Abstract
A micro free flow electrophoresis (μFFE) device was used to select DNA aptamers for human immunoglobulin E (IgE). The continuous nature of μFFE allowed 1.8×1014 sequences to be introduced over a period of 30 minutes, a 300-fold improvement in library size over capillary electrophoresis based selections (CE-SELEX). Four rounds of selection were performed within four days. Aptamers with low nM dissociation constants for IgE were identified after a single round of μFFE selection.
Introduction
Aptamers are single stranded DNA or RNA molecules that fold into unique structures that promote selective, high-affinity interactions with specific target molecules.1 Aptamers are isolated using an in vitro process commonly referred to as Systematic Evolution of Ligands by EXponential enrichment (SELEX).2, 3 Generally, the SELEX process consists of iterative rounds alternating between affinity enrichment of binding sequences and PCR amplification. Typically, aptamers with low nM to high pM dissociation constants are isolated after 10–15 rounds of selection. Aptamers have been selected for a variety of targets, including cells,4–6 proteins,7, 8 antibiotics,9–11 and small molecules such as amino acids12–14 and biological cofactors.15, 16 Aptamer applications range from purification,17–19 detection,20–23 and quantification22, 24, 25 of targets in basic research to diagnostic agents26, 27 and drug candidates28–32 in clinical applications.
Conventionally, affinity selections in SELEX are performed using nitrocellulose membranes or affinity chromatography, requiring manual manipulation of relatively large sample volumes and providing ample opportunity for non-specific interactions with solid surfaces. Typically 10–15 selection cycles are required, making the SELEX process labour intensive and time consuming. Since the earliest aptamer publications,2, 3 great efforts have been made to develop protocols that minimize solution volumes, are simple and fast, and are compatible with automated processes. Cox et al. were the first to automate the SELEX process by incorporating the separation, PCR and purification apparatus into a robotic pipetting worksurface.33, 34 Twelve cycles of selection were completed within 42 hours, reducing the selection time from several weeks to less than two days.
Recent developments in microfluidics provide further opportunities for enhancing the SELEX process. It is anticipated that a high through-put lab-on-a-chip device that combines high separation efficiency, increased PCR speed, low sample volume, minimal contamination, and automation will be possible.35–38 Towards this goal several microfluidic devices have been reported for isolating aptamers. A microfluidic device patterned with sol-gel droplets on individual microheaters was developed.39 This device can selectively generate aptamers for multiple targets simultaneously within the sol-gel droplets, significantly improving throughput. A continuous-flow magnetic activated chip-based separation (CMACS) device40 and a micromagnetic separation (MMS) chip41 have also been reported. Both devices can precisely control the path of aptamer bound magnetic beads with high separation efficiency. However, there are limitations to each of these strategies. There are concerns regarding protein stability and integrity through the multiple selection cycles in the sol-gel-microheater device; and complicated target immobilization procedures, elongated incubation, and the potential for non-specific interactions with surface structures are of concern in the CMACS and MMS devices.
Capillary electrophoresis based selections (CE-SELEX) address many common issues encountered in traditional selections.42–45 Selections are performed in free solution, reducing the opportunity for non-specific interactions and eliminating complicated immobilization strategies. The high resolving power of CE increases the rate of enrichment allowing high affinity aptamers to be obtained in 2–4 rounds of selection. Unfortunately, CE-SELEX is not without limitations of its own. Only several nL of library can be injected without causing unacceptable loss of resolution.45 This small volume limits the number of sequences that can be assessed and requires very high library concentrations. Fraction collection is also challenging since the abundance of aptamers in early rounds is often below the limit of detection and variability in mobilities requires collection windows to be adjusted on the fly.46
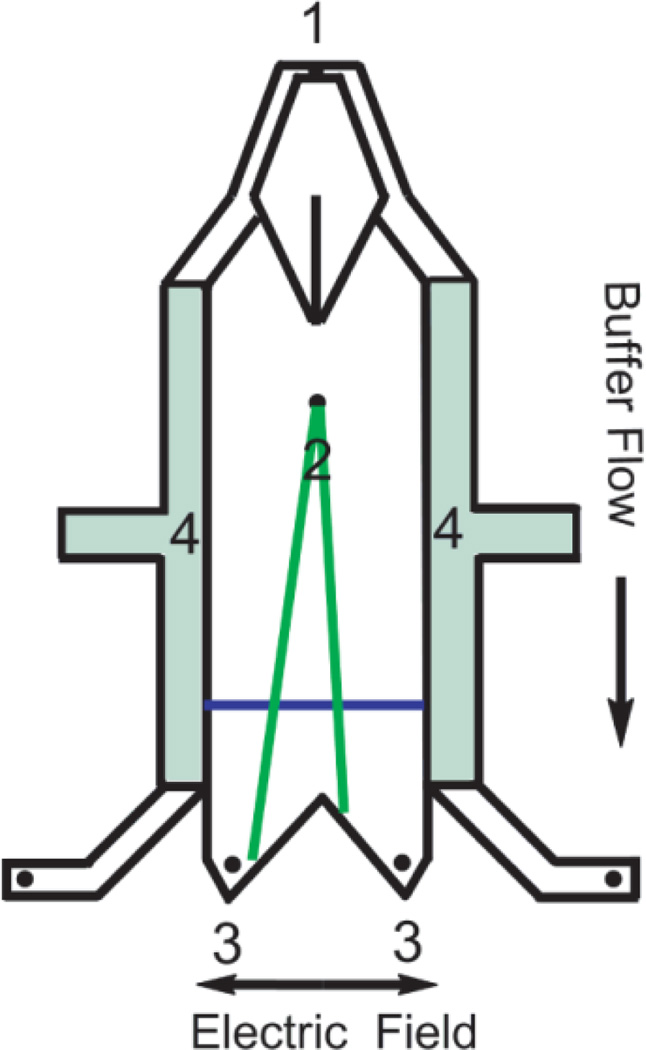
Micro Free Flow Electrophoresis (μFFE) provides a unique solution to the challenges presented by CE-SELEX. Contrary to many separations, μFFE can be used to continuously introduce, separate and collect analytes (see Fig. 1).47–50 Analyte is continuously streamed into a planar separation chamber. An electric field is applied perpendicularly to the pressure driven flow, deflecting analytes laterally according to their mobility. μFFE designs and separation conditions have been optimized allowing long term operation of the device.51, 52 μFFE separation theory and sources of band broadening have been characterized.53 Unique μFFE detection strategies have been demonstrated.54 Most interestingly, a gradient technique has been developed to measure aptamer-target equilibria, demonstrating the feasibility of using μFFE to separate binding sequences from non-binding sequences.55
Figure 1.
Schematic of a μFFE device demonstrating the buffer inlet (1), sample inlet (2), fraction collection outlets (3) and electrode channels (4). The blue line denotes the detection zone where the laser line is expanded across the separation channel.
In the current manuscript we report the first successful application of μFFE in a SELEX selection for DNA aptamers. IgE was chosen as a target since aptamers have previously been selected for this protein using both traditional SELEX56 and CE-SELEX42, 43 allowing a direct comparison with these approaches.
Experimental
Materials and Chemicals
Human myeloma IgE protein was purchased from Athens Research and Technologies (Athens, GA). Nuclease free H2O, forward primer 5’-FAM-AGC AGC ACA GAG GTC AGA TG-3’, reverse primer 5’-Biotin-TTC ACG GTA GCA CGC ATA GG-3’, the initial ssDNA library 5’-FAM-AGC AGC ACA GAG GTC AGA TG(N)40 CCT ATG CGT GCT ACC GTG AA-3’, and the selected aptamers were from Integrated DNA Technologies, Inc. (Coralville, IA). For PCR reactions, dNTPs and the 25 bp DNA ladder were from Invitrogen (Carlsbad, CA); Taq polymerase and ThermoPol buffer were from New England BioLabs (Ipswich, MA); and the gel loading dye was from Promega (Madison, WI). Other chemicals were purchased from Sigma Aldrich (St. Louis, MO) at the highest grade available, except Acetic acid (CH3COOH, 99.7%, Mallinckrodt Baker), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, 99%, Alfa Aesar), MgCl2 (99.8%, Mallinckrodt Baker), NaCl (99.0%, Spectrum), and KH2PO4 (99.9%, J. T. Baker). All buffers were prepared in nuclease free H2O, and filtered through 0.2 µm membrane filters before use.
μFFE Fabrication
μFFE devices were fabricated using previously reported procedures.55 Briefly, three photolithography steps were performed to define the electrode channels, the separation channel, and pattern the electrodes onto 1.1 mm borofloat wafer (Precision Glass & Optics, Santa Ana, CA) according to the design shown in Figure 1. The patterned chip was anodically bonded to a second borofloat wafer that had previously been drilled with inlet and outlet holes. Channel depths were approximately 20 µm in the separation channel and 100 µm in the electrode channels. The sample inlet hole had a diameter of 355 µm.
μFFE Separation
Before separation, the μFFE chip was coated with PEO to suppress the electroosmotic flow.57 Briefly, 1 M HCl was first pumped into the chip at 6 mL/min for 10 min, followed by 0.2 % PEO in 0.1 M HCl at 3 mL/min for 10 min. Finally, the separation buffer (25 mM HEPES, 300 µM Triton X-100, adjusted to pH 7.0 by 1 M NaOH) was pumped at 6 mL/min for 10 min to remove HCl and unbound PEO. Before the selection, the ssDNA library was heated to 72 °C for 5 min and then gradually cooled to room temperature. For the 1st round of selection, 100 µM random sequence ssDNA library was incubated with 10 nM IgE at room temperature for 20 min in the binding buffer (tris(hydroxyamino)methane-glycine-potassium TGK) buffer, composed of 25 mM tris(hydroxyamino)methane, 192 mM glycien, and 5 mM KH2PO4 at pH 8.3). The following IgE concentrations were used in subsequent selection rounds: 10 nM in the 2nd round, 1 nM in the 3rd round, and 100 pM in the 4th round. The mixture (40 µL in total) was then loaded into a 100 µL syringe (Hamilton Company, Reno, NV), and pumped into the μFFE chip (Figure 1, hole (2)) using a syringe pump (PicoPlus, Harvard apparatus, Holliston, MA) at 100 nL/min. The separation buffer was pumped into the device (Figure 1, hole 1) using a second syringe pump (pump 22, Harvard Apparatus) at 1mL/min. An electric field of 150 V/cm was applied across the chip so that the nonbinding ssDNA was separated from aptamer-IgE complexes. A stable current of 0.32 mA was observed over the 30 minute separation time. With EOF suppressed, the free ssDNA and the complex were split into two fractions (Figure 1, holes 3) and collected separately.
Laser Induced Fluorescence (LIF) Detection
The beam from a solid state laser (488 nm, 50 mW, Newport Corp, Irvine, CA) was expanded into a line and focused across the separation channel 1.5 cm downstream from the sample inlet. A microscope objective (3 × zoom) was positioned above the detection zone. Fluorescence images were recorded every second using a Cascade 512B CCD camera (Photometrics, Tucson, AZ) through an AZ100 stereomicroscope (Nikon Corp., Tokyo, Japan). An Endow GFP bandpass emission filter cube (Nikon Corp., Tokyo, Japan) consisting of a dichroic mirror (495 nm cutoff) and two bandpass filters (450–490 nm and 500–550 nm) were used for wavelength selection. Images were processed using MetaVue software (Downington, PA). Linescans were also recorded using by MetaVue, and later analyzed in Cutter 7.0.58
PCR Amplification and Purification
Collected sequences were PCR amplified immediately after each selection round. In the final reaction vials, there were 1 mM each of the four dNTPs, 7.5 mM MgCl2, 500 nM forward and reverse primers, 1 × ThermoPol buffer and 0.05 units/µL Taq polymerase. A negative control with all PCR reagents but no ssDNA was also performed every round to verify the absence of background contamination. 23 cycles of denaturation (94 °C, 30 s), annealing (55 °C, 30 s), and extension (72 °C, 20 s) were performed, with a final extension at 72 °C for 5 min. A 1.5 % agarose gel with ethidium bromide staining was used to confirm the presence of the desired PCR products. Single stranded FAM labeled DNA sequences were obtained using an on column purification followed by the ethanol precipitation.43
Dissociation constant Kd measurements
Affinities of the selected pools and individual aptamer sequences were measured using both affinity capillary electrophoresis (ACE) and fluorescence polarization (FP). Approximately 2.5 nM ssDNA samples were titrated with increasing concentrations of human IgE. A commercial CE system (P/ACE MDQ, Beckman Coulter, Inc., Fullerton, CA) equipped with laser induced fluorescence detection (λex = 488 nm, λem = 520 nm) was used to perform ACE experiments. Samples were injected into a 50 cm fused silica capillary (Polymicro Technologies, Phoenix, AZ) at 1psi for 4 sec. The separations were then performed at 30 kV in TGK buffer. The peak heights of the unbound ssDNA were used to calculate the bound fractions and estimate Kd using the following equation:59
| (2) |
in which fa is the bound fraction, [P]t, [D]t, and c are total IgE concentration, total DNA concentration, and maximum bound fraction, respectively. [P]t - 0.5([D]t + [P]t + Kd - (([D]t + [P]t + Kd)2 - 4[D]t[P]t)0.5) represents the free IgE concentration.
In FP experiments, 15 µL of the same samples used in ACE were loaded into a corning 3540 microplate (Corning Incorporated, Corning, NY) and experiments were performed on a Synergy™ 2 Microplate Reader (BioTek Instruments, Inc., Winooski, VT). Parallel and perpendicular intensities (λex = 485 ± 20 nm, λem = 528 ± 20 nm) were measured, and polarization values were calculated using Gen 5™ software (BioTek Instruments, Inc., Winooski, VT). The bound fraction was determined according to:
| (3) |
in which P, Po and Pm are the measured polarizations of a sample, free DNA, and the complex, respectively. Meanwhile, the overall fluorescence intensity was monitored and corrections to the bound fractions were made according to the intensity change as previously reported.60
DNA cloning and sequencing
A TOPO TA Cloning® Kit for Sequencing (Invitrogen, Carlsbad, CA) was used to perform cloning reactions and grow colonies. Briefly, 1 µL of the selected ssDNA from pool 1 through pool 4 was added to 99 µL PCR master mixes and amplified separately, as described above, except using unlabeled forward and reverse primers. 4 µL of the 100 µL fresh PCR products were incubated with TOPO® vector at room temperature for 5 min to allow the cloning reactions to take place. The inserted vectors were then chemically transformed into One Shot® TOP10 competent E.coli cells. The mixtures were incubated in a 37 °C shaking incubator for 30 min and then spread onto selective plates containing 50 µg/mL kanamycin. For each pool, two agar plates were used to grow colonies, which produced 300 to 400 colonies. Individual colonies were then randomly picked and cultivated in a 96 well plate with liquid LB containing 50 µg/mL kanamycin at 37 °C for another day. Cell pellets were sent to the Biomedical Genomics Center at the University of Minnesota for Sanger sequencing. Since some wells of the plate were used as controls, and some pellets were not sufficient to obtain confident sequencing information, 23, 28, 19, and 6 sequences were obtained from pools 1, 2, 3, and 4, respectively (See Table S1 for aptamer sequences).
Results and discussion
μFFE selection
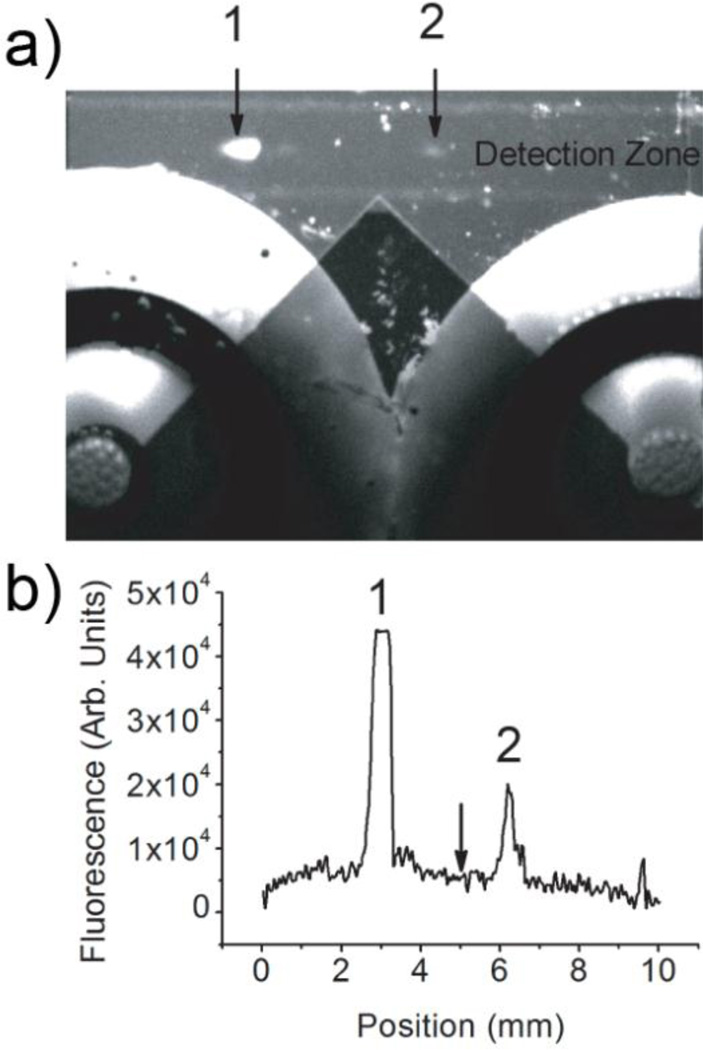
Figure 2 demonstrates the μFFE separation observed during the first round of selection. An electric field of 150 V/cm was applied across the flow channel. Unbound ssDNA sequences were deflected toward the anode due to the supressed electroosmotic flow (EOF). The aptamer–IgE complexes, which were only deflected minimally, were well separated from the unbound sequences. It should be noted that μFFE facilitates collection of analytes with low mobilities. Pressure rinses were necessary to observe24 or collect42, 43 aptamer-IgE complexes in previously reported CE separations. The flow was split into two streams at the exit of the μFFE separation chamber. The IgE-DNA complexes were collected into a centrifuge tube through the tubing connected to the right nanoport. This continuous collection strategy was much simpler than that of CE-SELEX, eliminating the complicated timing associated with collecting complexes as they migrated off the end of a capillary. A linescan (Figure 2b) recorded across the detection zone reinforces how well resolved the binding sequences are from the non-binding sequences. The high concentration of library (100 µM) used in the first round of selection caused the signal for the unbound sequences to go off scale but this was not a concern since the primary goal was to identify the position of the peaks for accurate fraction collection, not quantitation.
Figure 2.
(a) An image of a μFFE separation of free (1) and bound ssDNA (2) recorded during selection cycle #1. The fraction collection channels and outlet ports are clearly visible in this image. (b) A linescan across the detection zone imaged in (a). The arrow indicates the fraction cutoff point at the exit of the μFFE channel demonstrating clear separation of free (1) from bound (2) ssDNA. The anode is on the left in all images.
In the first round of selection, 100 µM of fluorescently tagged ssDNA library was incubated with 10 nM IgE in TGK binding buffer for 20 min. After incubation, 3 µL of the library-IgE mixture was introduced into the μFFE device over a period of 30 minutes, which corresponded to 1.8×1014 sequences. A typical CE separation only allows a discrete injection of approximately 8 nL.45 Over 300-fold more volume, and therefore sequences, could be introduced into the μFFE device. The number of sequences could easily be increased further by increasing the analyte flow rate, library concentration or collection time.
The multiple-depth μFFE design51 generated high flow over the electrodes to dissipate electrolysis bubbles while minimizing flow, and therefore dilution, in the separation chamber. The dilution factor is defined as the ratio of the collected sample volume to the introduced sample volume. In a μFFE device, the planar flow profile can be described using lubrication theory according to the following equation:61
| (1) |
in which q is the volumetric flow rate, ΔP is the pressure difference, H is the channel depth, w is the channel width, η is the buffer viscosity, and L is the channel length. Comparing the electrode and separation channels: the ratio of channel depths is 20 µm /100 µm = 0.2 (separation channel/electrode channel), the ratio of channel widths is 10 mm / (2 × 2 mm) = 2.5 (i.e. two electrode channels), and the ratio of channel lengths is approximately 1. The ratio of flow volume through the channels is 0.23 × 2.5 / 1 = 1: 50 (separation channel/electrode channel). Solution is only collected from half of the separation channel. As result, approximately 1% of the total volume through the μFFE device is collected. Over a period of 30 min, 30 mL separation buffer and 3 µL sample are introduced into the chip, resulting in a collected volume of 300 µL. This calculation agrees very well with experimental results. Comparing with the 3 µL of library injected yields a modest 100-fold dilution of the binding sequences during the μFFE separation. This compares favourably with dilution factors of 6,000 typically encountered in CE-SELEX.42–45 The higher DNA concentration in the collected fraction and increased volume collected facilitates PCR and decreases the potential for contamination or non-specific amplification. Furthermore, the transit time through the μFFE flow chamber is only 10–20 seconds, greatly decreasing the potential for dissociation during the separation when compared to CE, in which separations typically take 5–15 minutes. The electric field applied in μFFE (150 V/cm) is also lower than that typically used in CE-SELEX (~500 V/cm), further decreasing the potential for dissociation.
Aptamer Characterization
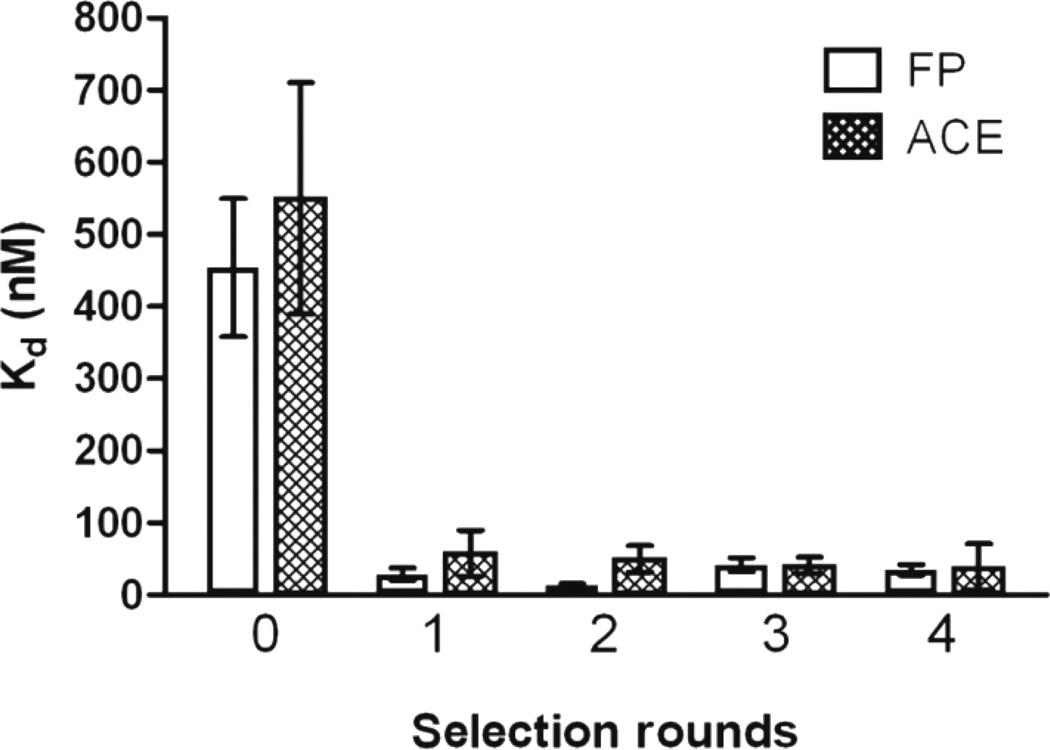
Four rounds of μFFE selection were performed. The affinity of the selected pools for IgE was monitored using two orthogonal methods62: fluorescence polarization (FP)60 and affinity capillary electrophoresis (ACE)42 (see Experimental, Dissociation constant Kd measurements). As shown in Figure 3, the initial library had a low affinity for IgE in TGK buffer, (Kd = 460 ± 160 nM and 550 ± 280 nM as measured by FP and ACE, respectively). A significant improvement in affinity was observed after a single round of μFFE selection, with Kd values of 29 ± 15 nM and 58 ± 55 measured by FP and ACE, respectively. No further improvement in affinity was observed after the second round of selection. IgE concentration was decreased by a factor of 10 (1 nM) in the third round of selection and by a factor of 100 (100 pM) in the fourth round of selection in an effort to improve the stringency of the selection. However, the enhanced stringency did not result in improved affinity for IgE, suggesting that the selection had converged after a single round of μFFE selection.
Figure 3.
Kd of the starting library and the selected pools measured by fluorescence polarization (FP) and affinity capillary electrophoresis (ACE). Error bars represent 95% confidence interval.
Aptamers were randomly cloned and sequenced from the DNA pools after each round of selection. 23 sequences were identified from round one, 28 sequences from round two, 19 sequences from round three, and 6 sequences from round four (see supplementary information for aptamer sequences). These sequences were analyzed to identify homologous sequences or motifs. As shown in Table 1, no identical or similar sequences were found in round 1; two identical sequences were found in rounds 2 and 3; three identical sequences were found in round 4 with a fourth sequence demonstrating 89% similarity as revealed by software ClustalW2®. It should be noted that sequences that appeared multiple times in one round did not share any conserved region or motifs when compared with sequences isolated from other rounds. Although the number of the clones was limited, identification of identical sequences in such a small sample size suggests a decrease in diversity as the pool converged. This is particularly evident in round four, where 4 out of 6 sequences isolated were very similar. The affinities of all sequences identified multiple times were measured by FP and ACE (see Table 1). The measured dissociation constants were remarkably similar and agreed well with the affinities measured for the bulk pools.
Table 1.
Kd of selected aptamers
| Aptamera) | FP Kdb) | ACE Kdb) |
|---|---|---|
| Multiple copy sequences identified | ||
| Clone 2.13 and 2.24 | 20 ± 4 nM | 136 ± 58 nM |
| Clone 3.5 and 3.14 | 29 ± 7 nM | 33 ± 21 nM |
| Clone 4.4, 4.5, and 4.6 | 39 ± 20 nM | 58 ± 17 nM |
| Clone 4.2 | 44 ± 15 nM | 62 ± 29 nM |
| Randomly chosen sequences from round 1 and round 3 | ||
| Clone 1.4 | 23 ± 6 nM | 58 ± 33 nM |
| Clone 1.13 | 22 ± 6 nM | 63 ± 17 nM |
| Clone 1.18 | 32 ± 11 nM | 66 ± 35 nM |
| Clone 3.2 | 20 ± 7 nM | 50 ± 27 nM |
| Clone 3.9 | 29 ± 11 nM | 62 ± 16 nM |
| Clone 3.11 | 28 ± 13 nM | 47 ± 17 nM |
Aptamers were given unique identifiers where the first number gives the selection round and the second number identifies a particular sequence cloned in that round.
Errors represent 95% confidence interval.
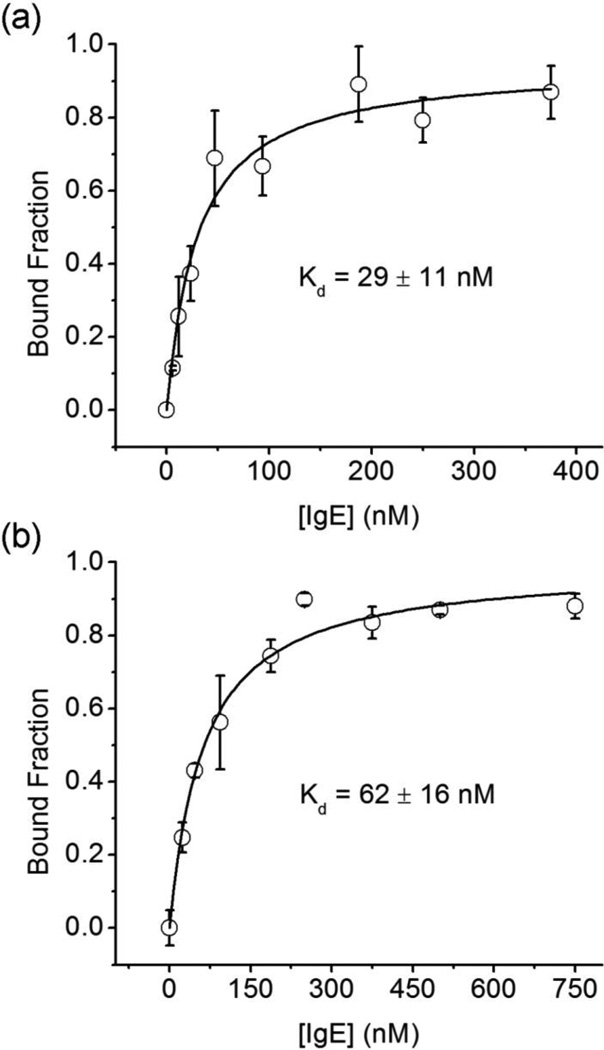
Finally, three aptamers were chosen from the cloned sequences obtained from rounds one and three using a random number generator. Dissociation constants for each of these sequences are shown in Table 1 (see Figure 4 for representative binding curves).
Figure 4.
Binding curves for sequence 3.9, obtained using (a) FP and (b) ACE. Three measurements were taken at every IgE concentration. Error bars represent standard deviation. Errors of the Kd values represent 95% confidence interval.
No statistical difference was observed between sequences obtained after one or three rounds suggesting that the selection had converged after a single round of μFFE selection. Kd values for sequences chosen randomly were statistically indistinguishable from those that appeared multiple times in the cloning results. This result is similar to previous CE-SELEX selections that yielded seemingly diverse pools of aptamers with similar affinities.42, 43 It should be noted that every sequence assessed demonstrated low nM affinity for IgE, even in the absence of negative selections, suggesting that similar to CE-SELEX, performing selections in free solution diminishes the opportunity for non-specific interactions. Dissociation constants measured using ACE were consistently higher than those measured using FP. Yang et al have demonstrated that the high electric field of CE can affect observed half-lives and binding constants of protein complexes.63 This raises a concern regarding whether the high field typically used in CE-SELEX and whether this field modifies the selection environment, affecting the applicability of the selected aptamers. Buchanan et al64 have demonstrated that a combination of low electric field and shorter time in the field minimize dissociation of aptamer-target complexes during a CE separation. In practice this can be difficult to balance in CE since separation time is inversely proportional to electric field. μFFE does not share this difficulty since electric field and separation time can be optimized independently. This unique property of μFFE is advantageous allowing more control over separation conditions when compared to CE.
Aptamers for IgE obtained using μFFE demonstrate similar dissociation constants to those obtained previously using CE-SELEX (~20 nM)42,43 and conventional SELEX (~10 nM)56. It should be noted that these aptamers were obtained after a single round of μFFE selection, which compared favorably with the 2–4 rounds required by CE-SELEX42, 43 or the 15 rounds required by conventional SELEX.56
Conclusions
In conclusion, we demonstrated the advantages of isolating aptamers using μFFE selections. Within 30 min of continuous μFFE separation and collection, 1.8×1014 sequences were assessed, a 300-fold improvement over CE-SELEX. μFFE also eliminated the complicated timing associated with fraction collection in CE-SELEX. Four selection cycles were completed within four days. Low nanomolar affinity sequences were identified after a single round of μFFE selection, suggesting that aptamers could be obtained even faster. Although the device was expected to have lower separation efficiency than CE, obtaining high affinity aptamers after a single selection round suggests a high rate of enrichment was achieved. The free solution μFFE separation simplifies the protocol, eliminating the need for target immobilization, elongated incubation, or negative selections. With these advantages, we believe that μFFE device can be readily adopted to select aptamers for a wide range of targets.
Supplementary Material
Acknowledgements
Meng Jing acknowledges Ryan T. Turgeon and Nicholas W. Frost for help during μFFE device fabrication. The authors acknowledge the National Institutes of Health (R01GM063533) for financially supporting this work. μFFE devices were fabricated using facilities at the University of Minnesota Nanofabrication Center, a NNIN funded site.
Footnotes
† Electronic Supplementary Information (ESI) available: aptamer sequences. See DOI: 10.1039/b000000x/
Notes and references
- 1.Syed MA, Pervaiz S. Oligonucleotides. 2010;20:215–224. doi: 10.1089/oli.2010.0234. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 4.Shangguan D, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, Sefah K, Yang CYJ, Tan WH. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang ZW, Shangguan D, Wang KM, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan WH. Analytical Chemistry. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 6.Shamah SM, Healy JM, Cload ST. Accounts of Chemical Research. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 7.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 8.Tuerk C, Macdougal S, Gold L. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallis MG, Streicher B, Wank H, vonAhsen U, Clodi E, Wallace ST, Famulok M, Schroeder R. Chemistry & Biology. 1997;4:357–366. doi: 10.1016/s1074-5521(97)90126-5. [DOI] [PubMed] [Google Scholar]
- 10.Patel DJ, Suri AK, Jiang F, Jiang LC, Fan P, Kumar RA, Nonin S. Journal of Molecular Biology. 1997;272:645–664. doi: 10.1006/jmbi.1997.1281. [DOI] [PubMed] [Google Scholar]
- 11.Berens C, Thain A, Schroeder R. Bioorganic & Medicinal Chemistry. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 12.Famulok M. Journal of the American Chemical Society. 1994;116:1698–1706. [Google Scholar]
- 13.Geiger A, Burgstaller P, vonderEltz H, Roeder A, Famulok M. Nucleic Acids Research. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarus M. Journal of Molecular Evolution. 1998;47:109–117. doi: 10.1007/pl00006357. [DOI] [PubMed] [Google Scholar]
- 15.Sassanfar M, Szostak JW. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 16.Burgstaller P, Famulok M. Angewandte Chemie-International Edition in English. 1994;33:1084–1087. [Google Scholar]
- 17.Romig TS, Bell C, Drolet DW. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 1999;731:275–284. [PubMed] [Google Scholar]
- 18.Deng Q, German I, Buchanan D, Kennedy RT. Analytical Chemistry. 2001;73:5415–5421. doi: 10.1021/ac0105437. [DOI] [PubMed] [Google Scholar]
- 19.Hutanu D, Remcho VT. Advances in Chromatography, Vol 45. 2007;45:173–196. doi: 10.1201/9781420018066.ch4. [DOI] [PubMed] [Google Scholar]
- 20.Hamaguchi N, Ellington A, Stanton M. Analytical Biochemistry. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 21.McCauley TG, Hamaguchi N, Stanton M. Analytical Biochemistry. 2003;319:244–250. doi: 10.1016/s0003-2697(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 22.Huang CC, Cao ZH, Chang HT, Tan WH. Analytical Chemistry. 2004;76:6973–6981. doi: 10.1021/ac049158i. [DOI] [PubMed] [Google Scholar]
- 23.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. Journal of the American Chemical Society. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 24.German I, Buchanan DD, Kennedy RT. Analytical Chemistry. 1998;70:4540–4545. doi: 10.1021/ac980638h. [DOI] [PubMed] [Google Scholar]
- 25.Mairal T, Ozalp VC, Sanchez PL, Mir M, Katakis I, O'Sullivan CK. Analytical and Bioanalytical Chemistry. 2008;390:989–1007. doi: 10.1007/s00216-007-1346-4. [DOI] [PubMed] [Google Scholar]
- 26.Proske D, Blank M, Buhmann R, Resch A. Applied Microbiology and Biotechnology. 2005;69:367–374. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- 27.Tombelli S, Minunni M, Mascini M. Biomolecular Engineering. 2007;24:191–200. doi: 10.1016/j.bioeng.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Fine SL, Martin DF, Kirkpatrick P. Nature Reviews Drug Discovery. 2005;4:187–188. doi: 10.1038/nrd1677. [DOI] [PubMed] [Google Scholar]
- 29.Nimjee SM, Rusconi CP, Sullenger BA. Annual Review of Medicine. 2005;56:555. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 30.Fichou Y, Ferec C. Trends in Biotechnology. 2006;24:563–570. doi: 10.1016/j.tibtech.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Lee JF, Stovall GM, Ellington AD. Current Opinion in Chemical Biology. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Kaur G, Roy I. Expert Opinion on Investigational Drugs. 2008;17:43–60. doi: 10.1517/13543784.17.1.43. [DOI] [PubMed] [Google Scholar]
- 33.Cox JC, Rudolph P, Ellington AD. Biotechnology Progress. 1998;14:845–850. doi: 10.1021/bp980097h. [DOI] [PubMed] [Google Scholar]
- 34.Cox JC, Ellington AD. Bioorganic & Medicinal Chemistry. 2001;9:2525–2531. doi: 10.1016/s0968-0896(01)00028-1. [DOI] [PubMed] [Google Scholar]
- 35.Hybarger G, Bynum J, Williams RF, Valdes JJ, Chambers JP. Analytical and Bioanalytical Chemistry. 2006;384:191–198. doi: 10.1007/s00216-005-0089-3. [DOI] [PubMed] [Google Scholar]
- 36.Gopinath SCB. Analytical and Bioanalytical Chemistry. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- 37.Xu YH, Yang XR, Wang EK. Analytica Chimica Acta. 2010;683:12–20. doi: 10.1016/j.aca.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Huang CJ, Lin HI, Shiesh SC, Lee GB. Biosensors & Bioelectronics. 2010;25:1761–1766. doi: 10.1016/j.bios.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 39.Park SM, Ahn JY, Jo M, Lee DK, Lis JT, Craighead HG, Kim S. Lab on a Chip. 2009;9:1206–1212. doi: 10.1039/b814993c. [DOI] [PubMed] [Google Scholar]
- 40.Lou XH, Qian JR, Xiao Y, Viel L, Gerdon AE, Lagally ET, Atzberger P, Tarasow TM, Heeger AJ, Soh HT. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian JR, Lou XH, Zhang YT, Xiao Y, Soh HT. Analytical Chemistry. 2009;81:5490–5495. doi: 10.1021/ac900759k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendonsa SD, Bowser MT. Journal of the American Chemical Society. 2004;126:20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 43.Mendonsa SD, Bowser MT. Analytical Chemistry. 2004;76:5387–5392. doi: 10.1021/ac049857v. [DOI] [PubMed] [Google Scholar]
- 44.Mendonsa SD, Bowser MT. Journal of the American Chemical Society. 2005;127:9382–9383. doi: 10.1021/ja052406n. [DOI] [PubMed] [Google Scholar]
- 45.Mosing RK, Mendonsa SD, Bowser MT. Analytical Chemistry. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 46.Mosing RK, Bowser MT. In: Nucleic Acid and Peptide Aptamers. Mayer G, editor. Humana Press; 2009. pp. 33–44. [Google Scholar]
- 47.Fonslow BR, Bowser MT. Analytical Chemistry. 2005;77:5706–5710. doi: 10.1021/ac050766n. [DOI] [PubMed] [Google Scholar]
- 48.Raymond DE, Manz A, Widmer HM. Analytical Chemistry. 1994;66:2858–2865. [Google Scholar]
- 49.Turgeon RT, Bowser MT. Analytical and Bioanalytical Chemistry. 2009;394:187–198. doi: 10.1007/s00216-009-2656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, Zhang CX, Janasek D, Manz A. Lab on a Chip. 2003;3:224–227. doi: 10.1039/b308476k. [DOI] [PubMed] [Google Scholar]
- 51.Fonslow BR, Barocas VH, Bowser MT. Analytical Chemistry. 2006;78:5369–5374. doi: 10.1021/ac060290n. [DOI] [PubMed] [Google Scholar]
- 52.Frost NW, Bowser MT. Lab on a Chip. 2010;10:1231–1236. doi: 10.1039/b922325h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fonslow BR, Bowser MT. Analytical Chemistry. 2006;78:8236–8244. doi: 10.1021/ac0609778. [DOI] [PubMed] [Google Scholar]
- 54.Turgeon RT, Bowser MT. Electrophoresis. 2009;30:1342–1348. doi: 10.1002/elps.200800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turgeon RT, Fonslow BR, Jing M, Bowser MT. Analytical Chemistry. 2010;82:3636–3641. doi: 10.1021/ac902877v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiegand TW, Williams PB, Dreskin SC, Jouvin MH, Kinet JP, Tasset D. Journal of Immunology. 1996;157:221–230. [PubMed] [Google Scholar]
- 57.Iki N, Yeung ES. Journal of Chromatography A. 1996;731:273–282. [Google Scholar]
- 58.Shackman JG, Watson CJ, Kennedy RT. Journal of Chromatography A. 2004;1040:273–282. doi: 10.1016/j.chroma.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 59.Hall KB, Kranz JK. Totowa, New Jersy: Humana Press; 1999. pp. 109–110. [Google Scholar]
- 60.Wei AP, Herron JN. Analytical Chemistry. 1993;65:3372–3377. doi: 10.1021/ac00071a007. [DOI] [PubMed] [Google Scholar]
- 61.Bird RB, Stewart WE, Lightfoot EN, editors. Transport Phenomena. New York: John Wiley and Sons; 2002. [Google Scholar]
- 62.Jing M, Bowser MT. Analytica Chimica Acta. 2011;686:9–18. doi: 10.1016/j.aca.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang P, Ma Y, Lee AWM, Kennedy RT. Electrophoresis. 2009;30:457–464. doi: 10.1002/elps.200800397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buchanan DD, Jameson EE, Perlette J, Malik A, Kennedy RT. Electrophoresis. 2003;24:1375–1382. doi: 10.1002/elps.200390176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.