Abstract
This article describes the use of non-enzymatic nucleic acid circuits based on strand exchange reactions to detect target sequences on a paperfluidic platform. The DNA circuits that were implemented include a non-enzymatic amplifier and transduction to a fluorescent reporter; these yield an order of magnitude improvement in detection of an input nucleic acid signal. To further improve signal amplification and detection, we integrated the enzyme-free amplifier with loop-mediated isothermal amplification (LAMP). By bridging the gap between the low concentrations of LAMP amplicons and the limits of fluorescence detection, the non-enzymatic amplifier allowed us to detect as few as 1,200 input templates in a paperfluidic format.
Introduction
Paper-based microfluidic devices (or paperfluidic devices) have been shown to be inexpensive and robust platforms for chemical and biochemical assays. Reaction components can be directly printed onto the paper,1 and reagents can be trapped on beads within the fiber matrix.2 Moreover, it is possible to create complex, three-dimensional topologies using paper and tape.3 These topologies define fluidic paths that carry samples to multiple analysis/detection regions, enabling multiplex detection.4 Assays implemented on paperfluidic platforms have included pH indicators, protein assays, and lipid sensors.5
Paperfluidic-based assays may prove to be especially useful in resource-limited settings. However, many bio-medically relevant diagnostics require antibodies and / or reporter enzymes, which can be expensive and may not be readily adapted to dehydration and long-term storage on paper.
Recently, alternative amplification assays based solely on reactions involving kinetically-trapped DNA conformers and toe-hold mediated strand displacement reactions have been developed.6 These designed chemical reaction networks are also known as DNA circuits. For example, an enzyme-free amplifier circuit based on catalytic hairpin assembly (CHA) has been described by Yin et. al.7 Our lab and others have previously adapted this amplifier to function with a variety of analytical readouts, including color, fluorescence, and electrochemistry.2, 8, 9
We now suggest that coupling paperfluidic devices with such circuits can potentially yield a robust analytical tool for resource-poor and point-of-care applications. Here we demonstrate that a DNA amplifier circuit can function directly in the context of a paperfluidics platform, and has a better signal-to-noise ratio (SNR) than sandwich-type, immobilization assays. We also demonstrate that the DNA circuits can be modularly integrated with upstream amplification methods such as loop-mediated isothermal amplification (LAMP) which is otherwise difficult to detect and analyze. By using DNA circuits to bridge the gap between low nanomolar amounts of product that result from LAMP and the hundreds of nanomolar amounts of fluorescent signaling molecules necessary to be read by eye, we were able to detect as few as 1,200 molecules of input template.
Results
Execution of DNA circuits during lateral flow
In order to create precisely defined devices on paper, we first define the regions in which water and sample will be confined. A design is generated via image editing software, and then printed on the paper using a commercially available wax printer1. The wax printer is similar to an inkjet printer in that it creates the printed image by dispensing precise droplets of a liquid pigment, molten wax, onto the page. Rather than drying, the wax solidifies as it cools. We anneal the paper strips by heating them to ~150 °C on a hot plate; this melts the wax into the fiber matrix. The annealed wax creates hydrophobic zones that confine water within the untreated paper. Buffer wicks into these regions by capillary action.
DNA circuits and samples can be introduced onto the paperfluidic platform either by spotting liquid or by drying reagents onto the paper itself, and are expected to react as the fluid phase wicks upwards by capillary action. As a proof-of-principle, we first designed a simple linear-fluorogenic circuit for the paperfluidic platform. As with the amplification cascade that will eventually be implemented, this reaction is based on a toehold-mediated strand displacement reaction that occurs between a single-stranded DNA (ssDNA) and a hemi-duplex, double-stranded DNA (dsDNA) with an overhanging single-stranded region (toehold).
Fluorogenesis occurs when domain 1 on strand S (the "sample" strand) forms a transient, weak interaction with the toehold subsequence 1* on strand Q (quencher strand). Hybridization to the toehold allows strand S to initiate branch migration between complementary regions 2 and 2*. The strand-displacement reaction proceeds because displacement of the short strand yields a longer, more stable double-stranded DNA (complex S-Q), concomitantly displacing the shorter strand F (fluorophore strand; Figure 1A). The formation of the double-stranded product is to a first approximation irreversible as there is no toehold for F to re-initiate branch migration.
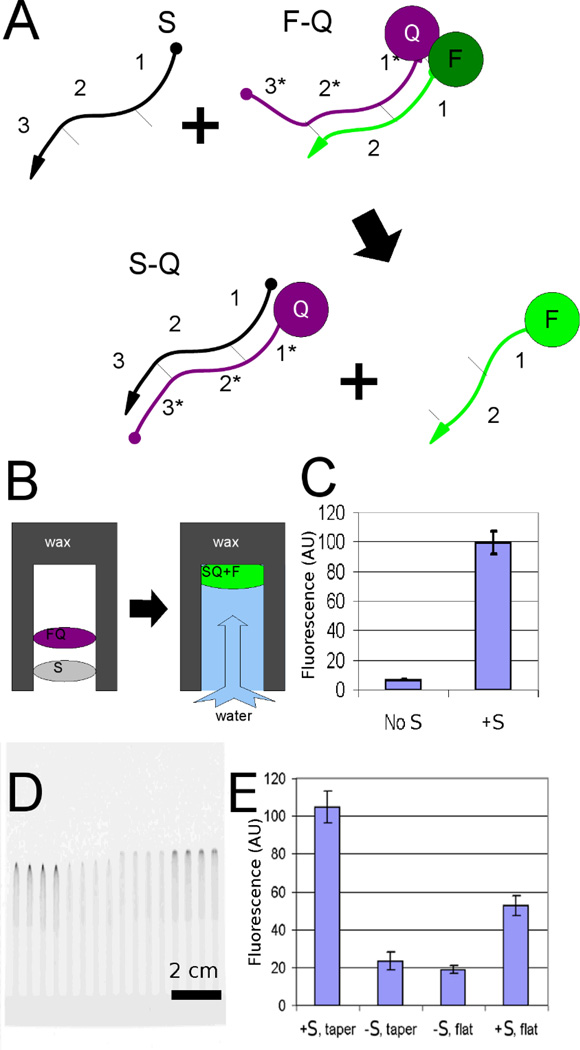
Figure 1. Detection of DNA circuits in paperfluidic devices.
(A) Schematic of a strand displacement reaction. A fluorogenic DNA strand displacement reaction involves displacement of a paired fluorescein-DNA (F) and quencher-DNA (Q) hybridized to one another by interaction of the quencher-bearing strand with a sample (S) via an engineered toehold (3'). (B) Schematic of the reaction in the context of the paperfluidic device. This reaction occurs while the DNA wicks through the paper, resulting in a moving, fluorescent spot. (C) The linear-fluorogenic circuit functions properly in homogeneous solution, as shown by plate reader measurements of the system. (D) Fluorescence image showing the raw results on the paper device; data from this image were processed to show (E) the fluorescence increase resulting from the linear-fluorogenic circuit operating within the paper. Signal detection is increased by a factor of 2 by merely introducing tapered geometry into the paperfluidic device.
By drying the F-Q complex onto the paper we can potentially create a fluorogenic detector for sample strand S (Figure 1B). The fluorescence increase in solution due to the addition of one mole-equivalent of S to F-Q is shown in Figure 1C. The final fluorescence image of the device run to completion is shown in Figure 1D. After incubation time of 20 minutes, the signal-to-noise ratio (SNR) was 12 and the signal-to-background ratio (SBR) was 15. In comparison, when S was applied to the same circuit pre-embedded in the paperfluidic device there was a SNR of 9.5 and a SBR of 2.5 (Figure 1E). This figure serves two purposes: to show the functionality of the linear-fluorogenic circuit and to show the enhancement due to the tip geometry.
We reasoned that by guiding signal into a smaller volume at the end of the channel we could more readily detect signals. We therefore tapered the end of the hydrophilic region of the paper test strip to various angles. In Figure 1E the original flat-ended geometry typical of most paperfluidic devices is compared with a more optimized angle geometry. The fluorogenic response was enhanced by the tapered regions (at left) relative to the flat strips (at right). Moving from a square detection region to a tapered detection region improved the signal by a factor of 2 and the SNR from 6.4 to 9.6.
Signal amplification on paper
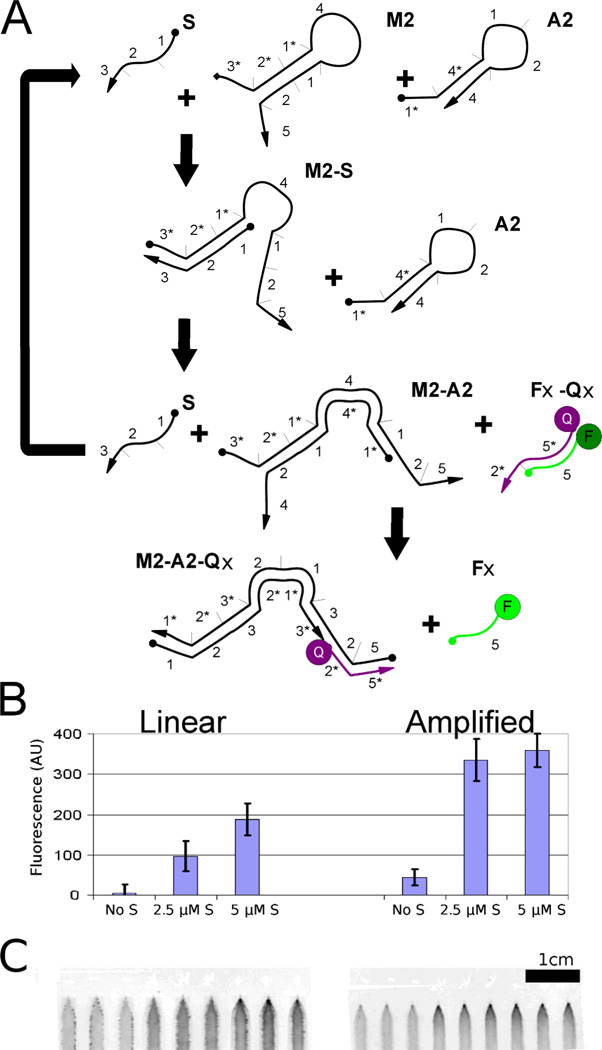
We previously designed a DNA-based amplification scheme based on the catalytic hairpin assembly (CHA) methods developed by Yin and Pierce10 (Figure 2A). In the system we will call the CHA-amplified-fluorogenic circuit, two DNA oligonucleotides form kinetically trapped hairpins that cannot initially react with one another. However, when the sample / catalyst strand S binds to the toehold of one the hairpins (M2), strand exchange leads to opening of the hairpin and the revelation of a single-stranded toehold (domain 1 on molecule M2). The open toehold of M2 interacts with the second hairpin (domain 1*, with the asterisk indicating complementarity, on molecule A2) to form a long region of dsDNA, again by strand exchange. The completion of the strand exchange reaction leads to displacement and release of the catalyst strand, which recycles to catalyze additional hairpin joining reactions.
Figure 2. Amplifier circuits in paperfluidic devices.
(A) Schematic of the CHA amplifier. A sample strand, S, acts as a catalyst that allows two complementary hairpins (A2 and M2) to form a double-stranded product, with concomitant release of a fluorescent oligonucleotide from Fx-Qx, similar to the strand-displacement reaction shown in Figure 2. (B) Fluorescent readout of the amplifier. The amplifier executed in the context of the paperfluidic device showed three-fold higher signal than the equivalent linear system described in Figure 2. (C) Shows the a fluorescence image of the paper strip from which data was collected
As can be seen in Figure 2B, reactions with the CHA-amplified-fluorogenic circuit yield significantly higher signal-to-background than with the linear-fluorogenic circuit. However, it is also clear that the apprehended signal is not linearly proportional to the analyte added: when 2.5 µM of the sample strand was added (one-half molar equivalent relative to the hairpin molecules), the amplifier still produced 95% of the full response. This differed from the previously described linear-fluorogenic circuit, in which larger inputs (one molecular equivalent relative to hairpin circuit concentrations) produced only 50% of the expected signal. It should be noted that the CHA-amplified-fluorogenic circuit may continue to react over time as all of the reagents are eventually all co-localized at the end of the strip. Thus, detection sensitivity and background will be time-dependent. Thus, in situ the CHA-amplified-fluorogenic circuit showed more than a 3-fold gain in raw signal, perhaps due to molecular crowding effects. Similarly, for an input of 2.5 µM S, the linear-fluorogenic circuit yielded a SNR of 2.5 while the CHA-amplified-fluorogenic gave a SNR of 5.5. Due to higher background (which may also result from molecular crowding), the SBR decreases from 21 in the linear-fluorogenic circuit to 7.5 for the CHA-amplified-fluorogenic circuit.
Altering the device to improve signal detection
One issue that became apparent early in the development of the platform was that the fluorescent signal that arose from strand displacement reactions was diffuse. In order to concentrate the signal, we attempted two modifications to the paperfluidic device: tapering the end of the wax-printed channel (discussed above), and embedding a DNA-coated bead within the paper to capture fluorescent oligonucleotides.
Allowing the reagents and products to migrate together to the end of the strip is simple, but requires that both a fluorophore and a quencher be present. In contrast, in other assays such as ELISA the detection region is washed free of unreacted signal transducers, further enhancing the signal relative to the background. Such a strategy is also possible for paperfluidic devices, but requires immobilized capture reagents11. While it might be possible to chemically modify a portion of the cellulose itself12, this would greatly increase the difficulty of fabrication. Instead, we embedded a suspension of micron-sized, DNA-functionalized beads directly within the paper matrix2.
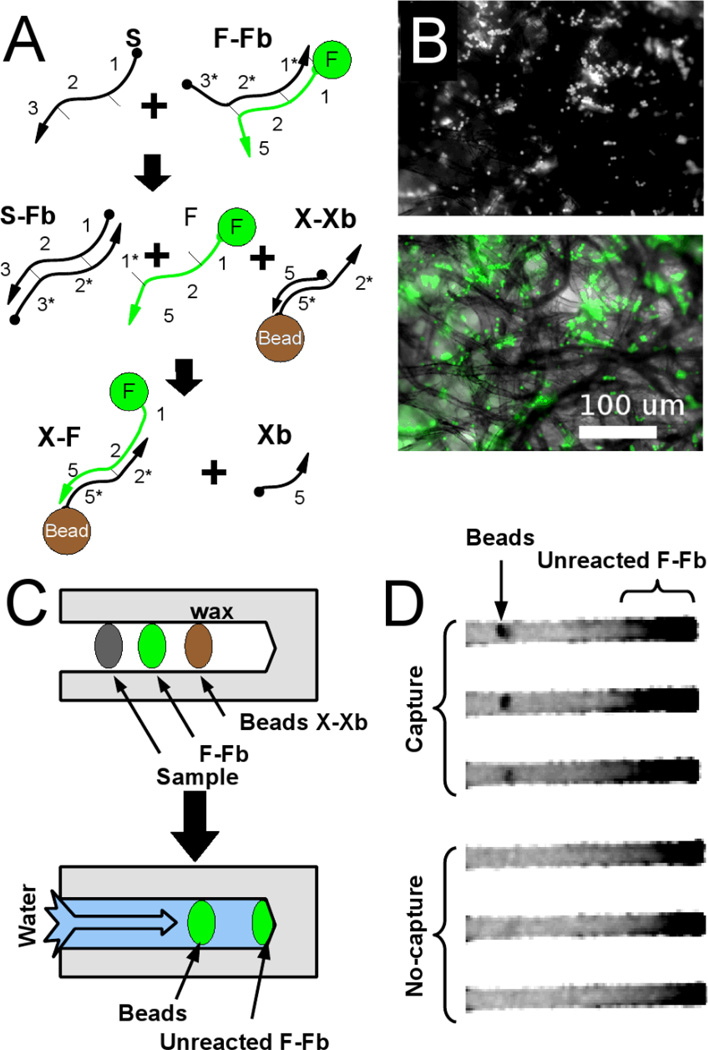
Strand capture on the immobilized beads occurs as outlined in Figure 3A which describes a linear-immobilization circuit. Sample strand S displaces the fluorophore strand F from a complementary oligonucleotide Fb, similar to the quencher strand previously described. Both the starting material and the product of this reaction are fluorescent, but only the free, single-stranded F product should specifically interact with the bead. F initiates a strand-displacement reaction with the immobilized hemi-duplex X-Xb to generate the final product X-F. In this design, un-reacted fluorescent material is carried past the beads while immobilized fluorescent DNA remains on the bead, reporting the presence of the original input sequence.
Figure 3. Signal immobilization in the paperfluidic device.
(A) Schematic of the reaction. Released fluorescent oligonucleotides can be captured at specific sites within the paperfluidic device, via beads containing antisense oligonucleotides. (B) Micrographs of immobilized beads. Beads caught in the paper fibers (fluorescence micrograph, top, brightfield, bottom) capture fluorescent oligonucleotide. (C) Schematic of paperfluidic device for detection by oligonucleotide capture on beads. A detection region (Beads X-Xb) is embedded into the paper just downstream of the DNA reagents. (D) Fluorescence image of successful capture showing residual, unreacted F-Fb downstream of bead spots.
It is also worth noting that the immobilized X-Xb construct contains both the capture oligonucleotide as well as a blocker. This design feature ensures that immobilization of F should only occur in the presence of free, fully single-stranded F. While other complexes in the system have partial complementarity to X, they lack the toehold contained in single-stranded F that allows displacement of Xb and concomitant immobilization.
We validated the linear-immobilization circuit in solution using gel electrophoretic analysis (data not shown) before applying it to the paperfluidic device. Simply pipetting the beads onto the strip led to their being physisorbed and embedded in the device. We initially used fluorescent beads to establish that flow through the paper strip did not cause the beads themselves to move. We attribute their relative immobility to physisorbtion because the beads are much smaller than the physical void spaces in the paper matrix (Figure 3B). For the purposes of our diagnostic assay, the DNA construct X-Xb is immobilized on beads by a biotin-avidin linkage. Any free F generated upstream of the beads should flow over the beads and become immobilized (Figures 3C,D).
Quantitative advantage of paper-based CHA circuits with embedded beads
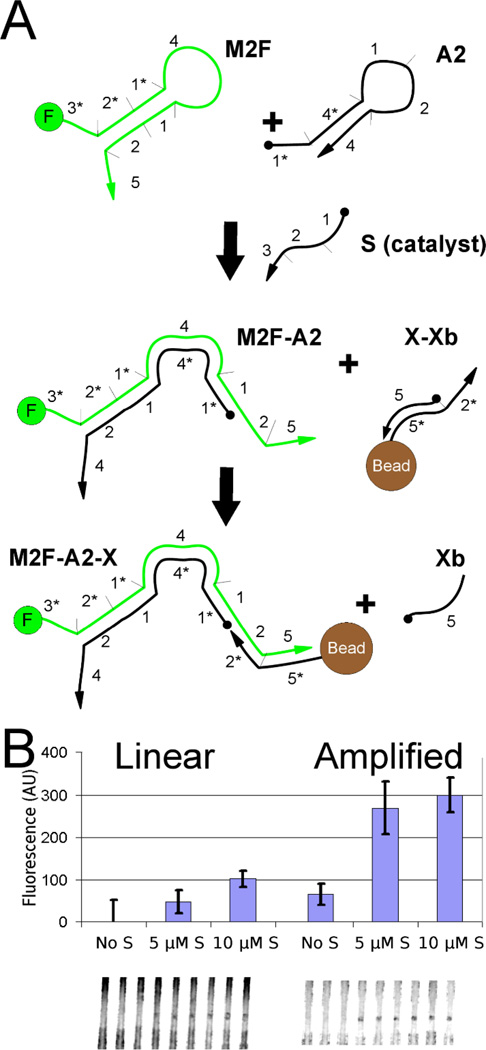
Having established the validity of the linear-immobilization circuit above, we proceeded to compare its performance to a CHA-amplified-immobilization circuit. This is very similar to the CHA-amplified-fluorogenic circuit presented above but detects an oligonucleotide sample (S) via the capture of the fluor on an embedded bead. Our strategy for applying the CHA-amplified-immobilization circuit is shown in (Figure 4A). The results indicate that both the linear-immobilization circuit and CHA-amplified-immobilization circuit can yield signals with embedded beads (Figure 4B). We found that the maximal fluorescence produced by the amplification circuit was higher by a factor of 3. At 5 µM of sample, the linear-immobilization circuit has a SNR of 2.3 while the CHA-amplified-immobilization circuit has a SNR of 3.2. This result was especially encouraging in that it showed that the DNA circuit can in fact turnover within the context of the paperfluidic device, and within the relatively short reaction time that is available during lateral flow (~5 minutes).
Figure 4. In situ CHA-amplification of signal.
(A) Schematic diagram of a second implementation of the amplifier. Again sample strand, S, acts as a catalyst that allows two complementary hairpins to hybridize to each other. However, in this design the full M2F-A2 duplex can now bind to immobilized beads. (B) Comparison of linear and amplified signals. In a direct comparison, the CHA-amplified-immobilization circuit shows five times higher signal than the linear-immobilization system described earlier.
The amplifier coupled with bead-based detection gave a non-linear response to the input sample strand (Figure 4B). Reducing the input sample by 50% (from 10 to 5 µM) caused only a 8% loss of fluorescent signal on the beads (within one standard deviation among triplicate measurements). Thus, the system is likely saturated, and the possible extent of CHA amplification may be much higher.
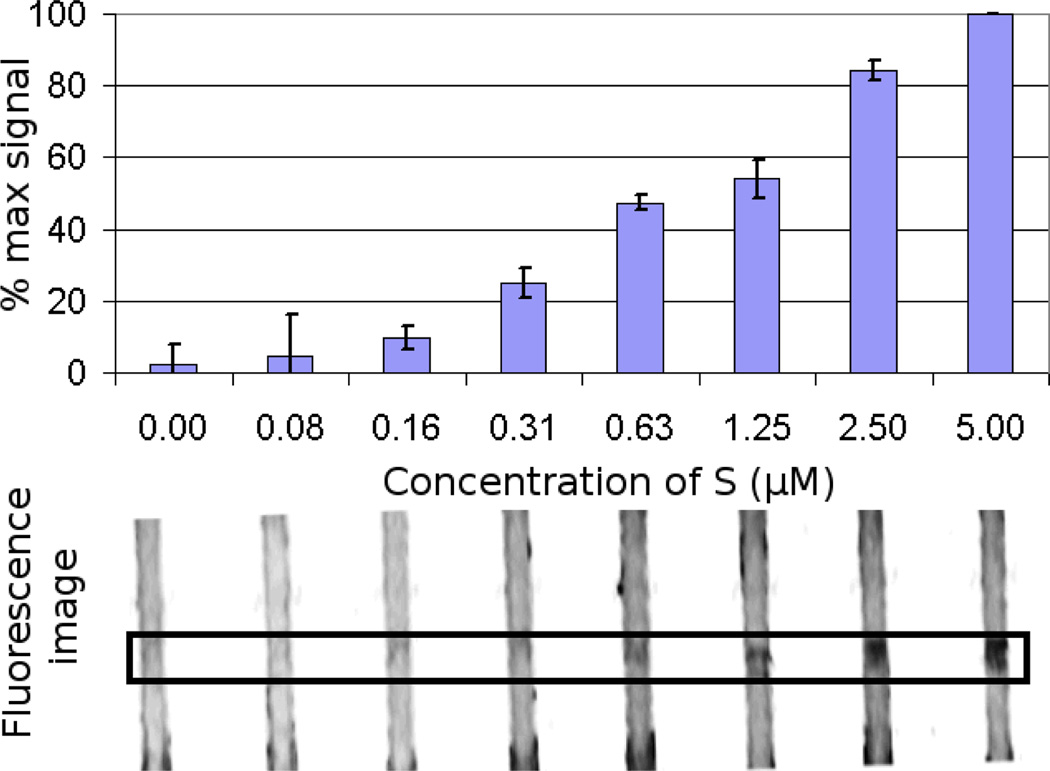
To more precisely determine the sensitivity for the CHA-amplified-immobilization circuit, a range of oligonucleotide sample (S) concentrations were assayed. Below 5 µM sample concentration, the input does not saturate the device, although the dose-response curve is still non-linear. The CHA-amplified-immobilization circuit maintained a SNR of 3 even at 0.3 µM sample concentration (Figure 5) indicating an improvement of 16-fold in the limit of detection (LOD) relative to the linear-immobilization circuit. Error bars in Figure 5 represent the standard deviation between devices (i.e. the deviation of the normalized results on separate printed paper strips run at different times), and indicate that at 310 nM, the signal can be discriminated above background with a >95% confidence.
Figure 5.
Limit of detection for the CHA-amplified-immobilization circuit on the paperfluidic device. The limit of detection (LOD) at 3 standard deviations above zero input is just above 0.3 µM.
Integration with Loop Mediated Isothermal Amplification (LAMP)
Loop-mediated isothermal amplification (LAMP) is a very sensitive amplification system that produces nanomolar concentrations of double-stranded DNA concatamers from even a few molecules of input template. Unfortunately, since our paperfluidic system had a detection limit on the order of hundreds of nanomolar, we needed to amplify the LAMP product (LP) signal by at least two orders of magnitude. In contrast to PCR, LAMP does not require thermal cycling. This is potentially advantageous for low-resource diagnostic as it does not require a sophisticated temperature control system. LAMP products can be difficult to detect, since the LP is not a single DNA species, but rather a collection of concatamers of different lengths. For example, on a gel, the LP appears as a periodic mixture of bands (see also Figure 6C).
Figure 6.
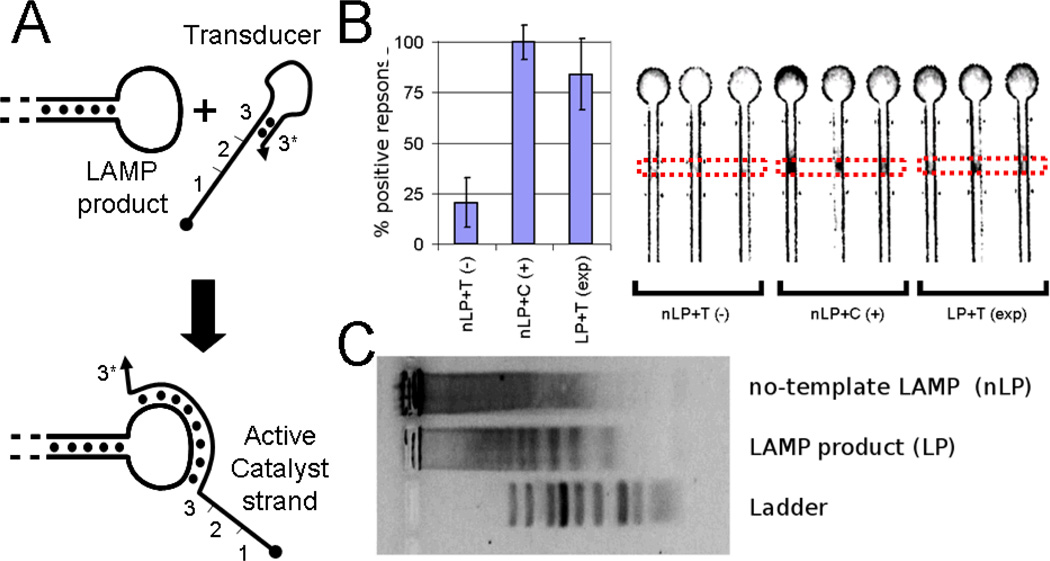
(A) Schematic showing strategy by which a transducer (T) binds to the ssDNA loop of the LAMP product (LP) to activate a DNA catalyst for CHA. (B) Fluorescence was measured in region indicated by dotted lines. The results show that the transduced output (LP+T) generates a clear output in the paperfluidic format comparable to the positive control, a LAMP reaction without template, and spiked with 5 nM active catalyst in place of the transducer (nLP+C) and well above the null template control (nLP+T). (C) A agarose gel shows that the LP gives a periodic series of concatamers while the null-template control (nLP) shows no specific product.
At each end of every LAMP concatamer is a single-stranded loop sequence (Figure 6A), which at the conclusion of the reaction is present at low nM concentrations. These single-stranded loops can in principle be used to open a synthetic hairpin DNA with a complementary loop sequence. This reaction would in turn reveal a single-stranded DNA that could function in a manner similar to the single-stranded catalyst described above. We refer to the hairpin as a “transducer,” since it takes the specific signal from the LP and creates a catalyst for CHA amplification circuits, as has been described elsewhere.13
In practice, we performed LAMP according to published protocols,14 incubated LP with the CHA reagents and transducer (T), then applied the product to the paper strip. CHA bridges the sensitivity gap between the high amplification, low concentration LAMP reaction and the lower-sensitivity paperfluidic detection. The LP (or a null-template control) was annealed with T at 5 nM final concentration. The CHA hairpin DNA was then added to 600 nM final concentration and allowed to incubate for several hours. Because the LAMP product was produced at low molar concentrations, the final concentration of the active catalyst for the CHA reaction was also low. The small number of active catalyst DNA molecules had to react for longer times (5 hours) to produce a significant quantity of product for detection by immobilization. This mixture was then applied to the paper upstream of the detection beads, as before.
While either LAMP or CHA on their own would have yielded too little amplification to allow observation on the paperfluidic platform, together they provided robust signaling (Figure 6B). Indeed, the LAMP-CHA-paperfluidics circuit proved to be extremely sensitive. A sample of 1200 molecules of input template (M13 viral DNA from a commercial source) was amplified by LAMP. After the LAMP reaction was complete, the product was successfully applied to the CHA-amplified-immobilization circuit that had been previously pre-fabricated on a paper device. In the final combination of LAMP and CHA with paperfluidic detection (LAMP-CHA-immobilization circuit), as little as 80 attomolar of template was detected.
Though LAMP is extremely sensitive, one of its disadvantages is that it is prone to amplifying non-specific sequences. When analyzed by gel electrophoresis, successful LAMP produces a characteristic periodic series of bands (Figure 6C). Unfortunately, even the no-template control shows amplified concatamers, likely due to self-priming by the primers. Diagnostic interpretation of the product-specific versus non-product-specific banding patterns would obviously be difficult or impossible. However, by using the transducer to a specific target sequence, the LP is rendered into a binary, easily read signal. Unlike current detection schemes15 based on total DNA yield, nonspecific amplification is filtered out by this system.
Similarly, while LAMP reactions have previously been detected using lateral flow and immuno-gold readouts16, 17, our technique does not require antibodies of any sort. All of the reagents on the test strip can potentially be chemically synthesized. This is important because proteins are generally not amenable to storage in dry, hot conditions, especially when dried onto paper, while DNA circuits are stable when heated and dried. We performed experiments that indicate reagents function properly after being allowed to dry ~5 min on the paper (data not shown) but we have not extensively studied the effects of long-term storage. Although we chose to use the protein avidin to immobilize the X-Xb DNA construct on the beads, this could also have been accomplished by EDC coupling of amine-DNA to carboxylate beads18, thereby avoiding proteins on the paper strip altogether.
Materials and Methods
Materials
All DNA was purchased from IDT (Coralville, Iowa) and PAGE purified unless otherwise stated in the text. Sequences for DNA circuit components are as follows:
S: CGA CAT CTA ACC TAG CTC ACT GAC;
F: 5’Fluorescein-AGA CGA CAT CTA ACC TAG CCC TTG TCA TAG AGC AC;
Fb: GTC AGT GAG CTA GGT TAG ATG TCG CCA;
Q: GTC AGT GAG CTA GGT TAG ATG TCG CCA-3’IowaBlack;
M2: TCA GTG AGC TAG GTT AGA TGT CGC CAT GTG TAG ACG ACA TCT AAC CTA GCC CTT GTC ATA GAG CAC;
M2F: 5’Fluorescein-TCA GTG AGC TAG GTT AGA TGT CGC CAT GTG TAG ACG ACA TCT AAC CTA GCC CTT GTC ATA GAG CAC;
A2: AGA TGT CGT CTA CAC ATG GCG ACA TCT AAC CTA GCC CAT GTG TAG A;
Fx: 5’Fluorescein-GTG CTC TAT GAC AAG GGC TAG GTT;
Qx: GCC CTT GTC ATA GAG CAC-3’IowaBlack
X: 5’Biotin-GTG CTC TAT GAC AAG GGC TAG GTT;
Xb: GCC CTT GTC ATA GAG CAC;
T: CGA CAT CTA ACC TAG CTC ACT GAC CGA TAT TCG TAA TCA TGG TCA TAG CTG TTA TCG GTC AGT GAG C
Primers for LAMP were also obtained from IDT and used as received. The sequences were the same as those described by Tomita et. al.14 and are as follows:
LAMP_M13BIP: CGA CTC TAG AGG ATC CCC GGG TAC TTT TTG TTG TGT GGA ATT GTG AGC GGA T
LAMP_M13FIP: ACA ACG TCG TGA CTG GGA AAA CCC TTT TTG TGC GGG CCT CTT CGC TAT TAC
LAMP_B3: ACT TTA TGC TTC CGG CTC GTA
LAMP_F3: GTT GGG AAG GGC GAT CG
Single stranded M13 template DNA was purchased from New England Biolabs (Ipswitch, MA) and used as received. Phosphate-buffered saline (PBS) was purchased from Thermo-Fisher (Waltham, MA). Binding buffer (BND) was prepared by adding magnesium chloride (Sigma-Aldrich St. Louis, MO) to PBS to a final concentration of 10 mM and sterile-filtering with a steri-flip filter (Millipore, Billerica, MA). All streptavidin-coated beads (10 µm non-fluorescent, and 1 µm non-fluorescent) were used as received (Bangs labs, Fishers, IN).
Fabricating paperfluidic devices
Wax printing was accomplished with a Xerox Phaser type printer (Xerox corp., Norwalk, CT) using only Xerox brand ink blocks. To create paperfluidic devices, Whatman brand 1 CHR type filter paper (obtained from Sigma-Aldrich St. Louis, MO) was loaded into the manual feed of the printer and the desired design was printed. The paper was then “annealed” by placing it on a heating block at 150 °C for 2 min. We confirmed the beads were distributed through the void volumes by using fluorescence microscopy (BX51 microscope, Center Valley, PA) and 7 um, green-fluorescent beads (Bangs labs, Fishers, IN). Beads were applied slowly to the paper with a pipetteman fitted with a 1–10 µl pipette tip (Rainin brand, Mettler-Toledo, OH).
Verification of the fluorogenic reaction
The fluorogenic reporter’s function was verified in a solution fluorescence assay. Briefly, the DNA displacer was added to a a pre-annealed solution of the fluorescein-quencher construct (FQ) and the increase in fluorescence was characterized. In more detail, both components of the FQ complex were added to BND buffer at 5 µm, annealed at 80 °C for 3 minutes, then cooled at 0.1 °C per second to room temperature. A mixture of all reaction components was prepared to a final volume of 10 µl in a 384 well plate. Triplicate samples were prepared with and without the sample “S” DNA (also at 1 µm). The plate was then read with a Safire plate reader (Tecan, Männedorf Switzerland). The final fluorescence was determined after 20 minutes and the standard deviation among the triplicates of the samples was calculated.
Preparation of linear fluorogenic reactions on a paper strip
The linear-fluorogenic circuit was prepared similarly for application to a paper strip. Briefly, we prepared the DNA species in BND buffer, pipetted them directly onto the strip in the appropriate locations and then dipped the strip into a dish of PBS and waited for the solution to wick to the top of the strip. The strip was then scanned.
For the linear-fluorogenic circuit, the “FQ” DNA construct was prepared as above by mixing fluorescein conjugated “F” and complementary, black-hole-quencher conjugated “Q” to a final concentration of 5 µM in BND buffer. We annealed at 80 °C for 3 minutes then cooled at 0.1 °C per second to room temperature.
When preparing paper strips, 1 µl of the 5 µM reporter complex solution (F-Q) was pipetted onto each lane of the paperfluidic device. Sample (“S”) at the appropriate concentration (5, 2.5 or 0 µM) was pipetted upstream of the fluorogenic reporter, just inside the open area through which buffer would wick.
The open end of the paper device (outside the lanes) was dipped into a crystallization dish of PBS. The strip was held in a vertical position with a binder clip. The PBS wicked up into the device through the spots of “S”, the other reagents, and reached the top of the strip after approximately 10 minutes. At this time the device was transferred to a fluorescence scanner, (Storm, GE Healthcare, Piscataway, NJ) covered with a sheet of clear acrylic to hold it flat and retain moisture, and scanned. It should be noted that fluorescein most fluorescent while it is hydrated. The average intensity standard deviation among triplicate measurements was measured with the free software package ImageJ. The fluorescent regions were identified by inspection, and regions of consistent size and shape were extracted. The numerical intensity data from these regions were averaged to produce a single measurement from each hydrophilic track on the device.”
Preparation of amplified fluorogenic reactions
The CHA-amplified-fluorogenic circuit was assembled and performed on the paper device very similarly to the strand-dispalcement case above. We made separate “M2,” and “A2,” solutions in BND at 5 µM. These separate solutions were then annealed at 80 °C for 3 minutes with rapid cooling (1 °C per second) to favor the hairpin conformation. We prepared a sample of “Fx-Qx” at 5 µM of each component in BND and annealed at 80 °C for 3 minutes with slow cooling (0.1 °C per second). We pipetted 1 µl of the fluorogenic Fx-Qx reporter complex into each lane. Just upstream of the reporter, 1 µl each of the CHA reagents “M2” and “A2” were pipetted onto each lane of the device. Just upstream of the CHA reagents we pipetted 1 µl of the appropriate sample (at 5, 2.5 or 0 µM). This was then dipped, allowed to wick, and scanned and measured as above.
Preparation of linear-immobilizad circuit and CHA-amplified-immobilization circuit
Bead based assays begin with the functionalizing of the beads with the X-Xb cosntruct. The DNA to be immobilized on the beads was prepared by mixing biotinylated “X” and complementary “Xb” (both used as-received without gel purification) to a final concentration of 100 µM in BND buffer. This solution was annealed at 80 °C for 3 minutes then cooled at 0.1 °C per second to room temperature. We added 0.2 nMol X-Xb of the annealed construct to 20 µl of the 10 µm, streptavidin-coated, non-fluorescent beads (as provided, 1% solids). The beads and biotinylated DNA were incubated ~10 minutes at room temperature then washed 3 times by centrifugation and resuspension in 20 µl of superblock (Thermo Fisher Scientific, Rockford, IL). One µl of the bead suspension was then pipetted onto each lane of the paperfluidic device very slowly to keep the beads in a small area. It should be noted that the beads were still suspended in superblock; this also serves to block the detector region of the paper and reduces background.
All other reagents are then spotted onto the paper and run the procedure in otherwise the same manner as for the fluorogenic assays. We annealed the complex “F-Fb” at final concentration 10 µM with 0.1 °C per second cooling. We annealed “S,” “M2F,” and “A2,” separately at 10 µM with 1 .1 °C per second cooling. 1 µl of either “F-Fb” complex (for linear reporter) or 1 µl each “M2F” and “A2” (for amplified reporter) were pipetted just upstream of the beads. Finally, the sample (“S”) at the appropriate concentration (10, 5 or 0 µM) was pipetted just inside the open area through which buffer would wick and the devices were dipped in PBS, allowed to wick, and were scanned and measured as above.
Determination of the limit of detection of the amplified immobilization-based assay
In order to determine the limit of detection of the CHA-amplified-immobilization circuit, we tested the response of the beads-immobilization detection system to catalytic sample DNA at decreasing concentrations. We annealed the sample DNA, “S,” at 5 µM then diluted samples 2:1 in BND to generate a series of concentrations from 5 µM to 80 nM. We prepared the “X-Xb” coated beads, M2F and A2 as above. We and added 1 µl of the beads and 1 µl of each of the CHA reactions to each lane as above. The strip was then dipped into PBS as above. To increase the incubation time, the device was held out of the PBS so that wicking was “paused” for 90 seconds then returned and allowed to wick normally. The devices were scanned as above. This was repeated three times with three separate paper devices. We used the program ImageJ to isolate circular regions corresponding to the locations of the beads. We extracted the average numerical intensity from each region of beads on each paperfluidic device. For each device, we normalized the regions’ average intensity values relative to the most intense region on the device. We then repeated this for the three devices. The error bars at each concentration represent the standard deviation of the average over the three devices.
LAMP reaction detected on paper
A loop mediated isothermal amplification (LAMP) reaction was prepared against M13 phage DNA as described elsewhere. Briefly, four primers specific to M13 phage DNA (see DNA sequence information, above) were added to LAMP buffer (1M betaine, 20 mM Tris, 10 mM KCl, 10 mM ammonium sulfate, 3 mM magnesium sulfate, and 0.1% Triton X-100 all acquired from Sigma Aldrich, St Louis, MO). Final concentrations were 0.8 µm for LAMP_M13BIP, and LAMP_M13FIP; 0.4 µm for LAMP_B3, and LAMP_F3. Finally, 1200 molecules of template were added to a 25 µl reaction. A no-template control was also prepared. Both were annealed at 80 °C then 1 µl of enzyme (bst polymerase, NEB, Ipswich, MA) was added and the solutions were incubated at 65 °C for 90 minutes. For paperfluidic analysis, 2.5 µl of the LAMP product (LP) and the null control (nLP) were then diluted into 7.5 µl of BND buffer and transducer (T) or active catalyst (S, for positive control) were added to 5 nm final concentration. This was then annealed at 95 °C for 5 min and cooled slowly (0.1 °C per second) to room temperature. Finally, M2 and A2 were annealed separately and added to a final concentration of 1.2 µM each. These samples were allowed to incubate for 5 hours at 37 °C then spotted onto test strips upstream of 1 µl, 1 µm avidinylated beads coated with X-Xb and dipped in PBS as above. Samples LP and nLP were also analyzed by agarose gel electrophoresis (2% SeaPlaque, Lonza, ME) in TBE with ethidium bromide. The 20 base pair dsDNA ladder was purchased from Jena Biosciences (Jena, Germany).
Conclusions
Paperfluidics provides many advantages as a platform for low-resource chemical or biochemical analysis. It is inexpensive, easily patterned, and physically robust. Detection can be accomplished by chemical changes in the detection reagents (e.g. de-quenching a fluorophore). This has been the dominant mode in the literature. Chemical changes inducing an observable, visual change in the color of the paper has been shown with a paper colorimetric device for measuring simultaneous glucose and protein in urine5. We designed five DNA circuits for detection of nucleic acids with a specific sequence by transduction to a fluorescent signal. These circuits were applied to linear or amplified, fluorogenic- or immobilization-based detection schemes that can be adapted to arbitrary targets including the product of an isothermal amplification step.
Our fluorogenic scheme is similar to previous colorimetric assays developed for paperfluidics. A optical change is caused by a specific target molecule. We note that in such schema, the detection is affected by the geometry of the detection point and that an optimal shape can improve the performance of the device. However, without a specific colorimetric or fluorogenic assay, such approaches will not work. Additionally, this outcome ot this approach is affected by the time between developing the strip and reading the response.
We also present an alternate approach where a fluorescent reporter is immobilized on beads contingent on the presence of the target molecules. The dynamics of water in paper is determined by the intrinsic properties of the device, rather than by sophisticated pumps and feedbacks as in other microfluidic systems19. This means that the time of the reaction can be defined by the geometry of the strip as the fluid moves past the detection point into a reservoir due to predictable rates of wicking. We show this in the immobilization-based detection mode which showed a detection limit of ~300 nM. In all of the schemes presented here, the final detection mode was fluorescence. Published reports have suggested that fluorescence readers can be made inexpensively20 and paper devices can be integrated with cellular phones21. Alternatively, colorimetric assays may be preferable and can also be adapted to the DNA circuit described here,8 an option that may be of particular use in low-resource settings.
Without transduction, our targets are currently limited to single stranded nucleic acids (e.g. mRNA, viral ssDNA, etc.). This may find uses in detecting specific single stranded markers for pathogens; sensitive detection of trypanosomes has been accomplished by targeting single-stranded mRNA22. It may also be possible to couple this or a similar strategy to immobilized antibodies or aptamer transducers (so-called “aptamer beacons”) in the future23. Thus, the enzyme-free CHA amplifier may be directly applicable even in the absence of LAMP.
We have demonstrated enzyme-free CHA-amplified circuits with limits of detection in the sub-micromolar range. Natural analytes may be present only in concentrations below this level. We suggest that improvements in DNA circuit design (such as nested amplifiers) can improve the limit of detection by several orders of magnitude7 and may eventually allow for practical use of a CHA-amplified paperfluidic system without LAMP or other pre-amplification.
By annealing the transducer with the target, we have demonstrated that we can broaden the type of analytes to include single stranded loops in the product of the LAMP reaction. LAMP has extremely high gain (in our case, greater than 108×) but relatively low molar output concentration of active, single-stranded, specific product (nM). If the catalytic amplifier circuit is given sufficient time to react, it can bridge the gap between the output concentration of LAMP and the detection limit of the paperfluidic strip for a combined sensitivity at the level of attomolar.
Acknowledgment
All or part of this work was funded by the National Institute of Health Fellowship (1 F32 GM095280-02), National Institute of Health (R21-HG005763-01, 1 R01 GM094933-02, and 5 R01AI092839-02), The Welch Foundation (F-1654), and National Security Science and Engineering Faculty Fellowship (FA9550-10-1-0169). The published material represents the position of the author(s) and not necessarily that of the sponsors.
Notes and references
- 1.Carrilho E, Martinez AW, Whitesides GM. Anal. Chem. 2009;81:7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- 2.Su S, Ali MM, Filipe CDM, Li Y, Pelton R. Biomacromolecules. 2008;9:935–941. doi: 10.1021/bm7013608. [DOI] [PubMed] [Google Scholar]
- 3.Martinez AW, Phillips ST, Nie Z, Cheng C-M, Carrilho E, Wiley BJ, Whitesides GM. Lab Chip. 2010;10:3428. doi: 10.1039/c0lc00021c. [DOI] [PubMed] [Google Scholar]
- 4.Fenton EM, Mascarenas MR, LoÌ pez GP, Sibbett SS. ACS Appl. Mater. Interfaces. 2009;1:124–129. doi: 10.1021/am800043z. [DOI] [PubMed] [Google Scholar]
- 5.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Angew. Chen. Int. Edit. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 7.Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Ellington AD, Chen X. Nuckeic Acids Res. 2011;39:e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dungchai W, Chailapakul O, Henry CS. Anal. Chem. 2009;81:5821–5826. doi: 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- 10.Eckhoff G, Codrea V, Ellington AD, Chen X. J. Syst. Chem. 2010;1:13–13. [Google Scholar]
- 11.Cheng C-M, Martinez AW, Gong J, Mace CR, Phillips ST, Carrilho E, Mirica KA, Whitesides GM. Angew. Chen. Int. Edit. 2010;49:4771–4774. doi: 10.1002/anie.201001005. [DOI] [PubMed] [Google Scholar]
- 12.Bora U, Sharma P, Kannan K, Nahar P. J. Biotechnol. 2006;126:220–229. doi: 10.1016/j.jbiotec.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Li B, chen X, Ellington A. Angew. Chen. Int. Edit. 2012 in submission. [Google Scholar]
- 14.Tomita N, Mori Y, Kanda H, Notomi T. Nat. Protocols. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 15.Tao Z-Y, Zhou H-Y, Xia H, Xu S, Zhu H-W, Culleton RL, Han E-T, Lu F, Fang Q, Gu Y-P, Liu Y-B, Zhu G-D, Wang W-M, Li J-L, Cao J, Gao Q. Parasite. Vector. 2011;4:115–115. doi: 10.1186/1756-3305-4-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiatpathomchai W, Jaroenram W, Arunrut N, Jitrapakdee S, Flegel TW. J. Virol. Methods. 2008;153:214–217. doi: 10.1016/j.jviromet.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Jaroenram W, Kiatpathomchai W, Flegel TW. Mol. Cell. Probes. 2009;23:65–70. doi: 10.1016/j.mcp.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Wittebolle L, Verstuyft K, Verstraete W, Boon N. J. Chem. Technol. Biot. 2006;81:476–480. [Google Scholar]
- 19.Allen PB, Milne G, Doepker BR, Chiu DT. Lab Chip. 2010;10:727–727. doi: 10.1039/b919639k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan H. DNA Computing. Springer Berlin / Heidelberg: 2005. pp. 653–658. [Google Scholar]
- 21.Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A. Lab on a Chip. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, Sugimoto C, Igarashi I. J. Clin. Microbiol. 2003;41:5517–5524. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamaguchi N, Ellington A, Stanton M. Anal. Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]