Abstract
Reactive oxygen species (ROS) generated by vascular endothelial and smooth muscle cells contribute to the development and progression of vascular diseases. We have recently shown that hyperoxia enhances NADPH Oxidase 4 (NOX4) expression, which regulates lung endothelial cell migration and angiogenesis. Regulation of NOX4 is poorly understood in the vasculature. The objective of this study is to identify transcriptional factor(s) involved in regulation of endothelial NOX4. We found that hyperoxia induced NOX4 expression was markedly reduced in Nrf2-/- mice, compared to Nrf2+/+ mice. Exposure of human lung microvascular endothelial cells (HLMVECs) to hyperoxia stimulated NRF2 translocation from the cytoplasm to the nucleus and increased NOX4 expression. Knock down of NRF2 expression using a siRNA approach attenuated basal NOX4 expression; however, it enhanced superoxide/ROS generation under both normoxia and hyperoxia. In silico analysis revealed presence of at least three consensus sequences for the antioxidant response element (ARE) in the promoter region of NOX4. In transient transfections, hyperoxia stimulated NOX4promoter activity in HLMVECs, and deletion of -438 to -458 and -619 to -636 sequences markedly reduced hyperoxia-stimulated NOX4 promoter activation. ChIP analysis revealed an enhanced recruitment of NRF2 to endogenous NOX4 promoter spanning these two AREs following hyperoxic insult. Collectively, these results demonstrate, for the first time, a novel role of NRF2 in regulating hyperoxia-induced NOX4 transcription via AREs in lung endothelium.
Introduction
Oxygen supplementation or hyperoxia is clinically practiced in treating premature babies and patients with cardiovascular and pulmonary diseases such as acute respiratory distress syndrome (ARDS) and emphysema [1]. However, prolonged exposure to hyperoxia in humans results in lung injury, pulmonary edema, inflammation and ultimately cell death [2]. Exposure of adult and neonatal animal models to hyperoxia mimics many of the pathological characteristics of human ARDS and bronchopulmonary dysplasia [3, 4]. However, preterm infants exposed to hyperoxia do not have alveolar cell death and hyperoxia does not cause injury for short periods of 3 days. Increased Reactive Oxygen Species (ROS) generation by hyperoxia is a major contributor to oxidant-induced lung injury. In the lung, several enzyme systems contribute to basal ROS production, including endothelial nitric oxide synthases, mitochondrial respiratory chain, cytochrome P450 monoxygenases, xanthine oxidase and NADPH Oxidase Nox proteins. Recently, we demonstrated increased expression of NOX2 and NOX4 in human lung endothelial cells by hyperoxia and blocking NOX2 or NOX4 attenuated hyperoxia-induced ROS/superoxide generation (O2-.)[5]. Further, in a mouse model of hyperoxia, deletion of NOX2 reduced lung inflammation and injury; interestingly, deletion of NOX2 up-regulated NOX4 expression in mouse lung [5]. At present, very little is known regarding regulation of NOX4 under basal condition or exposure of animals or cells to hyperoxia. In addition to increased ROS production, hyperoxia induces expression of several antioxidant enzymes and phase 2 detoxifying enzymes in the lung [6] and emerging studies both in vivo [7]and in vitro [8] support a critical role for nuclear factor-erythroid 2-related factor 2 (NRF2) in mediating the induction of antioxidant and phase 2 detoxifying enzymes.
NRF2 belongs to the Cap’n’collar/basic region leucine zipper (CNC-bZIP) transcription factor family that is activated by diverse oxidants, pro-oxidants, antioxidants, chemo preventive agents and electrophiles [9, 10]. Under basal conditions NRF2 levels are low as NRF2 is bound to Keap1 in the cytosol, ubiquitinated by E3 ubiquitin ligase and degraded by the 26S proteosome [11]. However, exposure to oxidative stress leads to dissociation of NRF2 from Keap1, stabilization and phosphorylation of NRF2 and translocation to the nucleus resulting in enhanced transcription of a number of target antioxidant genes via antioxidant response element (ARE) leading to cyto-protection [12]. In the lung, NRF2 confers a protective role against oxidative insults including hyperoxia, mechanical ventilation and cigarette smoke as lack of NRF2 exacerbates lung inflammation and injury [13]. Recent studies with Nrf2-/- mice suggest that the protective effects of NRF2 against hyperoxia-induced lung injury are possibly through transcriptional activation of antioxidant defense enzymes such as NAD(P)H:quinine oxidoreductase 1, glutathione transferase-Ya and –Yc, UDP-glycosyl transferase, glutathione peroxidase 2 and heme oxygenase 1 in the lung [14].
In an experimental acute lung injury model, we found that hyperoxia-induced NOX4 expression was significantly reduced in mice with genetic disruption of Nrf2 (Nrf2-/-) compared to wild type (Nrf2+/+) mice. As NRF2 regulates transcription of several antioxidant genes via the ARE[15], the present study was designed to test the hypothesis that hyperoxia-induced expression of NOX4 is in part regulated by NRF2 at the transcriptional level. Here we demonstrate for the first time the presence of three potential AREs located at -438 to -458, -619 to -636 and -665 to -677 in NOX4 promoter, and deletion of both -438 to -458 and -619 to -636 AREs, but not the -665 to -677 ARE, abolished hyperoxia-induced NOX4 promoter activity in human lung microvascular endothelial cells (HLMVECs). The binding of NRF2 to the endogenous NOX4 promoter in response to hyperoxia was demonstrated by ChIP assay. Further, hyperoxia stimulated translocation of NRF2 to the nucleus and the knockdown of NRF2 using siRNA approach attenuated hyperoxia-induced NOX4 expression. Finally, blocking ROS generation by p47phox or Rac1 siRNA attenuated hyperoxia-induced NRF2 translocation to cell nucleus and NOX4 expression. Our findings show for the first time a role for NRF2 in NOX4 regulation in hyperoxia and the influence of p47phox/ROS in NRF2 activation.
Experimental Procedures
Materials
HLMVECs, EBM-2 basal media, and Bullet kit were obtained from Lonza (San Diego, CA, USA). Phosphate-buffered saline (PBS) was from Biofluids Inc. (Rockville, MD, USA). Ampicillin, fetal bovine serum (FBS), trypsin, MgCl2, EGTA, Tris-HCl, Triton X-100, sodium orthovanadate, aprotinin and Tween 20, were obtained from Sigma-Aldrich (St. Louis, MO, USA). Dihydroethidium (hydroethidine) and 6-carboxy-2’,7’-dichlorodihydrofluorescein diacetate-di (acetoxymethyl ester) (DCFDA), were purchased from Life Technologies (Eugene, OR, USA). ECL kit was from Amersham Biosciences (Piscataway, NJ, USA). SMART Pool® small interfering RNA duplex oligonucleotides targeting p47phox were purchased from Dharmacon, Inc. (Lafayette, CO, USA). NOX2, Nrf2, p47phox, Rac1 and NOX4 siRNAs were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Polyclonal antibodies to antiNrf2 (ab-31163, Abcam, Cambridge, UK), NOX4 (sc-30141, Santa Cruz Biotechnology, USA) and NOX2 (BD 611414, BD Transduction Labs, USA) were purchased from respective companies.
Endothelial Cell Culture
HLMVECs, passages between 5 and 8, were grown in EGM-2 complete media with 10% FBS, 100 units/ml penicillin, and streptomycin in a 37 °C incubator under 5% CO2, 95% air atmosphere and grown to contact-inhibited monolayers with typical cobblestone morphology as described previously [16]. Cells from T-75 flasks were detached with 0.05% trypsin, resuspended in fresh complete medium, and cultured in 35- or 60-mm dishes or on glass cover slips for various studies under normoxia or hyperoxia.
Mouse experiments and Animal Care
All experiments using animals were previously approved by the Institutional Animal Care and Use Committee at University of Illinois at Chicago and The Johns Hopkins University. The Nrf2-sufficient (Nrf2+/+) and Nrf2-deficient (Nrf2–/–) CD-1/ICR strains of mice (6- to 8-wk-old female mice, 25–30 g body weight) were exposed to hyperoxia (100%O2) or room air as previously described (5). Nrf2 -/- mice were developed by Dr. Imamoto and submitted to RBRC 1997. Lung tissues were collected, homogenized and protein lysates were prepared for western blots.
Exposure of Cells to Hyperoxia
HLMVECs (~90% confluence) in complete EGM-2 medium were placed in a humidity-controlled airtight modulator incubator chamber (Billups-Rothenberg, Del Mar, CA, USA), flushed continuously with 95% O2, 5% CO2 for 30 min until the oxygen level inside the chamber reached ~95%. HLMVECs were then placed in a cell culture incubator at 37°C for the desired lengths of time (3-72 h). The concentration of O2 inside the chamber was monitored with a digital oxygen monitor. The buffering capacity of the cell culture medium did not change significantly during the period of hyperoxic exposure and was maintained at a pH ~7.4. In some experiments, cells were pretreated with PEG-conjugated SOD at 400 Units/mL for 1 h prior to addition of dihydroethidium or DCFDA and exposure to hyperoxia (3 h).
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from HLMVECs grown on 35-mm dishes using TRIzol® reagent according to the manufacturer’s instruction. iQ SYBR Green Supermix was used to do the real time using iCycler by Biorad. 18S rRNA (sense, 5’-GTAACCCGTTGAACCCCATT-3’, and antisense, 5’- CCATCCAATCGGTAGTAGCG-3’) was used as a housekeeping gene to normalize expression. The reaction mixture consisted of 0.3 μg of total RNA (target gene) or 0.03 μg of total RNA (18 S rRNA), 12.5 μl of iQ SYBR Green, 2 μl of cDNA, 1.5 μM target primers, or 1 μM 18 S rRNA primers, in a total volume of 25 μl. For all samples, reverse transcription was carried out at 25°C for 5 min, followed by cycling to 42 °C for 30 min and 85°C for 5 min with iScript cDNA synthesis kit. Amplicon expression in each sample was normalized to its 18 S rRNA content. The relative abundance of target mRNA in each sample was calculated as 2 raised to the negative of its threshold cycle value times 106 after being normalized to the abundance of its corresponding 18 S rRNA,(2-(primer Threshold Cycle)/2-(18 S Threshold Cycle)× 106). All primers were designed by inspection of the genes of interest and were designed using Beacon Designer 2.1 software. Negative controls, consisting of reaction mixtures containing all components except target RNA, were included with each of the RT-PCR runs. To verify that amplified products were derived from mRNA and did not represent genomic DNA contamination, representative PCR mixtures for each gene were run in the absence of the RT enzyme after first being cycled to 95 °C for 15 min. In the absence of reverse transcription, no PCR products were observed.
Preparation of Nuclear Extracts
Nuclear extracts were prepared from HLMVECs according to the manufacturer’s instructions (Active Motif North America, Carlsbad, CA). Briefly, cells were collected by scraping in ice-cold PBS containing phosphatase inhibitors and pelleted by centrifuging at 1000 × g for 5 min. The pellet was resuspended in 500 μl of 1X hypotonic buffer and incubated on ice for 15 min, followed by addition of 25 μl of detergent and high speed vortexing for 30 s as per the manufacturer’s recommendation. The suspension was centrifuged at 14,000 × g for 20 min in a micro centrifuge at 4 °C; the nuclear pellet was resuspended in 50 μl of lysis buffer A and incubated on ice for 15 min. This suspension was centrifuged for 10 min at 14,000 × g in a micro centrifuge, and the supernatant (nuclear extract) was aliquoted and stored at -80 °C for further analysis. Proteins in the nuclear extract were quantified by BCA protein assay.
Determination of Hyperoxia-induced Production of O2.-, and Total ROS
Two methods were used to measure total ROS or O2.- with spectrofluorimeter or fluorescence microscopy. Hyperoxia-induced O2.- release by HLMVECs was measured by hydroethidine fluorescence as described [17]. Total ROS production in HLMVECs exposed to either normoxia or hyperoxia was determined by the DCFDA fluorescence method. Briefly, HLMVECs (~90% confluent in 35-mm dishes) were loaded with 10 μM DCFDA in EGM-2 basal medium and incubated at 37°C for 30 min. Fluorescence of oxidized DCFDA in cell lysates, an index of formation of ROS, was measured with an Aminco Bowman series 2 Spectrofluorimeter using excitation and emission set at 490 and 530 nm, respectively, with appropriate blanks. Hyperoxia-induced ROS formation in cells was also quantified by fluorescence microscopy. HLMVECs (~90% confluent) in 35-mm dishes were loaded with DCFDA (10 μM) in EBM-2 basal medium for 30 min at 37°C in a 95% air, 5% CO2 environment. After 30 min of loading, the medium containing DCFDA was aspirated; cells were rinsed once with EGM-2 complete medium; cells were pre-incubated with agents for the indicated time periods followed by exposure to either normoxia (95% air, 5% CO2) or hyperoxia (95% O2, 5%CO2) for 3-72 h. At the end of the incubation, cells were washed twice with PBS at room temperature and were examined under a Nikon Eclipse TE 2000-S fluorescence microscope with Hamamatsu digital CCD camera (Japan) using a 20X objective lens and MetaVue software (Universal Imaging Corp., PA).
Determination of H2O2 by Amplex Red assay
H2O2 formation in the medium was determined by fluorescence method using an Amplex Red Hydrogen Peroxide Assay kit (Invitrogen). HLMVECs were transfected with scrambled or NRF2 siRNA for 48 h. Cells were exposed to normoxia (95% air–5% CO2) or hyperoxia (95% O2–5% CO2) for 3 h in 1.0 ml of phenol red free medium (GIBCO-BRL medium 199). At the end of incubation, the medium was collected and centrifuged at 4,000 g for 5 min, and fluorescence of the medium was measured according to manufacturer’s protocol on Bio-Rad iMark plate reader with absorption read at 560 nM.
Immunoflourescence Microscopy
HLMVECs grown on cover slips to ~95% confluence in EGM-2 complete medium as indicated, followed by exposure to either normoxia or hyperoxia for 3 h. Coverslips were rinsed with phosphate-buffered saline and treated with 3.7% formaldehyde in phosphate-buffered saline at room temperature for 20 min. After washing with phosphate buffered saline, cover slips were incubated in blocking buffer (1% bovine serum albumin in TBST) for 1 h, and cells were subjected to immunostaining with Nrf2 antibody (1:200) for 1 h and washed 3 times with TBST followed by staining with Alexa Fluor 488 (1:200 dilution in blocking buffer) for 1 h. After washing at least three times with TBST, the cover slips were mounted using commercial mounting medium (Kirkegaard and Perry Laboratories, Gaithersburg, MD) and were examined by immunofluorescence microscopy with Hamamatsu digital camera using a 60X oil immersion objective and MetaVue software.
In vitro capillary tube formation assay
HLMVECs transfected with scrambled or NRF2 siRNA (50 nM, 48 h) were plated on growth factor-reduced Matrigel (basement membrane extract, Trevigen Inc., Gaithersburg, MD), exposed to either normoxia or hyperoxia for 24 hours as described previously [5]. Capillary tube formation and completion of ring like structures were recorded and quantified using ImageJ program.
Endothelial Cell Migration
HLMVECs (control or transfected with siRNA) cultured in 6-well plates to ~95% confluence were starved in EBM-2 medium containing 1% fetal bovine serum 3 h. The cells were wounded by scratching across the monolayer with a 10 μl standard sterile pipette tip. The scratched monolayer was rinsed twice with serum-free medium to remove cell debris and incubated for 16 h in EBM-2 medium containing 1% fetal bovine serum under normoxia or hyperoxia. The area (~1 cm2 total) in a scratched area was recorded at 0 h and 16 h using a Hamamatsu digital camera connected to the Nikon Eclipse TE2000-S microscope with 10 X objective and MetaVue software (Universal Imaging Corp., PA, USA), and images were analyzed by the Image J software. The effect of hyperoxia and NRF2 siRNA on endothelial cell migration/wound healing was quantified by calculating the percentage of the free area not occupied by cells compared to an area of the initial wound that was defined as closure of wounded area.
Immunoblotting
HLMVECs grown on 100-mm dishes (~90% confluence) were rinsed twice with ice-cold phosphate-buffered saline and lysed in 100μl of modified lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4 , 1 mM NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 μg/ml pepstatin), sonicated on ice with a probe sonicator (three times for 15 s), centrifuged at 5000 × g in a micro centrifuge (4°C for 5 min), and protein concentrations of the supernatants were determined using Pierce protein assay kit. The supernatants, adjusted to 1 mg of protein/ml (cell lysates) were denatured by boiling in SDS sample buffer for 5 min, and samples were separated on 10% SDS-polyacrylamide gels. Protein bands were transferred overnight (24 V, 4°C) onto Nitrocellulose membrane (0.45μm) (BioRad, Hercules, CA, USA), probed with primary and secondary antibodies according to the manufacturer’s protocol, and immunodetected by using the ECL kit (Amersham Biosciences, Piscataway, NJ, USA). The blots were scanned (UMAX Power Lock II) and quantified by an automated digitizing system UN-SCAN-IT GEL (Silk Scientific Corp). HLMVECs were lysed in standard lysis buffer.
Transient Transfection of HLMVECs with NOX4 plasmid or NOX4 siRNA
HLMVECs grown to ~70% confluence in 6 well plates were transfected with Fugene HD Transfection Reagent (Roche Applied Science, IN) containing NOX4 wild type (1 μg cDNA), scrambled siRNA (50 nM) or siRNA for NOX4 (50 nM) in serum-free EBM-2 medium according to the manufacturer’s recommendation. After 3 h of transfection, the serum-free media was replaced by 1 ml of fresh complete EGM-2 medium containing 10% FBS and the growth factors, and cells were cultured for an additional 48 h prior to analysis for mRNA levels by real-time RT-PCR or protein expression by Western blotting.
Cloning of NOX4 sequence into pGL3-Luciferase Reporter
The proximal promoter (~3kb) of the human NOX4 was inserted into a pGL3 Luciferase vector. The promoter activity was calculated from the ratio of firefly luciferase to Renilla luciferase and expressed as arbitrary units.
Targeted deletions of NOX4 ARE
Deletions were introduced into the AREs of the 5’-flanking region of NOX4 using ClonEasy™ kit as per manufacturer’s recommendation. (GenScript, Piscataway, NJ) Three deletions of AREs were carried out with 7 possible combinations of the following sequences: del438-458, del619-636, del665-677.
del438-458 5’-ctatgtcattaaacaatctg---------------------acgcctgtaatcccagcag-3’
del619-636 5’-tgatcccagctactctac------------------agaattgcttgaacccag-3’
del665-677 5’-acccaggaggtggagg-------------agattacgacattgca-3’
Insertion of NOX4 wild type and deleted AREs into Luciferase reporter vector and activity assay
NOX4 promoter containing the three AREs and ARE targeted deletions of NOX4 plasmids were cloned into pGL3 backbone vectors (Promega). After cloning, all sequences were verified by University of Chicago sequencing Core facility and subsequently transfected into HLMVECs grown overnight in 35- mm dishes at a density of 2-3 × 105 per well using 1.6μg of each pGL3-luciferase reporter together with 0.1 μg of pRL-TK, an expression plasmid encoding Renilla luciferase (Promega), and Fugene HD transfection reagent. After 3h, the cells were grown in complete EGM-2 medium for 24h prior to exposure to normoxia or hyperoxia. The cells were then lysed with 150-200μl/per well of 1x Passive Lysis Buffer (Promega), and the luciferase gene activity (luc-activity) was measured using the Dual-Luciferase reporter assay system kit (Promega) using a Lumat LB 9507 Luminometer (Berthold Detection Systems, GmbH) to assess firefly and Renilla luciferase activity sequentially. Luciferase activities were normalized to Renilla internal control luminescence.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was carried out essentially as described previously [18]. Immunoprecipitations were performed using either control rabbit IgG or anti-NRF2 antibody. Immunoprecipitated material or 3 μl of 1:100 dilution of input (LDH) was subjected to 35 cycles of PCR using the following primers: the NRF2, 5- ATGGCCTTTCCTAGTTTGGG- 3 and 5- CCCAAGGGACAGAGCTAGTG- 3; the ldh promoter region, 5 -GCAAAGCCAGTACTCTTTCTG- 3 and 5 -CGGCCATGTTCTGAATCCAAG- 3.
Statistical Analysis
Analysis of variance and Student-Newman-Keul’s test were used to compare means of two or more different treatment groups. The level of significance was set to p<0.05 unless otherwise stated. Results are expressed as mean ± S.E.M.
RESULTS
NRF2 deficiency reduces hyperoxia-induced NOX4 expression in HLMVECs
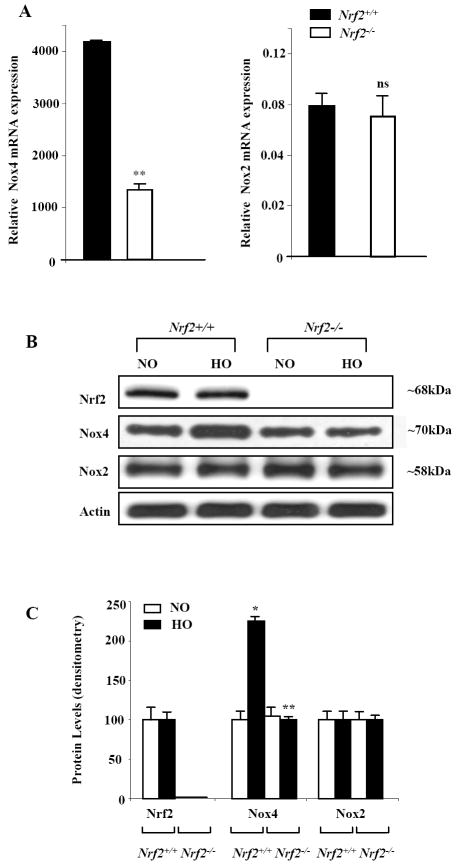
We have earlier demonstrated that exposure of HLMVECs to hyperoxia stimulated NOX4 mRNA and protein expression [5]. As hyperoxia activates NRF2 via ROS production [19], we investigated the role of NRF2 on NOX4 expression in mouse lung. Hyperoxia induced NOX4 but not NOX2 expression, which was significantly reduced in Nrf2-/- mice as compared to Nrf2+/+ (Figure 1A, 1B and 1C) suggesting a role for NRF2 in regulating NOX4 expression in response to hyperoxic insult. Further, exposure of Nrf2-/- and Nrf2+/+ mice to normoxia or hyperoxia did not affect mRNA expressions of p22phox and p47phox expression in the lung (Supplemental Figure 1).
FIGURE 1. Attenuation of hyperoxia-induced NOX4 expression in Nrf-/- mice.
C57 BL/6 and Nrf2-/- knockout mice in C57BL/6 background were exposed to either normoxia (room air) or hyperoxia (100% O2) for 48h. In A, total RNA was isolated and mRNA levels of NOX2 and NOX4 were quantified by real time RT-PCR and normalized to 18S rRNA. In B, cell lysates (30 μg of total proteins) were subjected to SDS-PAGE, as described shown under “Materials and Methods” and analyzed by Western blotting with anti-NOX4, anti-NOX2 (gp91phox) or anti-NRF2 antibodies. Shown is a representative blot from several experiments. Panel C shows relative expressions of NRF2, NOX4 and NOX2 proteins from the densitometry of Western blots depicted under Panel B. The values are mean ± S.E.M from three independent experiments in triplicate. *, significantly different from normoxia (p<0.05); **, significantly different from Nrf2+/+ wild type exposed to hyperoxia (p<0.01).
Nrf2 translocates to the nucleus in response to hyperoxia in HLMVECs
Earlier studies have shown that Nrf2 confers protection against hyperoxia-induced lung injury, in part, through the induction of antioxidant gene expression [14]. Since hyperoxia did not alter NRF2 protein expression in HLMVECs (Figure 1B and 1C), we investigated whether hyperoxia modulates sub-cellular localization of NRF2 from the cytoplasm to the nucleus. Fluorescent immunostaining of endothelial cells exposed to room air (normoxia) for various periods of time indicated that the NRF2 was predominantly localized in the cytoplasm (diffused staining); however, exposure to hyperoxia for 3 or 24 h induced NRF2 activation and translocation to the nucleus as evidenced by predominant fluorescent immunostaining of NRF2 in the nucleus (Figure 2A). The immunostaining result was further confirmed by NRF2 localization in nuclear extracts isolated from cells exposed to normoxia or hyperoxia. Western blotting of nuclear and cytoplasmic fractions, consistent with the immunoflourescence data, showed minimal amounts of NRF2 was present in the nuclei of cells exposed under normoxic conditions (Figure 2B). However, NRF2 levels in the nuclear fraction increased markedly after exposure of HLMVECs to hyperoxia (Figure 2B), suggesting a rapid nuclear accumulation of this transcription factor in response to hyperoxia. Further, Nox4 levels in nuclear extracts were increased with hyperoxia, a finding consistent with our earlier studies [5]. Expressions of LaminB1 and LDH were used as markers for the nuclear and cytoplasm preparations. Calreticulin served as a marker for endoplasmic reticulum to evaluate the quality of nuclear and cytoplasmic preparations (Figure 2B).
FIGURE 2. Hyperoxia induces Nrf2 translocation to the nucleus in HLMVECs.
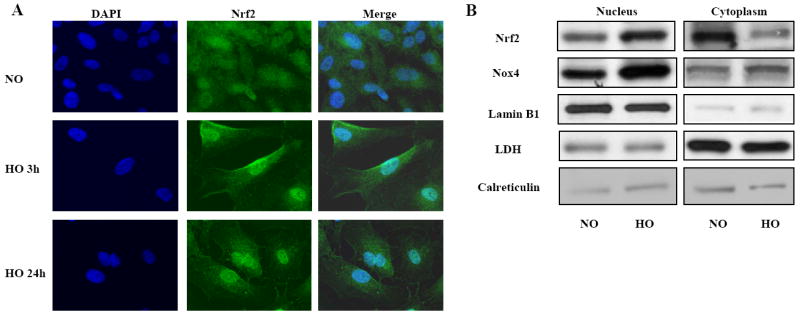
In Panel A, HLMVECs (~90% confluence) grown on glass cover slips were exposed to normoxia (NO) or hyperoxia (HO) for 3h or 24h, washed, fixed, permeabilized and probed with anti-NRF2 antibody. Shown is a representative immunofluorescence image from three independent experiments demonstrating translocation of Nrf2 to the nucleus after exposure to hyperoxia. In Panel B, HLMVECs in 100-mm dishes grown to ~90% confluence were exposed to normoxia (NO) or hyperoxia (HO) for 24h. Nuclear and cytoplasmic fractions were prepared using Active Motif Nuclear Isolation kit according to manufacturer’s protocol as described in “Materials and Methods”. Lysates from nuclear and cytoplasmic fractions (20μg proteins) were subjected to SDS-PAGE and immuno blotted with antibodies against Nrf2, NOX4, Lamin B1, Lactate dehydrogenase (LDH) and Calreticulin. Shown are representative blots from three independent experiments.
Silencing of NRF2 blocks cell migration and capillary tube formation of human lung microvascular endothelial cells
Hyperoxia stimulates cell migration and angiogenesis of endothelial cells [5]. Here, we studied the role of NRF2 in hyperoxia-induced endothelial cell migration. Down-regulation of NRF2 with siRNA blocked hyperoxia (16 h) induced endothelial cell migration in a scratch assay (Figure 3A). In addition to blocking cell migration, silencing NRF2 inhibited capillary tube formation in matrigel (Figure 3B). These results suggest a role for NRF2 in hyperoxia-induced cell migration and capillary tube formation in HLMVECs.
FIGURE 3. Down-regulation of NRF2 attenuates hyperoxia-induced endothelial cell migration and capillary tube formation.
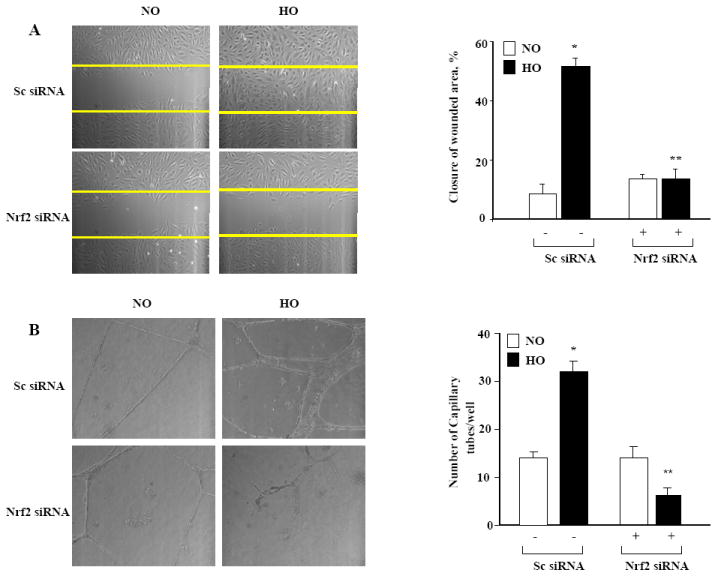
In Panel A, HLMVECs grown to ~50% confluence in 35-mm dishes were transfected with scrambled or NRF2 siRNA (50 nM, 48 h) in complete EGM-2 medium. Cells were scratched and exposed to either normoxia or hyperoxia (16 h) in EBM-2 medium containing 2% fetal bovine serum. Phase contrast images were captured at 0 and 16 h after the scratch and exposure and migration of cells into a “wound” was calculated as described in “Materials and Methods”. The values are the mean + S.E. for three independent experiments, and each experiment was carried out in triplicate. *significantly different from normoxia without NRF2 siRNA (P<0.05); **significantly different from cells exposed to hyperoxia without NRF2 siRNA (P<0.01). In Panel B, HLMVECs grown in 35-mm dishes to ~50% confluence were transfected with scrambled (sc) or NRF2 siRNA (50 nM, 48 h). After 48 h, cells were trypsinized and seed onto matrigel coated 35-mm dishes, as described in “Materials and Methods”. After 24 h of seeding, cells were either exposed to normoxia (NO) or hyperoxia (HO) for 24 h and formation of capillary tubes were visualized under phase-contrast microscope and quantified by counting the tubes formed in different areas. The values are mean + S.E. for three independent experiments. *significantly different from scrambled siRNA transfected cells under normoxia (P<0.05); **significantly different from scrambled siRNA transfected cells under hyperoxia (P<0.01).
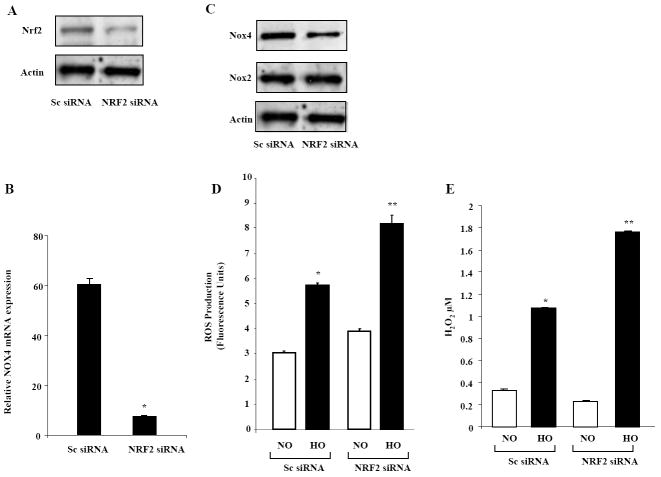
Knockdown of NRF2 mitigates hyperoxia-induced NOX4 expression and potentiates hyperoxia-induced ROS generation in HLMVECs
As NRF2 is a key transcriptional regulator of several antioxidant genes in response to hyperoxia, we investigated whether down-regulation of NRF2 altered hyperoxia-induced mRNA expressions of NOX2, NOX4 and ROS generation. HLMVECs were transfected with scrambled or NRF2 siRNA (50 nM, 48 h), and cell lysates were analyzed for NRF2, NOX2 and NOX4 expression. NRF2 siRNA significantly down-regulated NRF2 protein expression (Figure 4A) as well as mRNA and protein expression of NOX4 without altering NOX2 levels (Figure 4B and 4C). In parallel experiments, knockdown of NRF2 with siRNA augmented hyperoxia-induced ROS generation as measured by DCFDA oxidation (Figure 4D) or H2O2 released into the medium (Figure 4E) assessed by using Amplex Red Assay. These results suggest a role for NRF2 in regulating NOX4 expression and ROS generation in HLMVECs.
FIGURE 4. NRF2 siRNA increases hyperoxia-induced ROS production in HLMVECs.
Panel A confirms reduction in NRF2 protein expression by NRF2 siRNA. HLMVECs grown on 35-mm dishes to ~ 50% confluence were transfected with scrambled siRNA (Sc) or NRF2 siRNA (50nM, 48 h). Cell lysates (30 μg proteins) were extracted and subjected to SDS-PAGE, and analyzed by Western blotting with anti-NRF2 antibody as described shown under “Materials and Methods”. Shown is a representative blot from three independent experiments. In Panel B, HLMVECs grown on 35-mm dishes to ~ 50% confluence were transfected with scrambled siRNA or NRF2 siRNA (50nM) for 48 h, total RNA was extracted and NOX4 mRNA expression was quantified by real time RT-PCR and normalized to 18S rRNA. Values are average of three independent determinations. *, significantly different from scrambled siRNA transfected cells (P< 0.05). In Panel C, cell lysates (30 μg proteins) from HLMVECs grown on 35-mm dishes and transfected with scrambled or NRF2 siRNA (50 nM, 48 h) were subjected to SDS-PAGE, and Western blotted with anti-NOX4 or anti NOX2 antibody as described shown under “Materials and Methods”. Shown is a representative blot of several experiments. In Panel D, HLMVECs grown to ~90% confluence on 35-mm dishes were loaded with 10μM DCFDA for 30 min, washed once in basal EBM-2 medium and exposed to either normoxia (NO) or hyperoxia (HO) for 3 h. Total ROS accumulation was measured by Spectrofluorimeter as described in “Materials and Methods”. Values are mean ± S.E. from three independent experiments done in triplicate and normalized to total cell protein. *, significantly different from scrambled siRNA transfected cells exposed to normoxia (P<0.05); **, significantly different from scrambled siRNA transfected cells exposed to hyperoxia (P<0.01). In Panel E, HLMVECs grown to ~50% confluence in 100-mm dishes were transfected with scrambled or NRF2 siRNA (50nM) for 48 h. Cells were exposed to either normoxia (NO) or hyperoxia (HO) for 3 h and accumulation of H2O2 in the medium was measured using an Amplex Red assay. The values are mean ± S.E. from three independent experiments and expressed in concentration of H2O2. (μM). *, significantly different from scrambled siRNA transfected cells exposed to normoxia (P<0.05); **, significantly different from scrambled siRNA transfected cells exposed to hyperoxia (P<0.01).
Hyperoxia-induced ROS is necessary for Nrf2 translocation into nucleus in HLMVECs
Having demonstrated a role for Nrf2 in the regulation of NOX4, but not NOX2, expression in response to hyperoxia, we next investigated if ROS generated via NADPH oxidase regulates NRF2 activation. Down-regulation of NOX2 or NOX4 with siRNA (50 nM, 48 h) attenuated hyperoxia-induced NRF2 translocation to the nucleus in HLMVECs as determined by immunofluorescence microscopy (Supplemental Figure 2). As p47phox is a critical cytosolic component required for hyperoxia-induced ROS generation [16], HLMVECs transfected with p47phox siRNA down-regulated expression of p47phox (>80%) and almost completely blocked basal as well as hyperoxia-induced ROS/superoxide generation (data not shown) confirming the earlier results in human pulmonary artery endothelial cells (HPAECs) [16, 20]. Further, down-regulation of p47phox with siRNA abolished hyperoxia-induced NRF2 translocation from the cytoplasm to the nucleus (data not shown) suggesting that ROS production via NADPH oxidase pathway is essential for Nrf2 activation.
Hyperoxia Induced NOX4-promoter activity requires the antioxidant response element (ARE)
In silico analysis (Genomatix) revealed the presence of typical AREs in the promoter of human NOX4 gene [-438/-458; -619/-636; -665/-677]. To determine the functional role of the ARE in regulating NOX4 promoter activity, the proximal promoter of NOX4 with and without ARE binding elements was inserted into luciferase reporter vector. HLMVECs were transiently transfected with the reporter constructs; pGL3-NOX4-ARE Luc, pGL3-NOX4- minus ARE Luc, or and pGL3 vector for 48 h. As shown in Figure 5, hyperoxia significantly increased luciferase activity in cells transfected with NOX4-Luc (~ 2 fold, P<0.01) compared to normoxia. Furthermore, deletion of the entire ARE binding elements in the NOX4 promoter completely blocked hyperoxia-induced NOX4-Luc promoter activity (Figure 5). In addition, the Luciferase activity of the ARE-containing vector was higher in normoxia than the Luciferase reporter positive-control vector, which carries both the promoter and enhancer from SV40. These results support a role for ARE in hyperoxia-induced NOX4 promoter activity in human lung endothelial cells.
FIGURE 5. NRF2 siRNA attenuates hyperoxia-induced NOX4 promoter Luciferase activity.
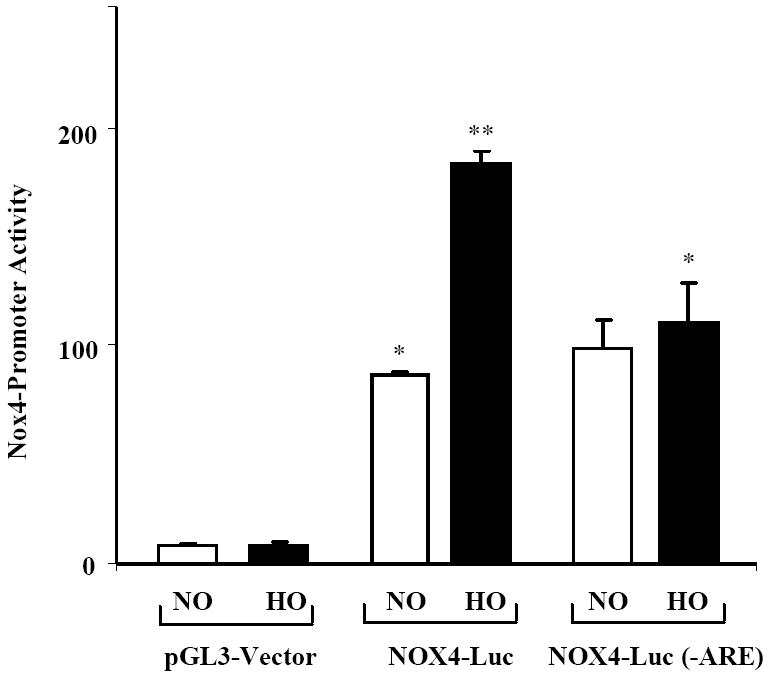
HLMVECs grown in 35-mm dishes to ~70% confluence were transfected with pGL3-vector, NOX4-Luc or NOX4-Luc(-ARE) plasmids (1 μg/ml) and exposed to either normoxia (NO) or hyperoxia (HO) for 3 h. Cells were lysed and analyzed by Luminometer using Promega Dual Luciferase kit as described in “Materials and Methods”. The values are mean + S.E. of three independent experiments in triplicate. *, significantly different from pGL3-vector transfected cells exposed to hyperoxia (P<0.05).
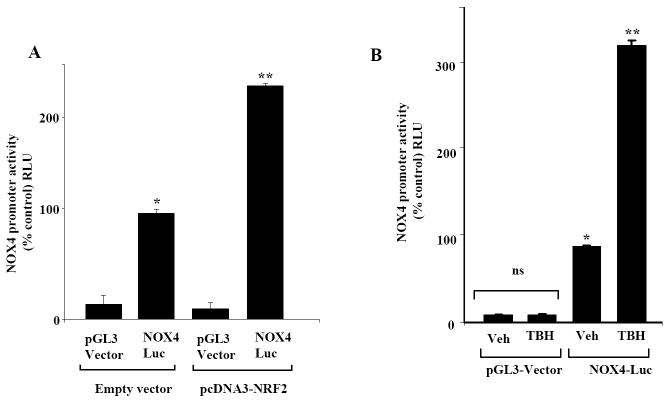
Over expression of Nrf2 or tert-butylhydroxyquinone (t-BHQ) treatment significantly enhances NOX4 promoter activity
NRF2 regulated gene expression and the response to the electrophilic agent, tert-butylhydroxyquinone, are mainly mediated by the ARE. To test effects of these two agents on the NOX4-promoter activity, HLMVECs were transfected with NOX4 luciferase reporter construct in the presence or absence of NRF2-cDNA vector. As shown in Figure 6A, over-expression of Nrf2 stimulated NOX4 promoter activity (2.3 fold) compared to cells transfected with empty vector. Similarly, treatment of cells with t-BHQ, a known activator of Nrf2 [21], also markedly (3.2 fold) increased NOX4 promoter driven luciferase activity (Figure 6B) compared to untreated cells. These results further suggest that the regulation of NOX4 gene expression by NRF2 via ARE occurs in HLMVECs.
FIGURE 6. Over expression of NRF2 and stimulation by TBH increases NOX4-promoter activity in HLMVECs.
In Panel A, HLMVECs grown on 35-mm dishes (~70% confluence) were transfected with vector control, NRF2 plasmid, pGL3 vector, and NOX4-Lucerifearse reporter plasmid for 24 h. Cell lysates were analyzed for luciferase activity using Promega Dual Luciferase kit as described in “Materials and Methods”. The values are mean + S.E. of three independent experiments in triplicate. *significantly different from pGL3-vector (p<0.05); **significantly different from NOX4-Luc cells challenged with vehicle (p<0.01). In Panel B, HLMVECs (~70% confluence) were transfected with pGL3-vector or NOX4-Luc reporter plasmid for 24 h prior to treatment with vehicle or tertiary butyl hydroquinone (TBH) (160 μM) for 3 h. Cells were lysed and analyzed for luciferase activity using Promega Dual Luciferase kit as described in “Materials and Methods”. The values are mean ± S.E. for three independent experiments. *significantly different from pGL3-vector (p<0.05); **significantly different from NOX4-Luc cells challenged with vehicle (p<0.01).
Rac1 and p47phox siRNA block hyperoxia-induced NOX4 ARE-Luc Promoter activity
We have earlier demonstrated that hyperoxia-induced ROS generation is linked to activation of Rac1 and p47phox in human lung endothelial cells [20]. As hyperoxia-induced NRF2 activation is ROS dependent [7], we next evaluated the role of Rac1 and p47phox in enhancement of NOX4 ARE-luciferase promoter activity. Down-regulation of Rac1 or p47phox expression with siRNA completely attenuated hyperoxia-induced NOX4 ARE-luciferase promoter activity (Fig. 7). The role of hyperoxia-induced ROS in NRF2 activation was also confirmed after silencing p47phox with siRNA. As shown in Supplemental Figure 3, knockdown of p47phox with siRNA attenuated NRF2 binding to NOX4 promoter in response to hyperoxia as assessed by ChIP assay. These results further confirm a role for Rac1 and p47phox in driving the NOX4 promoter activity via ROS signaling in HLMVECs.
FIGURE 7. Attenuation of hyperoxia-induced NOX4-promoter activity by NRF2, Rac1 siRNA or p47phox siRNAs in HLMVECs.
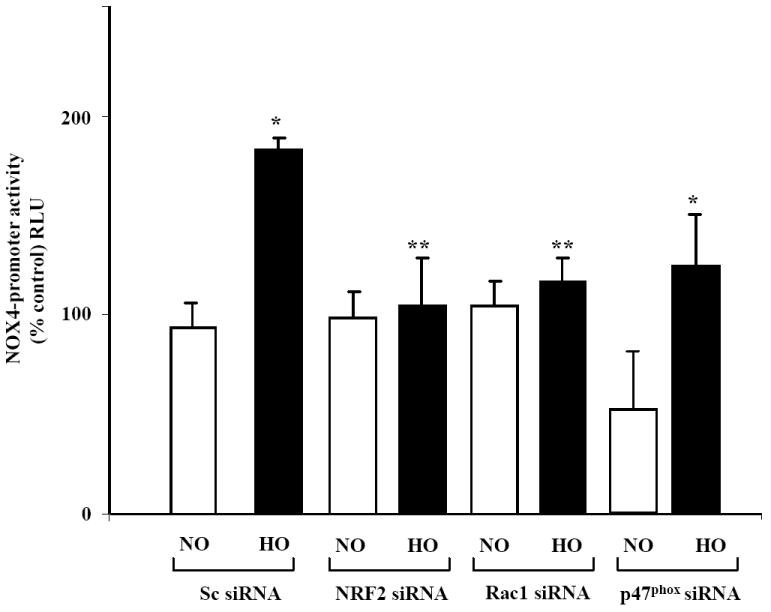
HLMVECs were grown on 35-mm dishes (~50% confluence) were transfected with scrambled (50 nM), NRF2 (50 nM), Rac1 (50 nM) or p47phox (50 nM) siRNA using transfection reagent from Genlantis for 48 h as described in “Materials and Methods”. After 48 h, HLMVECs were transfected with pGL3-vector or NOX4-Luc reporter plasmid and cells were allowed to grow for an additional 24 h prior to exposure to normoxia (NO) or hyperoxia (HO) for 3 h. Cells were lysed and luciferase activity was measured using a luminometer as per manufacturer’s guidelines. The values are mean ± S.E from three independent experiments. *, significantly different from scrambled siRNA transfected cells exposed to normoxia (P<0.05); **significantly different from scrambled siRNA transfected cells exposed to hyperoxia (P<0.001).
NRF2 binds to the NOX4 ARE of promoter in HLMVECs
To further evaluate the biological significance of enhanced NOX4 promoter activity by hyperoxia, ChIP experiments were performed. Immunoprecipitation with anti-NRF2 antibody was carried out on formaldehyde fixed protein-DNA complexes isolated from normoxia and hyperoxia treated HLMVECs. The association of the protein of interest with the NOX4 ARE was assessed by PCR using specific primers to amplify the NOX4-ARE containing region. As shown in Figure 8, the quality of the ChIP DNA immunoprecipitated by NRF2 was confirmed by the amplification of human NOX4 ARE-containing region. The ChIP data were confirmed by real time RT-PCR. In NRF2 immunoprecipitation experiments, a shift in the amplification curve to the left after exposure to hyperoxia was observed (Ct value shift of 4 ± 0.18), (Fig 8B) whereas rabbit IgG failed to pull down the NOX4 ARE and did not show any shift in hyperoxia (Fig. 8A). Sheared chromatin as input also did not show any shift (Fig. 8C). Also, PCR reactions using primers specific for ldh (lactate dehydrogenase) promoter did not amplify the product ensuring no genomic DNA contamination of the samples (Figure 8D). These ChIP results show the recruitment of NRF2 to the NOX4 ARE in human lung endothelial cells.
FIGURE 8. Hyperoxia enhances NRF2 binding to ARE cis-elements in NOX4 promoter.
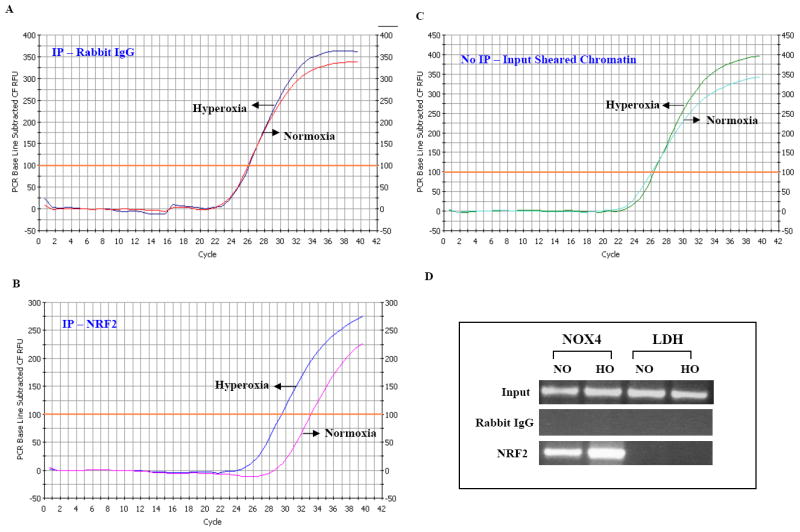
HLMVECs grown in 100 mm dishes (~80% confluence Cells were exposed to normoxia and hyperoxia (3h) before proteins were cross-linked to DNA through addition of formaldehyde directly to the culture medium. Enriched ChiPed DNA was analyzed using quantitative PCR. Panels A, B, and C show data from the q-PCR. Chromatin Immunoprecipitation Assay (ChIP) was performed with EZ ChIP (Upstate) as per manufacturer’s instructions. A, shows the IP of rabbit IgG as a control. Normoxic and Hyperoxic samples are on the same curve not showing any shift. In Panel B, as an additional control, IP of sheared chromatin was used and the normoxia and hyperoxia samples do not show any shift. In Panel C, NRF2 immunoprecipitates show that hyperoxia induces a shift of the curve to the left indicating the binding of NRF2. D, shows the gel of samples from Normoxia and hyperoxia with LDH used as an internal control. Rabbit IgG served as a control for all samples. Experiments were done in triplicates and shown is a representative gel image.
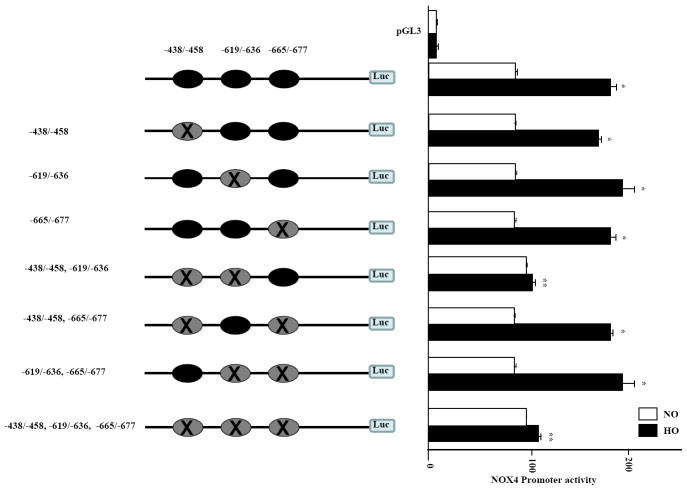
Deletion of ARE sequences -438-458 and -619-636 ARE in NOX4 promoter are critical for hyperoxia induced NOX4 promoter activity in endothelial cells
To determine the requirement of ARE sequences, we deleted the three ARE regions ( -438/-458; -619/-636; -665/-677) either alone or in combination of NOX4 ARE 3.2 kb plasmid to yield a total of seven deleted mutants using ClonEasy™ by GenScript as per manufacturer’s guidelines. The deletions at: -438/-458; -619/-636; -665/-677; -438/-458, -619/-636; -438/-458, -665/-677; -619/-636, -665/-677; -438/-458, -619/-636, -665/-677 were confirmed by sequencing and comparing to the original plasmid, and subsequently, the seven deleted NOX4 promoter sequences were inserted into the pGL3 vector for Luciferase Reporter analysis. HLMVECs were transfected with these deleted mutants for 24 h prior to hyperoxia exposure for 3 h. As shown in Figure 9, deletion of all three ARE regions (-438/-458; -619/-636 and -665/-677) almost completely attenuated hyperoxia-induced NOX4 ARE-Luciferase reporter activity. Interestingly, among the rest of the deleted plasmids, tandem deletions at -438/-458 and -619/-636 blocked the Luciferase reporter activity in response to hyperoxia compared to deletions at -438/-458; -619/-636; -665/-677; -438/-458, -665/-677; -619/-636 and -665/-677. These results from the deleted ARE sequences in NOX4 promoter show that 2 tandem sequences at -438/-458 and -619/-636 at the 5’ end of NOX4 promoter are critical for hyperoxia induced NOX4 promoter activity (Figure 9).
FIGURE 9. Correlation between deletion of ARE sequences in NOX4 promoter and NOX4-Luciferarse reporter activity.
Seven deletions of ARE sequences in NOX4 promoter were carried out using Genscript’s (Piscataway, NJ) proprietary ClonEasy technology. The seven NOX4-Luciferase deleted reporter sequences were cloned into pGL3 vector, and sequences were confirmed with the gene sequencing facility at the University of Chicago. HLMVECs (~80% confluence) were transfected with these 7 ARE deleted plasmids (~1μg with Fugene HD transfection reagent) for 24 h as described in “Materials and Methods”. After 24 h, the cells were exposed to normoxia or hyperoxia for 3 h and cell lysates were analyzed for luciferase activity using Promega Dual Luciferase kit. Values are mean ± S.E.M for three independent experiments in triplicate. *, significantly different from normoxia (p<0.05); **, significantly different from NOX4-Luciferase (-438-458, -619-636 and -665-677) transfected cells exposed to hyperoxia (p<0.01).
DISCUSSION
In this study, we provide evidence for the first time demonstrating a role for NRF2, a critical transcriptional regulator of antioxidant genes, in up-regulating NOX4 expression in the mouse lung and human lung endothelium in response to hyperoxia. Results from this study show that: (i) the presence of functional cis-acting ARE sequences at positions -438-458 and -619-636 in the 5’-flanking region of NOX4 gene, (ii) recruitment of NRF2 to the endogenous NOX4 promoter encompassing ARE in human lung endothelial cells following hyperoxia exposure, and (iii) lack or diminished levels of hyperoxia induced NOX4 expression in human lung endothelial cells or mouse lung with reduced levels of NRF2.The works from various laboratories and ours have clearly established increased ROS/O2-. production during hyperoxia [17, 20], and ROS generated through NADPH oxidase to be a key component of hyperoxia-induced lung inflammation and injury [5]. Mechanisms that regulate hyperoxia-induced NADPH oxidase activation are complex involving phospholipase D-mediated and Src-dependent tyrosine phosphorylation of p47phox and cortactin [16] and recruitment of Rac1, IQGAP1and other cytoskeletal as well as oxidase components to caveolin-1 rich lipid microdomains [22]. Also, we have recently reported that hyperoxia up-regulates expression of NOX4 and NOX2, but not NOX1 or NOX3, proteins in human pulmonary artery endothelial cells and HLMVECs, and knockdown of NOX4 expression using an siRNA approach markedly attenuated hyperoxia-induced ROS/ O2-. production and migration of human lung endothelial cells [5]. Although polymerase [DNA-directed] delta-interacting protein 2 positively regulates NOX4 activity [23] via interaction with p22phox in smooth muscle cells, very little is known regarding the regulation of NOX4 expression in lung cell types in response to stressful stimuli such as hyperoxia. Other transcriptional factors like E2F1, Smad binding elements, HIF-1a and NF-kB were also shown to be regulating NOX4 promoter. In smooth muscle cells, E2F family of transcriptional factors is known to be binding to NOX4 promoter positively regulating NOX4 transcription [24]. In freshly isolated human pulmonary artery smooth muscle cells (HPASMC), TGF-β1 initially promoted differentiation, with upregulated expression of smooth muscle contractile proteins. TGF-β1 also induced expression of NOX4, the only NAD(P)H oxidase membrane homolog found in HPASMC, through a signaling pathway involving Smad 2/3 but not mitogen-activated protein (MAP) kinases [25]. In another study [26] in pulmonary artery smooth muscle cells, ROS generated either by exogenous H2O2 or by a NOX4-containing NADPH oxidase stimulated by thrombin activated or induced NF-κB and HIF-1α. The reactive oxygen species-mediated HIF-1α induction occurred at the transcriptional level and was dependent on NF-κB activation. Most of these studies emphasized on the regulation of NOX4 in smooth muscle cells; however a little is known about the regulation of NOX4 in human lung microvascular endothelial cells upon exposing them to hyperoxia. Our current findings are consistent with the earlier studies [27] demonstrating a predominant role for NRF2 localization in cytoplasm of endothelial cells under normoxia, and nuclear translocation and accumulation, rather than induction of its mRNA, in hyperoxic condition.
While redox imbalance due to excess ROS generation has been implicated in the pathogenesis of hyperoxia- and oxidant-induced lung injury [5], hyperoxia activates expression of endogenous antioxidant enzymes, such as superoxide dismutase, glutathione peroxidase, heme oxygenase 1 and catalase to offset the oxidant burden in the lung [14]. Emerging evidence from in vivo and in vitro studies have identified NRF2 as a key regulator in mediating the induction of genes encoding several antioxidant enzymes via the ARE in response to stressful stimuli, including hyperoxia [13]. Genetic disruption of Nrf2 rendered mice highly susceptible to hyperoxia-induced lung injury compared to Nrf2+/+ mice and this susceptibility was mainly attributed to decreased expression levels of several antioxidant genes [28]. We observed that induction of NOX4 expression by hyperoxia was impaired in Nrf2-null mice compared to Nrf2+/+ mice (Figure 1), suggesting a potential role of Nrf2 in regulating NOX4 expression in response to hyperoxia in vivo. A direct role for NRF2 in regulating NOX4 expression was confirmed in human lung endothelial cells. Knockdown of NRF2 expression using siRNA approach markedly attenuated NOX4 expression, without altering NOX2 levels (Figure 4). These results suggest that NRF2 specifically regulates NOX4 expression in lung endothelium. In silico analysis of 5’-upstream region of NOX4 promoter revealed at least two conserved NRF2 binding AREs, at position -619-/636 and -665/-677 and an imperfect ARE at -438-458; with ~60% sequence homology to the consensus ARE (5’-GCTGAGNNN-3’). We found through deletion analysis of AREs on NOX4 that both -438-458 ARE-Like sequence and -619-636 ARE in NOX4 (Figure 9) are critical for hyperoxia induced NOX4 promoter activity in endothelial cells. Deletion of either -438/-458 ARE-like sequence or the -619/-636 ARE did not significantly affect hyperoxia induced NOX4 promoter activity; however, disruption both of these sequences crippled hyperoxia induced NOX4 promoter activity. Thus, it is likely that these two AREs (-438-458 and -619-636) compensate each other function in the absence of other and regulate NOX4 promoter activity. ChIP analysis clearly shows that NRF2 binds to the endogenous NOX4 promoter in intact living cells and the binding was further enhanced in response to hyperoxia. Consistent with this result, we found that knockdown of NRF2 expression suppresses both basal and hyperoxia inducible NOX4 expression. Further studies are warranted to define the exact nature of transcriptional factor binding at these two functional AREs and the molecular basis underlying the compensatory mechanisms that operate to regulate NOX4 expression in response to hyperoxia and other pro-oxidant and toxic stimuli.
The physiological and pathophysiological significance of NRF2/ARE mediated up-regulation of NOX4 in hyperoxia is unclear. Although NRF2 is the master transcriptional factor that regulates expression of >300 genes encoding antioxidant and phase 2 detoxifying enzymes [29], using a integrated computational system, a set of polymorphic AREs in the human genome with little known antioxidant function has been reported [30]. Our results show for the first time that the NRF2 transcription factor, which is known to up-regulate antioxidant gene expression, also selectively regulates one member of NOX family of proteins that is involved in ROS production in vascular cells. Previously, it is shown that hyperoxia-induced NRF2 activation and translocation to nucleus is mediated by ROS signaling in pulmonary epithelial cells [19] and our present studies with human lung endothelial cells suggesting interdependence between the ROS and NRF2 signaling in NOX4 up-regulation. NOX4 is predominantly localized in the nucleus of the human lung and umbilical vein endothelial cells [31, 5] and generates H2O2 [32], an important signaling molecule in the lung. Recent studies have shown an important role for localized H2O2 in cellular signaling and its regulation of peroxiredoxins [33] that are known to be regulated by NRF2 regulated cellular stress. It is thus conceivable that NRF2 mediated up-regulation of NOX4 may be a cellular response to offset some of effects of antioxidant gene(s) activation and to maintain a cellular balance of H2O2 production required for endothelial cell function and signaling. Further experiments are necessary to define a physiological role for ROS → NRF2 → NOX4 signaling in lung endothelial function under normal and pathological conditions.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants HL08553 and HL58064 to V.N, HL66109 to SPR.
The abbreviations used are
- ROS
Reactive Oxygen Species
- Nox
NADPH Oxidase
- Scramb
Scrambled small interfering RNA
- siRNA
small interfering RNA
- DHE
Dihydroethidium
- HPAECs
Human Pulmonary artery endothelial cells
- HLMVECs
Human Lung Microvascular endothelial cells
- ECs
Endothelial Cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chabot F, Mitchell JA, Gutteridge JM, Evans TW. Reactive oxygen species in acute lung injury. Eur Respir J. 1998;11:745–757. [PubMed] [Google Scholar]
- 2.Ryter SW, Kim HP, Hoetzel A, Park JW, Nakahira K, Wang X, Choi AM. Mechanisms of cell death in oxidative stress. Antioxid Redox Signal. 2007;9:49–89. doi: 10.1089/ars.2007.9.49. [DOI] [PubMed] [Google Scholar]
- 3.Rahman I. Redox signaling in the lungs. Antioxid Redox Signal. 2005;7:1–5. doi: 10.1089/ars.2005.7.1. [DOI] [PubMed] [Google Scholar]
- 4.Tao F, Gonzalez-Flecha B, Kobzik L. Reactive oxygen species in pulmonary inflammation by ambient particulates. Free Radic Biol Med. 2003;35:327–340. doi: 10.1016/s0891-5849(03)00280-6. [DOI] [PubMed] [Google Scholar]
- 5.Pendyala S, Gorshkova IA, Usatyuk PV, He D, Pennathur A, Lambeth JD, Thannickal VJ, Natarajan V. Role of Nox4 and Nox2 in hyperoxia-induced reactive oxygen species generation and migration of human lung endothelial cells. Antioxid Redox Signal. 2009;11:747–764. doi: 10.1089/ars.2008.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy NM, Kleeberger SR, Kensler TW, Yamamoto M, Hassoun PM, Reddy SP. Disruption of Nrf2 impairs the resolution of hyperoxia-induced acute lung injury and inflammation in mice. J Immunol. 2009;182:7264–7271. doi: 10.4049/jimmunol.0804248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 8.Papaiahgari S, Kleeberger SR, Cho HY, Kalvakolanu DV, Reddy SP. NADPH oxidase and ERK signaling regulates hyperoxia-induced Nrf2-ARE transcriptional response in pulmonary epithelial cells. J Biol Chem. 2004;279:42302–42312. doi: 10.1074/jbc.M408275200. [DOI] [PubMed] [Google Scholar]
- 9.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. Embo J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong M, Kan YW, Chan JY. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in gamma-gcs(l) and gss expression in mouse fibroblasts. J Biol Chem. 1999;274:37491–37498. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- 11.Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 Antioxidant Response by the Ubiquitin Proteasome System: An Insight into Cullin-Ring Ubiquitin Ligases. Antioxid Redox Signal. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vargas MR, Johnson JA. The Nrf2-ARE cytoprotective pathway in astrocytes. Expert Rev Mol Med. 2009;11:e17. doi: 10.1017/S1462399409001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho HY, Kleeberger SR. Nrf2 protects against airway disorders. Toxicol Appl Pharmacol. 244:43–56. doi: 10.1016/j.taap.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Cho HY, Reddy SP, Kleeberger SR. Nrf2 defends the lung from oxidative stress. Antioxid Redox Signal. 2006;8:76–87. doi: 10.1089/ars.2006.8.76. [DOI] [PubMed] [Google Scholar]
- 15.Lee JM, Johnson JA. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J Biochem Mol Biol. 2004;37:139–143. doi: 10.5483/bmbrep.2004.37.2.139. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury AK, Watkins T, Parinandi NL, Saatian B, Kleinberg ME, Usatyuk PV, Natarajan V. Src-mediated tyrosine phosphorylation of p47phox in hyperoxia-induced activation of NADPH oxidase and generation of reactive oxygen species in lung endothelial cells. J Biol Chem. 2005;280:20700–20711. doi: 10.1074/jbc.M411722200. [DOI] [PubMed] [Google Scholar]
- 17.Parinandi NL, Kleinberg MA, Usatyuk PV, Cummings RJ, Pennathur A, Cardounel AJ, Zweier JL, Garcia JG, Natarajan V. Hyperoxia-induced NAD(P)H oxidase activation and regulation by MAP kinases in human lung endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;284:L26–38. doi: 10.1152/ajplung.00123.2002. [DOI] [PubMed] [Google Scholar]
- 18.Kalinina N, Agrotis A, Tararak E, Antropova Y, Kanellakis P, Ilyinskaya O, Quinn MT, Smirnov V, Bobik A. Cytochrome b558-dependent NAD(P)H oxidase-phox units in smooth muscle and macrophages of atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2002;22:2037–2043. doi: 10.1161/01.atv.0000040222.02255.0f. [DOI] [PubMed] [Google Scholar]
- 19.Papaiahgari S, Zhang Q, Kleeberger SR, Cho HY, Reddy SP. Hyperoxia stimulates an Nrf2-ARE transcriptional response via ROS-EGFR-PI3K-Akt/ERK MAP kinase signaling in pulmonary epithelial cells. Antioxid Redox Signal. 2006;8:43–52. doi: 10.1089/ars.2006.8.43. [DOI] [PubMed] [Google Scholar]
- 20.Usatyuk PV, Gorshkova IA, He D, Zhao Y, Kalari SK, Garcia JG, Natarajan V. Phospholipase D-mediated activation of IQGAP1 through Rac1 regulates hyperoxia-induced p47phox translocation and reactive oxygen species generation in lung endothelial cells. J Biol Chem. 2009;284:15339–15352. doi: 10.1074/jbc.M109.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury I, Mo Y, Gao L, Kazi A, Fisher AB, Feinstein SI. Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radic Biol Med. 2009;46:146–153. doi: 10.1016/reeradbiomed.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singleton PA, Pendyala S, Gorshkova IA, Mambetsariev N, Moitra J, Garcia JG, Natarajan V. Dynamin 2 and c-Abl are novel regulators of hyperoxia-mediated NADPH oxidase activation and reactive oxygen species production in caveolin-enriched microdomains of the endothelium. J Biol Chem. 2009;284:34964–34975. doi: 10.1074/jbc.M109.013771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyle AN, Deshpande NN, Taniyama Y, Seidel-Rogol B, Pounkova L, Du P, Papaharalambus C, Lassegue B, Griendling KK. Poldip2, a novel regulator of Nox4 and cytoskeletal integrity in vascular smooth muscle cells. Circ Res. 2009;105:249–259. doi: 10.1161/CIRCRESAHA.109.193722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Sheppard OR, Shah AM, Brewer AC. Positive regulation of the NADPH oxidase NOX4 promoter in vascular smooth muscle cells by E2F. Free Radic Biol Med. 2008;45:679–685. doi: 10.1016/j.freeradbiomed.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Sturrock A, Cahill B, Norman K, Huecksteadt TP, Hill K, Sanders K, Karwande SV, Stringham JC, Bull DA, Gleich M, Kennedy TP, Hoidal JR. Transforming growth factor-beta1 induces Nox4 NAD(P)H oxidase and reactive oxygen species-dependent proliferation in human pulmonary artery smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2006;290:L661–L673. doi: 10.1152/ajplung.00269.2005. [DOI] [PubMed] [Google Scholar]
- 26.Bonello S, Zahringer C, BelAiba RS, Djordjevic T, Hess J, Michiels C, Kietzmann T, Gorlach A. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler Thromb Vasc Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 27.Papaiahgari S, Yerrapureddy A, Hassoun PM, Garcia JG, Birukov KG, Reddy SP. EGFR-activated signaling and actin remodeling regulate cyclic stretch-induced NRF2-ARE activation. Am J Respir Cell Mol Biol. 2007;36:304–312. doi: 10.1165/rcmb.2006-0131OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho HY, Kleeberger SR. Genetic mechanisms of susceptibility to oxidative lung injury in mice. Free Radic Biol Med. 2007;42:433–445. doi: 10.1016/j.freeradbiomed.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 29.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroda J, Nakagawa K, Yamasaki T, Nakamura K, Takeya R, Kuribayashi F, Imajoh-Ohmi S, Igarashi K, Shibata Y, Sueishi K, Sumimoto H. The superoxide-producing NAD(P)H oxidase Nox4 in the nucleus of human vascular endothelial cells. Genes Cells. 2005;10:1139–1151. doi: 10.1111/j.1365-2443.2005.00907.x. [DOI] [PubMed] [Google Scholar]
- 32.Park SJ, Chun YS, Park KS, Kim SJ, Choi SO, Kim HL, Park JW. Identification of subdomains in NADPH oxidase-4 critical for the oxygen-dependent regulation of TASK-1 K+ channels. Am J Physiol Cell Physiol. 2009;297:C855–864. doi: 10.1152/ajpcell.00463.2008. [DOI] [PubMed] [Google Scholar]
- 33.Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.