Abstract
We have previously reported that the BRAFV600E signaling causes genome-wide aberrations in gene methylation in melanoma cells. To explore the potential molecular mechanisms for this epigenetic effect of BRAFV600E, in this in silico study we analyzed 11 microarray datasets retrieved from NCBI GEO database and examined the relationship of the expression of the epigenetic genes (genes involved in epigenetic regulation) with BRAFV600E signaling, methylation and expression of tumor-suppressor genes (TSGs) in melanoma, and patient survival with this cancer. Among 273 epigenetic genes examined, 12 genes were down-regulated (named DD genes) and 16 were up-regulated (UU genes) by suppression of the BRAFV600E signaling using inhibitors. While the expression of 245 non-DD/UU genes overall had no correlation with the expression and methylation of a set of potential TSGs, the expression of DD genes was significantly correlated negatively with the TSG expression and positively with TSG methylation. Expression of UU genes was positively, albeit weakly, associated with the TSG expression. Overall, no correlation was found between UU gene expression and TSG methylation. Importantly, the expression of DD genes, but not UU genes, was significantly associated with decreased survival of patients with melanoma. Interestingly, the promoters of DD genes contain more binding motifs of c-fos and myc, two BRAFV600E signaling-related transcription factors, than those of UU and non-DD/UU genes. Thus, these results link epigenetic genes to methylation and suppression of tumor suppressor genes as a mechanism involved in BRAFV600E-promoted melanoma tumorigenesis and uncover a novel molecular signature that predicts a poor prognosis of melanoma.
Keywords: BRAFV600E, Tumor-suppressor genes, Melanoma, Epigenetic regulation, Gene methylation
1. Introduction
Melanoma is a highly aggressive skin tumor that originates from melanocytes and its incidence has been increasing over the past decades worldwide [11,13]. Metastatic capacity of melanoma is extremely high, and most patients with distant metastases from melanoma die within 5 years [11,13]. A prominent oncogenic genetic event in melanoma is the BRAF T1799A mutation, which is detected in 50–70% of melanomas and results in the constitutively activated BRAFV600E kinase [4,22]. Through driving the Ras/Raf/MEK/ERK signaling pathway (MAPK pathway), BRAFV600E signaling plays a fundamental role in the tumorigenesis of melanoma [4,22].
Epigenetic modifications, which include DNA methylation, histone modifications, and chromatin remodeling, have been recognized as important factors that regulate gene expression [15]. While epigenetic mechanisms play a major role in normal physiological events, such as embryogenesis and gene imprinting, aberrant epigenetic events are involved in many critical pathways and steps in human tumorigenesis, including that of melanoma [15]. To date, more than 70 genes have been identified that are hyper- methylated at their promoter regions and many genes that are involved in histone modification and chromatin remodeling have been demonstrated to be aberrantly regulated in melanoma [20]. These epigenetic alterations lead to the aberrant regulation of many fundamental pathways that regulate cell cycle, apoptosis, invasion, and growth of cells [20].
Through a genome-wide methylation microarray analysis, we recently showed that a wide range of genes were linked to BRAFV600E signaling through their hyper- or hypo-methylation [7]. In the present in silico study, we further explored the potential molecular mechanisms for the epigenetic effects of BRAFV600E. We found the epigenetic genes down-regulated by BRAFV600E signaling inhibitors were closely associated with the methylation and silencing of tumor-suppressor genes (TSGs) in melanoma and patient survival.
2. Materials and methods
2.1. Definition of epigenetic genes
We defined epigenetic genes as those being involved in DNA methylation, histone modification or chromosome remodeling (Supp. Table 1). The Gene Ontology (GO) term and GO ID that we used to search for human epigenetic genes in GO (www.geneontology.org) were listed in Supp. Table 2. Additional epigenetic genes were added from previous literature when the GO annotation was missing (see references in Supp. Table 1). Genes that were down-regulated and up-regulated by suppression of the BRAFV00E signaling were named DD genes and UU genes, respectively.
2.2. Microarray dataset and data analysis
All the datasets were downloaded from NCBI GEO database (www.ncbi.nlm.nih.gov/geo). For Affymetrix array-based datasets, the signal values from the raw datasets were background corrected and normalized with MAS5.0. For gene expression datasets from other platform-based arrays, the processed data was downloaded and the signal values were normalized by using the median array as reference array. In all datasets, the expression values of all probes for a given gene were reduced to a single value by taking the maximum expression value for each sample [18].
To identify epigenetic genes that are regulated by BRAFV600 signaling pathway, we analyzed two microarray datasets GSE10086 and GSE20051, in which melanoma cells were treated with two BRAFV600 signaling inhibitors. Significance analysis of microarrays (SAM) (http://www-stat.stanford.edu/~tibs/SAM/) was used to extract differently expressed genes with a false discovery rate (FDR) of 0.08. Only genes that were differently expressed at the same direction in both datasets were regarded as genes regulated by BRAFV600 signaling pathway.
To examine the correlation between the expression of epigenetic genes and TSGs in clinical melanoma samples, we use the 11 frequently hyper-methylated genes in melanoma cells [3] as representatives of TSGs, and calculated the Pearson correlation coefficient (Pearson’s r) of the expression of each of the 273 epigenetic gene to the expression of each of the 11 hyper-methylated genes. These 11 genes, most of which were previously demonstrated to be potential TSGs [1,3,10,19], have a methylation level ≥60% that is correlated with a 4-fold decrease in mRNA levels and an average post-demethylation re-expression fold-change of >4 in at least 2 of 11 melanoma lines [3]. The melanoma microarray datasets that satisfy the following two criterions were included for this analysis: (1) having a melanoma sample size ≥30; (2) the average of Pearson’s r between the expression of 11 potential TSGs themselves >0.2 (low or negative correlation between the expression of 11 genes will cover up the real correlation of epigenetic genes to the group of the 11 genes). Seven melanoma datasets from the GEO database, including GSE3189, GSE4841, GSE4843, GSE7553, GSE10916, GSE19234 and GSE22153, fulfill the two requirements.
Dataset GSE28356, in which gene methylation from 11 melanoma cell lines were analyzed, has its matched gene expression dataset GSE7127 in GEO database. The gene expression and methylation data from these two matched datasets was used to analyze the correlation between the expression of the epigenetic genes and methylation of TSGs. The calculation of gene methylation level was described previously [3].
2.3. Kaplan–Meier analysis
The patient’s survival information was extracted from GEO database as sample descriptions of the corresponding datasets. Survival probabilities were calculated with the Kaplan–Meier method and differences between survival probabilities were analyzed with the log-rank test. All survival analyses were done using the entire follow-up time. Based on the method described previously [8], we categorized tumors into two groups characterized by high (sum > 0) or low (sum < 0) expression of DD genes according to the sum of relative expression levels of the 12 DD genes in each tumor. Similarly, two groups of tumors were also categorized according to the sum of expression values of the 16 UU genes. The expression values of each DD and UU genes were median-centered across all samples before the sum was calculated.
3. Results
3.1. Identification of epigenetic genes regulated by the BRAFV600 signaling pathway
By searching Gene Oncology and literature, we obtained a total of 273 epigenetic genes, which are presented in Supplemental Table 1. The 273 epigenetic genes are distributed in all of 23 chromosomes. Positional gene enrichment analysis of the epigenetic genes showed that some of the genes are enriched in several regions of chromosomes, such as 3q26 and 12q12 (Supp. Fig. 1).
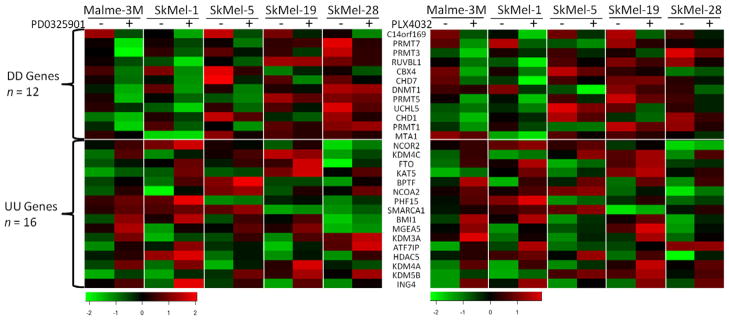
To identify which epigenetic genes are regulated by BRAFV600 signaling pathway, we analyzed two microarray datasets, GSE10086 and GSE20051, in which five melanoma cell lines harboring BRAFV600E were treated with the MEK inhibitor PD0325901 or the BRAFV600E-specific inhibitor PLX4032 [9,16]. Forty and 58 epigenetic genes were differently expressed after treatment with PD0325901 or PLX4032 respectively. Among them, 12 (DD genes) were down-regulated by both treatments and 16 (UU genes) genes were up-regulated by both treatments (Fig. 1). The DD and UU genes in the two datasets showed similar expression patterns across all the melanoma cell lines (Pearson’s r = 0.88), indicating that these results on the expression of DD and UU genes are highly consistent and reproducible.
Fig. 1.
Differentially expressed epigenetic genes in melanoma cells after inhibition of BRAFV600 signaling. The differently expressed genes were identified by SAM procedure, and the heatmaps was generated by R program.
3.2. The promoter regions of DD genes are enriched with the binding motifs of c-fos and myc
To provide further evidence that the expression of DD and UU genes were regulated by BRAFV600 signaling, the 273 epigenetic genes were analyzed for enrichment of consensus binding motifs of well-characterized BRAFV600 signaling transcription factors, including CREB1, Elk-1, c-Fos, myc, STAT1, STAT2 and PPARγ [17], within their promoter regions using the RSA tools (legend to Supplemental Fig. 2). No difference was observed for the number of binding motifs of the seven transcription factors between UU and non-DD/UU genes, suggesting that MAPK pathway might regulate the expression of UU genes at the post-transcription level or through other transcription factors. On the other hand, DD genes harbor more binding motifs of c-Fos and myc, two well-known proto-oncogenes, than non-DD/UU genes (p < 0.02 for c-Fos and p < 0.002 for myc, permutation test) (Supplemental Fig. 2). The percentages of genes that harbor the c-Fos or myc binding motifs in promoter region are also higher for DD genes than for non-DD/UU genes (p < 0.05, Fisher exact test) (Supp. Fig. 2).
3.3. High expression of DD genes is associated with the silence of 11 potential TSGs in melanomas
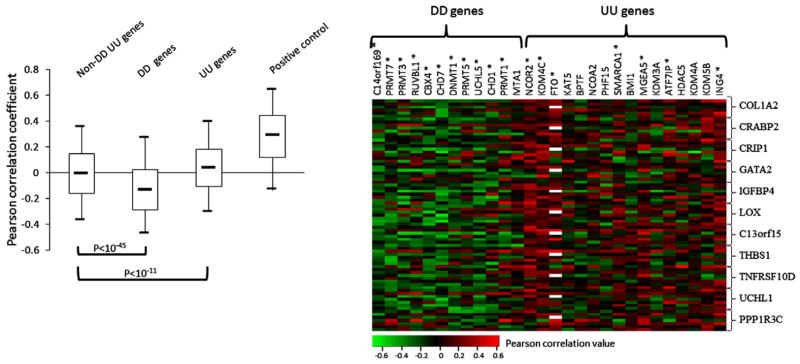
We previously found that BRAFV600E caused hyper- or hypo-methylation of a wide range of genes in melanoma cells [7]. In the present study we asked whether the DD or UU genes may be related to such epigenetic impact of BRAFV600E signaling. We examined the correlation of the expression of the 273 epigenetic genes and 11 potential TSGs that are frequently methylated in melanomas [3] in the 7 microarray datasets that all contain gene expression data from ≥30 clinical melanoma samples. The Pearson’s r between the expression of each epigenetic gene and each TSG were calculated and their distribution is shown in Fig. 2A. The median value of the Pearson’s r in non-DD/UU gene group is close to 0, indicating that, overall, the expression of these genes has no correlation with the 11 potential TSGs (Fig. 2A). In contrast, the median value of the Pearson’s r in UU and DD gene group is 0.041 and −0.133, indicating positive and negative correlation, respectively (p < 10−11, Welch t-test, compared with the non-DD/UU genes) (Fig. 2A). Fig. 2B shows the individual Pearson’s r of the expression of each DD and UU genes to the 11 methylated genes. The vast majority of data points are green (representing negative correlation) for DD genes except MTA1. In contrast, the data points from only half of UU genes are mostly red (positive correlation). Randomization test showed that 11 of 12 of the DD genes were significantly correlated with the expression of 11 potential TSGs negatively, while only 7 of 16 UU genes were positively correlated with the 11 genes significantly (Supp. Fig. 3A).
Fig. 2.
Pearson correlation analysis of the expression of epigenetic genes to the expression of the 11 potential TSGs in melanoma. (A) Box-Whisker plots of Pearson’s r. The Pearson’s r between the expression of 11 potential TSGs themselves were used as control of positive correlation. The box-plot shows the five statistics (lower whisker is 5% minimum, lower box part is the 25th percentile, solid line in box presents the median, upper box part is 75th percentile, and upper whisker is 95% maximum). Welch t-test was used to analyze the difference of Pearson’s r between the three groups of epigenetic genes. (B) Individual Pearson’s r of the expression of each DD and UU gene to the expression of 11 potential TSGs. Each row represents Pearson’s r of the DD and UU genes to a TSG from one microarray dataset. The DD or UU genes whose Pearson’s r values are statistically significant from the whole epigenetic genes were indicated with symbol *(Randomization test, Supp. Fig. 3A). The missing values were shown with white color.
3.4. High expression of DD genes is associated with hyper-methylation of 31 potential TSGs in melanomas
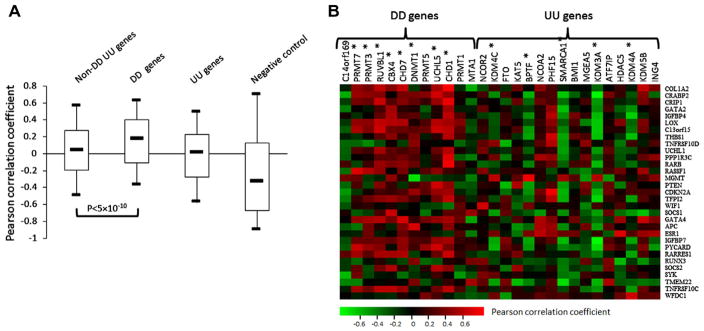
We further examined the correlation of the epigenetic genes to the methylation level of TSGs in melanoma cells by analyzing a gene expression microarray dataset and its matched gene methylation dataset as described in Methods. To avoid potential false positive results for the analysis of this small sample data, we included another 20 TSGs that were frequently methylated in melanomas [20] (totally 31 genes when added to the previous 11 hyper-methylated genes) for correlation analysis. The Pearson’s r values were calculated between the expression and methylation level of each TSG, and its median value is −0.334 (Fig. 3A), indicating negative correlation between the expression and methylation of the 31 TSGs. The median value of Pearson’s r between the expression of the 12 DD genes and the methylation level of the 31 TSGs is 0.173, indicating a positive correlation between them (Fig. 3A). In contrast, the median values of the Pearson’s r in both UU and non-DD/UU groups are close to 0, suggesting that, overall, there is no correlation between the expression of these genes with the methylation of the 31 TSGs (Fig. 3A). The individual Pearson’s r of the expression of each DD and UU genes to the 11 methylated genes were shown in Fig. 3B. Supplemental Fig. 3B shows that 8 of 12 DD genes were positively while 5 of 16 UU genes were negatively correlated with the methylation of TSGs significantly. These results further indicated that the DD genes are better correlated with silence of TSGs in melanoma cells than the UU genes.
Fig. 3.
Pearson correlation analysis of the expression of epigenetic genes to the methylation level of the 31 potential TSGs. (A) Box-Whisker plots of Pearson’s r. The Pearson’s r of the expression of the 31 methylated genes to their own methylation level were used as control of negative correlation. The five statistics of the box-plot and statistic method were described in legend. (B) Individual Pearson’s r of the expression of each DD and UU gene to the methylation of the 31 methylated genes. The DD or UU genes whose Pearson’s r values are statistically significant from the whole epigenetic genes were indicated with symbol *(Randomization test, Supp. Fig. 3B).
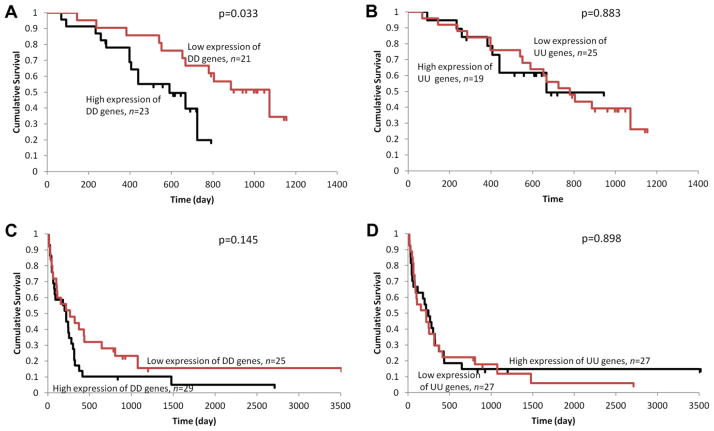
3.5. High expression of DD genes is associated with decreased survival of patients with melanoma
Two of the seven microarray datasets used in gene expression correlation analysis contain the patient’s survival information. Kaplan–Meier analysis was performed to determine the correlation of the expression of DD and UU genes to patient’s prognosis. We categorized all the patients into two groups according to the sum of the relative expression level of either the 12 DD genes or the 16 UU genes as described in Methods. In dataset GSE19234, the patients with low expression of DD genes had a significantly longer survival than patients with high DD gene expression (p = 0.03, log-rank test) (Fig. 4A). Among the 15 patients who had survived for more than 2 years after excision of the metastatic lesion, 14 patients were from the group with low expression of DD genes. By contrast, no survival difference was observed between patients with low and high expression of UU genes (Fig. 4B). Similarly, in the dataset GSE22153, the patients with low DD gene expression had better prognosis than patients with high DD gene expression, although it is not statistically significant (Fig. 4C). Again, no survival difference between patients with low and high expression of UU genes was observed (Fig. 4D).
Fig. 4.
Survival analysis in patients with melanoma. Kaplan–Meier analysis were performed for groups based on the sum of the relative expression level of DD (A, C) or UU (B, D) genes. The survival information was retrieved from dataset GSE19234 (A, B) and GSE22153 (C, D).
4. Discussion
It is well known that the epigenetic modulators interact with each other. For example, histone methyltransferases can regulate DNA methylation levels through modulating the stability of DNMT proteins [5]. In addition, it can also direct DNA methylation to specific genomic targets by recruiting DNMTs [23]. On the other hand, DNA methylation can direct histone modifications, which was supported by evidences such as methylated DNA mediates H3K9me through MeCP2 recruitment [6]. Numerous studies showed the synergistic effects of DNA methylation and histone deacetylation inhibitors on restoration of hyper-methylated gene expression in cancer cells [14], further illustrating the close cooperation of the epigenetic modulators. Therefore, it is anticipated that besides DNA methylation, histone modification and chromosome remodeling are also involved in the regulation of methylation and expression of the 31 potential TSGs examined in this study. This is the basis for us to use the correlation between the expression of the 273 epigenetic genes and the expression and methylation of the 31 TSGs to evaluate the epigenetic effects of DD and UU genes in melanoma cells.
Data from this study also supports the notion that the epigenetic modulators cooperate to regulate gene expression. For example, expression of the 11 DD genes negatively correlated with the TSG expression in melanoma was all positively correlated with the TSG methylation, with the only exception of C9orf169. Moreover, the DD genes that showed higher negative correlation to the TSG expression tend to have higher positive correlation with the TSG methylation levels (Supp. Fig. 4). On the other hand, while the expression of 11 of 12 DD genes were significantly correlated with the expression of the TSGs, only the expression of 8 of 12 DD genes were significantly correlated with the TSG methylation level (Supp. Fig. 3). Expression of C9ORF169, a histone demethylase, showed negative correlation with the TSG expression while it also had negative correlation with the TSG methylation (Supp. Fig. 4), suggesting that epigenetic modulators do not always play in a coordinated manner.
Among the 7 BRAFV600E signaling-related transcription factors, DD genes are more enriched with the binding sites for myc and c-fos, two well clarified proto-oncogenes. In addition, all of the 12 DD genes except PRMT3, CBX4 and CHD1, are potential oncogenes since they either promoted cell proliferation and transformation or caused resistance of cancer cells to anti-cancer drugs, and overexpression of these genes were frequently observed in some cancers (Supp. Table 3). Interestingly, 7 of the 16 UU genes, including BMI1, KDM3A, KDM4C, KDM5B, BPTF, SMARCA1 and ATF7IP, were reported to have proto-oncogenic effects, while only 4 of the UU genes, including ING4, KAT5, NCOA2 and NCOR2, showed tumor-suppressing functions in previous studies (Supp. Table 3). Thus, it is not surprising that overall the DD genes are better correlated than the UU genes with the methylation and expression of TSGs in melanoma cells and prognosis of patients with melanoma.
As inhibition of the BRAFV600 signaling pathway induces growth arrest and even cell death in BFARV600E melanoma cells, it is anticipated that inhibitors of the pathway will likely up-regulate TSG expression while down-regulate the oncogene expression in melanoma cells. Therefore, it is slightly confusing that nearly half of the UU genes turn out to have oncogenic effects. Previously it was reported that low level of ERK activation promotes DNA synthesis and cell cycle progression whereas high level of ERK activation cause cell cycle arrest and senescence by inducing the expression of various TSGs [12]. Similarly, one can hypothesize that moderate inhibition of MAPK pathway causes cell cycle arrest through down-regulating the expression of some oncogenes, while over inhibition of the pathway may induce the expression of some other oncogenes directly or indirectly, which may be an instinct of cells formed in a long evolutionary process to maintain their survival under extreme environments.
Protein arginine methyltransferases (PRMTs) catalyze the methylation of arginine in various proteins including histones H2A, H3 and H4 [2]. In mammalian cells, 10 PRMTs have been classified into Type I (PRMT1, 3, 4, 6 and 8) and Type II (PRMT5, 7 and FBXO11) depending on their specific catalytic activity [2]. No activity has yet been demonstrated for PRMT2 and PRMT9 [2]. Interestingly, in our study, 4 PRMTs including PRMT1, 3, 5 and 7, which accounts for the one third of 12 DD genes, were down-regulated by BRAFV600E signaling inhibition, and their expressions were negatively correlated with the TSGs expression in melanoma cells significantly. In addition, PRMT3 and 7 were also positively correlated with the methylation level of the TSGs significantly. Three of the 4 PRMTs showed oncogenic effects in previous studies [2,21]. These results suggested that histone methylation by PRMTs might play an important role in the epigenetic silence of TSG by BRAFV600E signaling in melanoma cells.
In summary, in this study we found that the expression of 12 epigenetic genes, which are regulated by BRAFV600E signaling, were closely associated with the methylation and silence of TSGs in melanoma cells, and furthermore, associated with short survival of patients with melanoma. In addition, the 12 genes are enriched with the binding sites for proto-oncogenes myc and c-fos, suggesting that myc and c-fos mediated transcription regulation of the expression of epigenetic genes might be involved in the BRAFV600 signaling-induced TSG silence in melanoma cells.
Supplementary Material
Acknowledgments
This work was supported, in part, by an ATA Young Investigator Award to DL and the NIH R01 CA134225 to MX.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbrc.2012.07.046.
References
- 1.An X, Jin Y, Guo H, et al. Response gene to complement 32, a novel hypoxia-regulated angiogenic inhibitor. Circulation. 2009;120:617–627. doi: 10.1161/CIRCULATIONAHA.108.841502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonazzi VF, Nancarrow DJ, Stark MS, et al. Cross-platform array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as genes frequently silenced by methylation in melanoma. PLoS One. 2011;6:e26121. doi: 10.1371/journal.pone.0026121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies MA, Samuels Y. Analysis of the genome to personalize therapy for melanoma. Oncogene. 2010;29:5545–5555. doi: 10.1038/onc.2010.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteve PO, Chin HG, Benner J, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Nat Acad Sci USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuks F, Hurd PJ, Wolf D, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 7.Hou P, Liu D, Dong J, et al. The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells. Cell Cycle. 2012;11:286–295. doi: 10.4161/cc.11.2.18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonsson G, Busch C, Knappskog S, et al. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res. 2010;16:3356–3367. doi: 10.1158/1078-0432.CCR-09-2509. [DOI] [PubMed] [Google Scholar]
- 9.Joseph EW, Pratilas CA, Poulikakos PI, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Nat Acad Sci USA. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneda A, Wakazono K, Tsukamoto T, et al. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res. 2004;64:6410–6415. doi: 10.1158/0008-5472.CAN-04-1543. [DOI] [PubMed] [Google Scholar]
- 11.Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol. 2004;150:179–185. doi: 10.1111/j.1365-2133.2004.05708.x. [DOI] [PubMed] [Google Scholar]
- 12.McMahon M, Woods D. Regulation of the p53 pathway by Ras, the plot thickens. Biochim Biophys Acta. 2001;1471:M63–M71. doi: 10.1016/s0304-419x(00)00027-5. [DOI] [PubMed] [Google Scholar]
- 13.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 14.Momparler RL. Cancer epigenetics. Oncogene. 2003;22:6479–6483. doi: 10.1038/sj.onc.1206774. [DOI] [PubMed] [Google Scholar]
- 15.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 16.Pratilas CA, Taylor BS, Ye Q, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Nat Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebolt-Leopold JS, English JM. Mechanisms of drug inhibition of signalling molecules. Nature. 2006;441:457–462. doi: 10.1038/nature04874. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno K, Hirata H, Majid S, et al. IGFBP-4 activates the Wnt/beta-catenin signaling pathway and induces M-CAM expression in human renal cell carcinoma. Int J Cancer. 2011;129:2360–2369. doi: 10.1002/ijc.25899. [DOI] [PubMed] [Google Scholar]
- 20.van den Hurk K, Niessen HE, Veeck J, et al. Genetics and epigenetics of cutaneous malignant melanoma: a concert out of tune. Biochim Biophys Acta. 1826;2012:89–102. doi: 10.1016/j.bbcan.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Verbiest V, Montaudon D, Tautu MT, et al. Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 2008;582:1483–1489. doi: 10.1016/j.febslet.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 22.Vultur A, Villanueva J, Herlyn M. Targeting BRAF in advanced melanoma: a first step toward manageable disease. Clin Cancer Res. 2011;17:1658–1663. doi: 10.1158/1078-0432.CCR-10-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Q, Rank G, Tan YT, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.