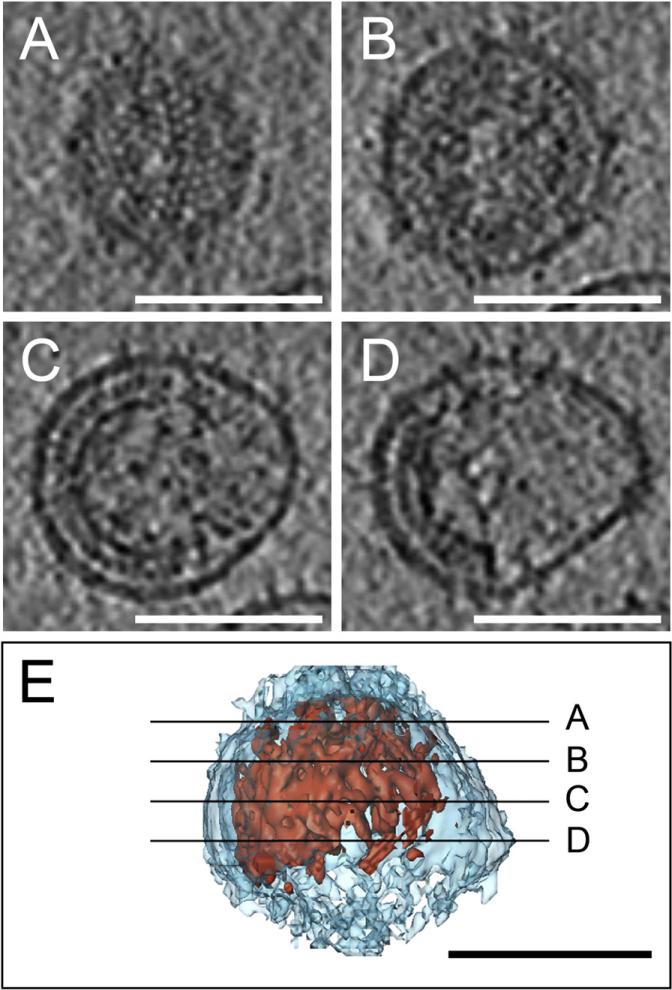
Immature HIV-1 particles were produced in infected MT-4 cells cultured in the presence of a specific inhibitor of the viral protease, and complete inhibition of Gag processing was verified on silver stained gels (Figure S1). Purified particles were vitrified by plunge-freezing into liquid ethane and subjected to cryo-ET. Nine tilt series were recorded on two independent virus preparations. From visual inspection of the resulting tomograms, the Gag density appeared as one continuous Gag layer in each virion, typically covering between half and roughly three-quarters of the membrane surface (see Movie S1). Figure 1 shows a rendering of a typical particle and sections through different parts of the virion. The Gag lattice appears complete at the top (Figure 1A) while being clearly incomplete at the center (Figure 1C) and lower part (Figure 1D) of the same particle. Thirty virions reconstructed with sufficient quality were selected for further quantitative analysis.
Figure 1. Gag Organizes as a Truncated Sphere in Cryo-Electron Tomograms of Immature HIV-1.
(A–E) Four computational slices through a cryo-electron tomogram of an immature HIV-1 particle are shown (A–D, top) along with a surface model showing the position of the slices in the 3D volume. The slices are 2 nm thick and placed 20 nm apart. The membrane is depicted in light blue, Gag in red. Scale bars are 100 nm.
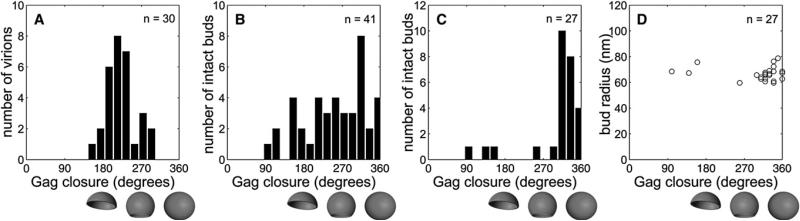
Since the Gag layer appeared continuous in all reconstructions, its closure can be described by an angle ranging from 0° to 360°, with 180° representing a half sphere and 360° representing a closed sphere (Figure 2A). The average closure of the 30 selected immature virions was 229° ± 35° (equivalent to a completeness of 70% ± 13%) (Table 1 and Figure 2A). No particles with a Gag layer closure >300° were detected. Knowing the closure and radius of the Gag layer, its area can be derived. The lattice arrangement and spacing of the immature Gag lattice has been measured (Briggs et al., 2004), providing the number of Gag proteins that are required to cover a given area of membrane. The resulting number of Gag proteins per virion is highly dependent on the particle diameter, and HIV-1 sizes were found to be variable in different preparations. A virus of 73 nm radius with a complete Gag shell would thus contain ~4900 Gag proteins, as reported in our previous study (Briggs et al., 2004). If the Gag layer is only 70% complete, 3500 Gag proteins would be required for a virus with the same radius. The 30 HIV-1 particles measured here had a radius of 63 ± 5 nm, corresponding to an average Gag content of 2400 ± 700 molecules for a 70% complete Gag shell (Table 1). To confirm that the incomplete Gag shell is a not a specific feature of virions produced from MT4 cells, we also performed cryo-ET on immature virions purified from HeLa cells transfected with a complete HIV-1 proviral plasmid. These cells produce lower amounts of HIV-1, but the purified virions exhibited a similarly incomplete Gag lattice (Movies S2 and S3).
Figure 2. The Gag Layer Is Less Closed in Released Virions Than in Late-Budding Sites.
(A–C) Gag closure measurements for released immature particles (A), cellular budding sites from the release competent (B), and budding arrested groups (C) (see Table 3 for constructs used).
(D) The radius of the bud (from the center to the plasma membrane) was plotted against the closure of the Gag layer for the 27 fully contained buds from the budding arrested group. The radius of curvature was similar in early and late buds. A closure of 180° corresponds to a half-sphere and 360° to a full sphere, as depicted underneath the panels.
Table 1.
Quantitative Data from Tomograms of HIV-1 Budding Sites and Released Immature Virions
| Released immature virions (n = 30) | Buds in release-competent group (n = 41) | Buds in budding-arrested group (n = 27) | |
|---|---|---|---|
| Radius (nm) | 63 ± 5 | 75 ± 7 | 67 ± 5 (66 ± 5) |
| Closure (degrees) | 229 ± 35 | 253 ± 73 | 306 ± 66 (328 ± 22) |
| Number of Gag molecules | 2400 ± 700 | 4100 ± 1700 | 3600 ± 1100 (3800 ± 770) |
Radius of curvature and Gag shell closure was measured for particles and for buds fully contained in the sections, and the number of Gag molecules was calculated from these numbers and the known lattice constant (see Supplemental Data). For definition of release-competent and budding-arrested groups, see Table 3. In the budding-arrested group, the numbers in parentheses indicate the values when the three budding sites that did not have a late bud morphology (i.e., Gag closure >180°) were discarded.
As an independent complementary approach, mass measurements of individual immature and mature HIV-1 virions were performed using STEM. This technique permits the mass determination of individual freeze-dried particles (Vogt and Simon, 1999; Wall et al., 1998). Tobacco mosaic virus (TMV) was analyzed in the same preparations to calibrate all measurements. Parallel HIV-1 preparations were produced in MT-4 cells in the presence and absence of a specific HIV protease inhibitor, and the purified virus was inactivated with either paraformaldehyde (PFA) or 2-aldrithiol (AT-2) (Rossio et al., 1998). Since the mode of inactivation had no significant influence on virus mass (Table 2), the data sets were pooled. The TMV-corrected mass for mature HIV-1 was 276 ± 88 MDa (n = 258) and for immature HIV-1 was 281 ± 82 MDa (n = 84) (Table 2). Thus, the mass (and presumably Gag content) of immature and mature HIV-1 was not significantly different, and the data sets were pooled to give a mean mass of 277 MDa (Table 2 and Figure S2). From the mass, an estimate of the total number of Gag proteins contained within a particle can be derived using available information about the stoichiometry of other known components that have been quantified relative to Gag in previous studies (see Table S1 for details). From this calculation, we estimate Gag to contribute slightly less than 50% of total virus mass, corresponding to 133 MDa in these preparations. This equals approximately 2400 copies of Gag and is in excellent agreement with the calculation from the cryo-ET measurements described above. Aliquots of the same preparations as analyzed by STEM were also imaged by cryo-EM, and a mean particle radius of 66 nm was measured directly from the images (Table 2). Based on the known packing density of Gag (Briggs et al., 2004), approximately 3900 copies of Gag would be required to form a completely closed Gag lattice within a particle of this radius. The measured 2400 copies of Gag correspond to 61% of this number (Figure S2) or a closure of the Gag layer of 209° for these preparations.
Table 2.
STEM Mass and Cryo-EM Size Measurements on Released Immature and Mature HIV-1
| Immature + PFA | Mature + PFA | Mature + 2AT | Total | |
|---|---|---|---|---|
| Cryo-EM radius (nm) | 65 ± 17 (n = 150) | 65 ± 8 (n = 150) | 69 ± 16 (n = 150) | 66 ± 17 (n = 450) |
| STEM radius (nm) | 64 ± 16 (n = 84) | 60 ± 18 (n = 162) | 61 ± 17 (n = 96) | 61 ± 17 (n = 342) |
| STEM HIV-1 mass (Mda) | 319 ± 93 (n = 84) | 292 ± 89 (n = 162) | 304 ± 103 (n = 96) | 302 ± 94 (n = 342) |
| STEM TMV mass (kDa/Å) | 14.86 ± 1.63 (n = 168) | 14.06 ± 0.81 (n = 324) | 14.11 ± 0.90 (n = 192) | 14.27 ± 1.14 (n = 684) |
| Corrected STEM HIV-1 mass (Mda) | 281 ± 82 (n = 84) | 272 ± 83 (n = 162) | 282 ± 96 (n = 96) | 277 ± 86 (n = 342) |
Immature and mature HIV-1 (NL4-3) particles were purified from infected MT-4 cells and inactivated with either PFA or 2AT. Size measurements by cryo-EM were performed on the same virus preparations used for STEM. Since there was no significant change in mass or size due to the method of inactivation or proteolytic maturation, these data were pooled to yield the numbers in the last column (total). The true TMV mass per length is 13.1 kDa/Å, and the ratio of measured and true TMV mass was used for calibration to calculate the corrected HIV mass.
Electron Tomography of HIV-1 Budding Sites
Next, we wanted to study the morphology of the cellular HIV-1 budding sites. Quantitative analysis of a sufficient number of such structures is currently only feasible using sectioned material, and we chose electron tomography of plastic embedded sections for this part of the study to allow a higher throughput and more flexibility in the choice of cell types. Electron tomography was performed on thick sections (300 nm nominal thickness) of fixed, resin-embedded cells producing HIV-1. Thirty-six tilt series were recorded, containing 145 budding sites in total, of which 68 were fully contained within the section (i.e., not partially cut away in the sectioning process). Producer cells were either MT-4 cells infected with HIV-1 or HeLa cells transfected with the noninfectious HIV-1 expression plasmid pΔR (Gottwein and Kraüsslich, 2005), which contains all viral open reading frames and yields particles morphologically indistinguishable from HIV-1. To induce a late-budding arrest, we either used a pΔR variant that does not recruit ESCRT due to mutation of the PTAP motif (Gottwein and Kraüsslich, 2005) or performed cotransfection of pΔR with an expression vector for dominant-negative VPS4A (dnVPS4A) (von Schwedler et al., 2003) (Table 3).
Table 3.
Data Collected on Budding Sites in Resin-Embedded Sections
| Virus or construct | Cell type | Number of tomograms | Number of budding sites | Number of budding sites fully contained within sections |
|---|---|---|---|---|
| HIV-1, strain NL-4-3 | MT-4 cells | 19 | 78 | 36 |
| pΔR | HeLa | 2 | 6 | 5 |
| Total, release-compentent group | 21 | 84 | 41 | |
| pΔR-PTAP(–) | HeLa | 7 | 39 | 16 |
| pΔR + dnVPS4A | HeLa | 8 | 22 | 11 |
| Total, budding-arrested group | 15 | 61 | 27 |
The number of recorded tomograms is stated for each construct, along with the total number of budding sites in these tomograms, and the number of budding sites that were fully contained within the sections, i.e., not sliced off in the sectioning. All data analysis was performed on this subset of fully contained structures. See the Experimental Procedures section for details of data-collection parameters.
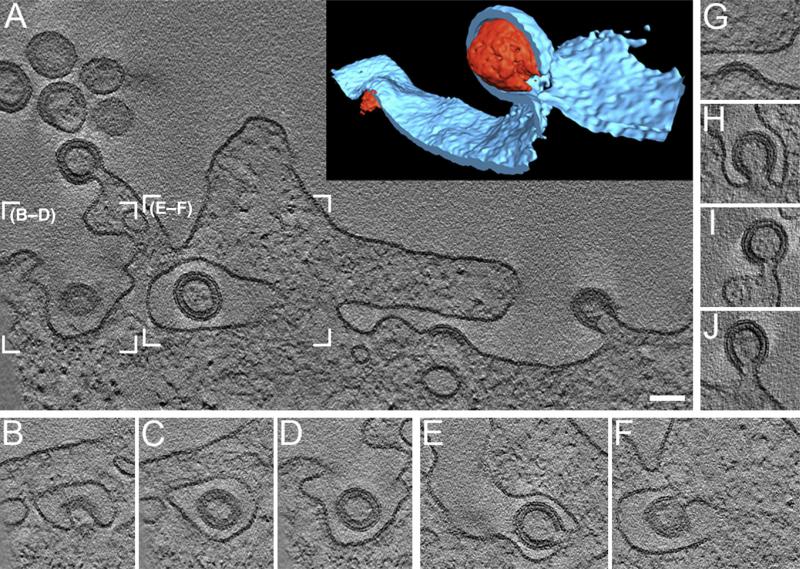
Figure 3 and Movie S4 show a representative tomogram from a HeLa cell transfected with PTAP-deficient pΔR. The tomogram was recorded on an area of the plasma membrane with high budding activity. Such areas were often found to be clustered on small parts of the cell surface, separated by larger areas where no budding was detected. While the integrity of actin filaments and other cytosolic components was compromised by the fixation and staining, membranes and Gag protein layers appeared intact and well preserved. All HIV-1 budding sites contained a single, continuous, membrane-apposed Gag layer. Since the tomograms used for quantitative analysis included the entire HIV-1-specific structure, possible membrane continuity with the host cell and the relative budding stage (early/late) could be unequivocally determined, avoiding the ambiguity present in studies of thin sections. For example, Figures 3B–3F show computational z sections through two parts of the tomogram in Figure 3A; Figure 3B clearly reveals a late HIV-1 bud, while the same structure appears like a released, immature virion in the other sections (Figures 3C and 3D). Furthermore, this seemingly extracellular particle appears to be localized within an intracellular compartment in Figure 3C, but this is unequivocally identified as invagination of the plasma membrane in Figure 3D. A similar situation was observed for the adjacent structure in the tomogram, where two late buds protrude into a membrane invagination. In Figure 3A, this appears as an immature particle within an intracellular compartment, but in the section displayed in Figure 3F, this particle is identified as a membrane-connected late bud, and Figure 3E reveals the continuity with the plasma membrane, along with the second late-budding structure.
Figure 3. Tomograms of Cellular Budding Sites Reveal Gag Ultrastructure and Membrane Connectivity.
(A) A 2 nm thick computational slice through a tomogram of a HeLa cell expressing a budding-arrested (PTAP mutation) HIV-1 construct. The inset in the upper right corner is a surface model of the two budding sites in the right part of the slice, with the membrane (cut open) in light blue and Gag in red. (A movie of the entire tomogram is available as Movie S1).
(B–D) Slices through the tomogram at the position indicated by the left box in (A), at different z positions (–82 nm, –61 nm, and –18 nm with respect to A, respectively).
(E–F) Slices through the tomogram at the position indicated by the right box in (A), at –69 nm and +23 nm, respectively.
(G–J) Central slices through four of the fully contained budding sites in this tomogram, rotated so that the center of the Gag density points upwards. Scale bar is 100 nm.
The stalk regions of the late buds were carefully examined for potential differences, dependent on the construct(s) used for transfection. According to theory, the PTAP-deficient Gag molecules would be expected to be incapable of ESCRT recruitment, while cotransfection of dnVPS4A might prevent disassembly of ESCRT complexes at the budding site; however, no consistent morphological difference between these two experimental conditions could be identified at the resolution of the images analyzed in this study. Therefore, the tomograms were grouped as release competent (HIV-1 infection or pΔR transfection) and budding arrested (PTAP-deficient or dnVPS4A cotransfection), and fully contained budding sites in each of these groups were pooled for quantitative analysis (41 and 27 buds, respectively) (Table 3).
As described for the released particles, each fully contained budding site was characterized by its radius of curvature and the closure of the Gag layer (Table 1). Both early- and late-budding structures were detected in the tomograms, providing a sequential three-dimensional view of this process (Figures 3G–3J). Plotting radius of curvature against the degree of closure for the budding-arrested group (Figure 2D) revealed that the few early-budding sites observed had a similar radius of curvature as the nearly closed spheres. This indicates that the radius of curvature is determined early in the budding process.
The 41 budding sites in the release-competent group showed a broad distribution of closures from early-budding sites to almost closed spheres (average closure 253°) (Table 1 and Figure 2B). Further, they had a larger radius than both the arrested buds and the released immature particles (Table 1). On the other hand, the 27 budding sites comprising the budding-arrested group exhibited a significantly more closed Gag shell than the extracellular virions (Wilcoxon rank sum test, p = 7 × 10–7). The arrested late-budding structures had a radius of 66 ± 5 nm, similar to the immature particles, but an average closure of the Gag layer of ~330° (Figure 2C and Table 1). This corresponds to 3800 Gag molecules in the arrested late-budding structures for these preparations. To determine whether the opening in the Gag layer detected in all HIV-1 budding sites analyzed could have been caused by the fixation and embedding procedure, we performed tomography of thick sections of cells expressing a budding-arrested variant (PSAP deficient) of Mason-Pfizer Monkey Virus (M-PMV) (Gottwein et al., 2003). M-PMV is a beta-retrovirus whose Gag polyproteins assemble in the cytoplasm into spherical procapsids, which are subsequently enveloped at the plasma membrane. In this case, a completely closed spherical Gag shell was observed for 10 of the 11 fully contained budding sites in the tomograms (data not shown), thus indicating that the incomplete Gag shells detected in the case of HIV-1 were not the result of sample preparation.
DISCUSSION
Combining structural studies on cellular HIV-1 budding sites with structural data and mass measurements of released virions, we have focused our attention on the assembly of the Gag layer at the budding site and how this is linked to viral release. The results of this three-dimensional quantitative study of HIV-1 morphogenesis yield a refined quantitative estimate for virion composition and suggest that HIV-1 release results from concerted rather than sequential action of Gag and ESCRT.
Our tomographic analysis of immature HIV-1 particles confirms the recent findings by Wright et al. (Wright et al., 2007), who reported that the Gag lattice is incomplete. We found, however, that Gag molecules arrange as a continuous truncated sphere, typically covering approximately two-thirds of the viral membrane, while Wright et al. reported ~40% coverage and a patchy distribution of ordered Gag segments. Incompleteness of the Gag lattice in the immature virion could be due either to budding of the virus before the Gag shell is closed or to partial disassembly of the Gag shell after release. The two possibilities can be distinguished by mass determination, since disassembled Gag molecules would remain enclosed in the extracellular virion. STEM analysis of individual freeze-dried HIV-1 particles revealed virtually identical masses for immature and mature virions, corresponding to a Gag membrane coverage of ~61% and an average of 2400 Gag molecules per virion (with an average radius of 66 nm). The close correspondence of these measurements with the cryo-ET data shows that postrelease disassembly cannot be the reason for the incomplete immature Gag lattice.
Given that the ~70% Gag coverage derived from our cryo-ET measurements is consistent with that from STEM-based mass determination, it appears likely that the lower coverage observed by Wright et al. is due to experimental differences and the measurement of different properties. Whereas we have manually fitted a truncated sphere model to the part of the membrane covered by Gag density, Wright et al. imposed the stricter criterion of density with local six-fold symmetry. Given the noisy reconstructions of cryo-ET, this criterion can be expected to yield a lower coverage. Second, Wright et al. used noninfectious HIV-1 particles purified from 293T cells, while we have analyzed inactivated (and thereby briefly fixed) HIV-1 from MT-4 cells. Both the PFA fixation as well as differences in the lipid composition of the viral envelope dependent on the producer cell (B.G. and H.-G.K., unpublished data) may have increased the stability of the Gag lattice in our case. A less stable lattice resulting in partial disassembly during sample preparation could also explain the patchy appearance of the Gag lattice described by Wright et al.
It should be noted that an incomplete Gag layer and the resulting lower Gag stoichiometry are more compatible with previous biochemical measurements than the large number of Gag molecules required to form a complete shell. Incompleteness of the lattice was not detected in the previous study because of inherent limitations of projection images, and in vitro-assembled particles were used as a proxy for the Gag shell in the virion (Briggs et al., 2004). More recent three-dimensional analysis of such in vitro-assembled particles confirmed that they are more complete than immature virus particles (data not shown), thus validating the previously reported stoichiometry for these particles. One should also emphasize that the revised number of Gag molecules is still double the 1200–1500 CA molecules needed for formation of the mature cone-shaped capsid shell (Briggs et al., 2003). This corresponds well to results of the hydrogen-deuterium exchange experiment, which indicated that ~50% of CA in the mature virion shows rapid exchange and may thus not be part of the capsid shell (Lanman et al., 2004). Furthermore, the copy numbers of other virion constituents (current estimates summarized in Swanson and Malim [2008]) need to be revised based on these results, since they are generally defined in relation to Gag. As already stated for Gag, these numbers are dependent on virion size, which is highly variable in the case of HIV-1, and we therefore provide estimates for different virion diameters in Table S1. Virions with a radius of 66 nm (or 73 nm) and a 61% complete Gag shell would thus contain ~340 (~420) molecules of Vpr (1:7) (Müller et al., 2000), ~240 (~300) molecules of cyclophilin A (1:10) (Franke et al., 1994), and ~120 (~150) molecules of integrase and reverse transcriptase (1:20) (Welker et al., 1996) (see Table S1).
The result that the Gag shell in released immature HIV-1 is only approximately two-thirds closed already indicates that the well-described late-budding structures displaying “lollipop” morphology may not be direct precursors of the extracellular virus. This was confirmed by quantitative three-dimensional analysis of budding-arrested structures. Their Gag layers were almost completely closed, independent of whether release was blocked by mutations in Gag preventing recruitment of ESCRT or mutations in VPS4A preventing ESCRT disassembly and recycling. Thus, the so-called late-budding structure contains many more Gag molecules than the released virus and can therefore not be an intermediate in HIV-1 morphogenesis. The almost complete spherical Gag shell found in in vitro-assembled particles and arrested budding sites may represent the equilibrium geometry, which is reached due to a prolonged lifetime of arrested or dead-end structures at the membrane. Consequently, wild-type HIV-1 release must occur before the assemblage reaches its steady state geometry, most likely by action of the ESCRT machinery. Notably, budding sites in the release-competent group exhibited a different geometry from both budding-arrested structures and released immature virions. Their Gag layers were found in a broader continuum of states, with many buds having a more closed Gag shell than the immature virions. This, combined with their slightly larger average radius, prompts us to speculate that those “wild-type” budding structures observed by EM (with a calculated average Gag content of 4100 molecules) may represent a mixture of productive budding events and a class of delayed budding structures that are not part of the productive release pathway.
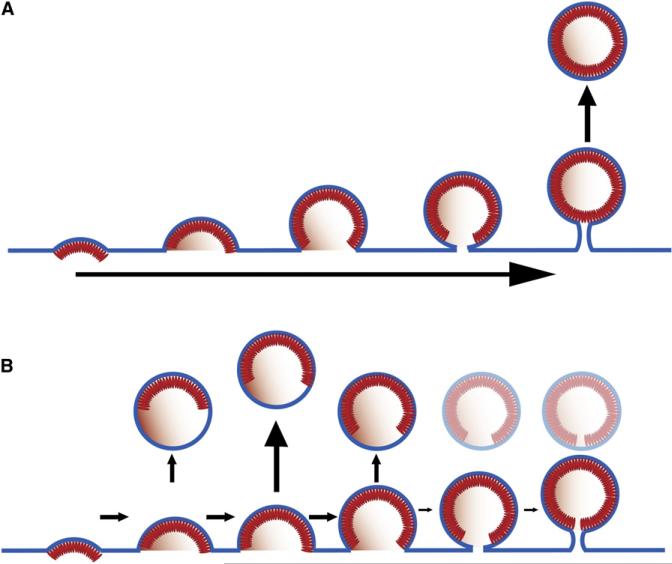
The current model of HIV-1 morphogenesis suggests that an almost closed spherical Gag shell assembles at the plasma membrane concomitant with induction of curvature (budding). The resultant structure is thought to be connected to the cell by a thin membrane tether, which is subsequently severed by the ESCRT machinery (abscission; Figure 4A). The results of this study indicate that the model needs to be revised and that release is triggered by the ESCRT machinery much earlier in the budding process. The average diameter of the membrane at the rim of the Gag layer in the released immature virus preparation was 109 ± 19 nm, compared to only 36 ± 21 nm in the arrested buds. We therefore speculate that ESCRT functions by closing the bulged membrane early in the budding process (Figure 4B) rather than pinching off a “ready-made virus” (Figure 4A). ESCRT function in HIV-1 release is thus more similar than previously thought to its role in cellular vesicle formation where ESCRT is involved in all stages of sorting and vesiculation and is clearly not limited to the final step of abscission (Hurley, 2008; Williams and Urbe, 2007). It is also tempting to speculate that the accessible rim of the incomplete Gag layer in the immature virus is the site where proteolytic cleavage of Gag and maturation commences.
Figure 4. Two Alternative Models for HIV-1 Assembly and Release.
(A) HIV-1 release occurs after complete assembly of the Gag shell.
(B) Release occurs concomitant with formation of the Gag shell by simultaneous assembly and ESCRT-mediated vesiculation, as suggested by this study.
According to the revised model of HIV-1 morphogenesis, Gag recruits ESCRT-I, and consequently ESCRT-III, in a PTAP-dependent manner to the emerging viral bud; and ESCRT-III, in conjunction with VPS4, serves to constrict the wide membrane rim induced by Gag assembly. Thus, ESCRT-mediated release would occur concomitant—and in kinetic competition—with Gag assembly. The revised model suggests that ESCRT-III contributes to membrane bending to complete the budding process, while Gag assembly drives early budding. This role of ESCRT is consistent with the recently solved structure of the ESCRT-III component CHMP3 (Muziol et al., 2006) and supported by two recent studies of the mechanism of ESCRT-III. The first of these reported plasma membrane deformation by ESCRT-III, showing that overexpression of CHMP4A/B together with inactive VPS4B induced bending of the plasma membrane away from the cytoplasm to form buds and tubules protruding from the cell surface (Hanson et al., 2008). The second study reported that CHMP2A and CHMP3 form helical tubes in vitro, with the membrane-binding sites facing the outside (Lata et al., 2008). These tubes could be disassembled by the action of VPS4. A similar membrane-bending function could be envisioned in virus release to complete the budding process. The principle of virus budding proposed for HIV-1 in this report may also apply to other viral systems.
EXPERIMENTAL PROCEDURES
Sample Preparation
HeLa and MT-4 cells were maintained in DMEM and RPMI-1640, respectively, and supplemented with 10% fetal calf serum and antibiotics. Transfections were performed using the calcium phosphate method or FuGENE 6 (Roche). The HIV-1 proviral plasmid pNL4-3 (Adachi et al., 1986), plasmids pDR and its PTAP-deficient variant (Gottwein and Kräusslich, 2005), as well as the expression plasmid for dnVPS4A have been described. The expression plasmid for dnVPS4A was a kind gift of W.I. Sundquist, University of Utah (von Schwedler et al., 2003). Infection of MT-4 cells with HIV-1 strain NL43 by coculture and purification of the virus from tissue culture supernatant by Iodixanol gradient centrifugation was performed as described (Welker et al., 2000), and the same purification protocol was used for virus from transfected HeLa cells. Immature particles were obtained by adding a specific HIV protease inhib itor (either lopinavir at a concentration of 2 μM or saquinavir at a concentration of 5 μM) 6 hr after infection. Samples for cryo-ET or STEM were inactivated using PFA or 2-AT as previously described (Rossio et al., 1998). Samples for electron tomography were fixed and EPON-embedded using standard procedures.
Transmission Electron Microscopy
Data were recorded using a Philips CM300 or a FEI Tecnai F30 Polara transmission electron microscope (FEI; Eindhoven, The Netherlands), both equipped with 300 kV field emission guns; Gatan GIF 2002 Post-Column Energy Filters; and 2k × 2k Multiscan CCD Cameras (Gatan; Pleasanton, CA). All data collection was performed at 300 kV, with the energy filter operated in the zero-loss mode. Cryotilt series were typically recorded from –63° to +63° with an angular increment of 3°. The total dose was between 45 and 70 electrons Å–2. The defocus was between –4.5 and –5.5 μm. For the cryotilt series, the pixel size at the FEI Tecnai F30 Polara was 0.49 nm at the specimen level. Cellular tilt series were typically recorded from –65° to +65°, with an increment of 1.0° and defocus of –3 to –4 μm. For the cellular tilt series, the pixel sizes at the specimen level were 0.821 and 0.805 nm at the Philips CM300 and the FEI Tecnai F30 Polara, respectively.
STEM Mass Measurements
STEM was carried out essentially as described in Vogt and Simon, 1999. TMV as a control with known mass per length was included in all samples for calibration. For each HIV-1 particle analyzed, we measured the mass/Å of two segments of TMV from the same image. There was no clear correlation between TMV mass/Å and the mass of the virus particle from the same image. The corrected virus mass was therefore calculated by dividing by the TMV mass/Å for the data set and multiplying by the known mass/Å (13.1 kDaÅ–1). The error in the estimate of the mean of the TMV mass/Å was considered to be small compared to the variation in HIV-1 virus particle mass. The mass/Å of TMV for the complete pooled data set was 14.3 ± 1.1 kDa/Å (n = 684). The contribution of different virion constituents to total HIV-1 mass was estimated based on published reports, where relative amounts of different components were estimated in relation to the HIV-1 CA protein, and this calculation is shown in Table S1.
Data Analysis
Three-dimensional reconstructions from tilt series were performed with the weighted back-projection method, using the TOM toolbox (Nickell et al., 2005) for MATLAB (Mathworks; Natick, MA) or IMOD (Kremer et al., 1996). Further analysis of the tomograms was done using the TOM toolbox and SPIDER (Frank et al., 1996). Briefly, the Gag shell closures were estimated by transforming the extracted subvolumes—containing single virions or budding sites—to spherical polar coordinates and estimating the extent of the Gag layer in this representation, whereas the radii were measured by locating the membrane peak in a radial density plot (see Supplemental Experimental Procedures). Visualizations of the tomograms were done with Amira (http://www.amiravis.com).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to V. Vogt, M. Lehmann, and S. Jäger for help with some of the experiments and to S. Prinz, A. Habermann, H. Oberwinkler, and B.Y. Lin for technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft within SPP1175 to H.-G.K and to K.G. The BNL STEM facility was an NIH-supported resource center, NIH 5 P41 EB2181, with additional support provided by DOE, OBER.
Footnotes
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, two figures, one table, and four movies, and can be found online at http://www.cell.com/cellhostandmicrobe/supplemental/S1931-3128(08)00367-3.
REFERENCES
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- Briggs JAG, Wilk T, Welker R, Kräusslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JAG, Simon MN, Gross I, Kräusslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li YH, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Kräusslich HG. Analysis of human immunodeficiency virus type 1 Gag ubiquitination. J. Virol. 2005;79:9134–9144. doi: 10.1128/JVI.79.14.9134-9144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Bodem J, Müller B, Schmechel A, Zentgraf H, Kräusslich HG. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 2003;77:9474–9485. doi: 10.1128/JVI.77.17.9474-9485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE., Jr. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 2004;11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Göttlinger HG, Weissenhorn W. Helical Structures of ESCRT-III Are Disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucić V, Förster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Müller B, Tessmer U, Schubert U, Kräusslich HG. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than gag and is phosphorylated in infected cells. J. Virol. 2000;74:9727–9731. doi: 10.1128/jvi.74.20.9727-9731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Nickell S, Förster F, Linaroudis A, Del Net W, Beek F, Hegerl R, Bau-meister W, Plitzko JM. TOM software toolbox: acquisition and analysis for electron tomography. J. Struct. Biol. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Rossio JL, Esser MT, Suryanarayana K, Schneider DK, Bess JW, Vasquez GM, Wiltrout TA, Chertova E, Grimes MK, Sattentau Q, et al. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 1998;72:7992–8001. doi: 10.1128/jvi.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CM, Malim MH. SnapShot: HIV-1 proteins. Cell. 2008;133:742. doi: 10.1016/j.cell.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Vogt VM, Simon MN. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JS, Hainfeld JF, Simon MN. Scanning transmission electron microscopy of nuclear structures. Methods Cell Biol. 1998;53:139–164. doi: 10.1016/s0091-679x(08)60878-x. [DOI] [PubMed] [Google Scholar]
- Welker R, Kotler H, Kalbitzer HR, Kräusslich HG. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology. 1996;219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- Welker R, Hohenberg H, Tessmer U, Huckhagel C, Kräusslich HG. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.