Abstract
Background and Purpose
Gender is suggested to be an important determinant of ischemic stroke risk factors, etiology and outcome. However, the basis for this remains unclear. The Y chromosome is unique in males. Genes expressed in men on the Y chromosome that are associated with stroke may be important genetic contributors to the unique features of males with ischemic stroke, which would be helpful for explaining sex differences observed between men and women.
Methods
Blood samples were obtained from 40 males at ≤ 3, 5 and 24 hours following ischemic stroke and from 41 male controls (July 2003- April 2007). RNA was isolated from blood and processed on Affymetrix Human U133 Plus 2.0 Arrays. Y chromosome genes differentially expressed between male stroke and male control subjects were identified using an analysis of covariance (ANCOVA) adjusted for age and batch. A p<0.05 and fold change (FC) > ∣1.2∣ were considered significant.
Results
Seven genes on the Y chromosome were differentially expressed in males with ischemic strokes compared to controls. Five of these genes (VAMP7, CSF2RA, SPRY3, DHRSX, PLCXD1,) are located on pseudoautosomal regions (PARs) of the human Y chromosome. The other two genes (EIF1AY and DDX3Y) are located on the non-recombining region of the human Y chromosome (NRY). The identified genes were associated with immunology, RNA metabolism, vesicle fusion and angiogenesis.
Conclusions
Specific genes on the Y chromosome are differentially expressed in blood following ischemic stroke. These genes provide insight into potential molecular contributors to sex differences in ischemic stroke.
Keywords: gene expression, ischemic stroke, gender, blood, Y chromosome
Introduction
Ischemic stroke is influenced by ones sex, with suggested differences in risk factors, outcomes and etiology between men and women [1-5]. For instance, men have increased stroke risk in middle age compared to women [1, 6], while women tend to have strokes at a later age and have more cardioembolic stroke [1, 3, 7]. These differences have been explained by both hormone dependent and hormone-independent mechanisms [8]. Peripheral and brain immune and inflammatory responses, cell apoptosis and cell death may contribute to the gender differences in stroke [9, 10]. The Y chromosome is unique in males, and is a clear genetic difference that exists between males and females. Previous studies have demonstrated the importance of the Y chromosome in brain function and disease. The Sex-determining region Y (SRY ) gene on the Y chromosome modulates brain dopamine concentrations and has been linked to increased risk in men of dopamine related disorders such as schizophrenia and Parkinson’s disease [11]. Whether there is a similar association between Y chromosome gene expression and ischemic stroke is unknown. Thus we set out to discover genes expressed in men on the Y chromosome that are associated with stroke which may identify genetic contributors to the unique features of males with ischemic stroke, and may be help for explaining sex differences observed between men and women.
Though the Y chromosome has few genes, it is complex. It can be divided into two regions, the pseudoautosomal region (PAR) and the non-recombining region of the Y chromosome (NRY). PAR accounts for 5% of the Y chromosome, and encodes genes that are identical to those present on the X chromosome. Gene expression levels for these PAR genes can be different in males and females, which may result in a variety of dose-dependent effects [12-14]. NRY accounts for the remaining 95% of the Y chromosome. Genes on NRY are considered as Y specific since they are not recombined with X chromosome. Some of these are expressed mainly in testes (testis-specific genes), and some have non-identical homologues on the male X chromosome(Y-linked homologues). It remains unclear if the functions for these X and Y homologues are interchangeable[15], but some studies suggested they have different function [16, 17]. Thus, genes expressed on the Y chromosome may contribute to sex differences in stroke.
In the current study, we compared Y chromosome gene expression in males with ischemic stroke to male controls. A number of Y chromosome genes were differentially expressed following stroke, suggesting that Y chromosome gene expression may contribute to sex differences in stroke.
Materials and Methods
Subjects
Male subjects with acute ischemic stroke were recruited through the CLEAR trial - a multi-center, randomized, double-blind safety study of recombinant tissue-plasminogen activator (rt-PA) and eptifibatide [18] (NCT00250991 at Clinical-Trials.gov) (July 2003-April 2007). This is part of the NINDS-funded Specialized Programs of Translational Research in Acute Stroke (SPOTRIAS) Network. The internal review board at each site approved the study, and written informed consent was obtained from all patients. Stroke was defined as an acute onset of neurological deficits lasting longer than 24 hours with corresponding infarction present on CT or MRI, Patients had a National Institutes of Health Stroke Scale (NIHSS) > 5 and were 18–80 years of age. The blood samples of each subject were drawn at ≤ 3, 5 and 24 h following their stroke. The first blood sample was drawn prior to 3 h following stroke onset and before treatment. Controls were male subjects similar in age without symptomatic cardiovascular disease who were recruited from outpatient neurology, internal medicine and vascular clinics at the University of Cincinnati, University of California Davis, University of California San Francisco and Wake Forest University. Subjects with prior history of stroke, myocardial infarction or peripheral vascular disease were not included as controls. CLEAR trial study inclusion and exclusion criteria are previously described [18] .
Sample processing and data analysis
Blood sample collection, RNA isolation and processing on Affymetrix U133 Plus 2.0 expression arrays were performed as previously described [19]. Whole blood (15ml) was collected from each subject via antecubital fossa venipuncture into six PAXgene Vacutainer tubes (Qiagen, Valencia, CA, USA). These tubes contain a solution that immediately lyses all of the cells in whole blood and stabilizes the RNA without measurable degradation. The RNA represents genes expressed in all white blood cells, immature red blood cells and immature platelets. Blood samples were stored frozen at −80°C until processed.
Total RNA was isolated using the PAXgene Blood RNA Kit (Qiagen) according to the manufacturer’s protocol. RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Foster City, CA, USA) and quantified with fiberoptic spectrophotometry using the Nanodrop ND-1000 (Nanodrop Inc., Wilmington, DE, USA). Reverse transcription, amplification and sample labeling were carried out using Nugen’s Ovation Whole Blood reagents (Nugen Technologies, San Carlos, CA). Each RNA sample was hybridized and scanned according to Nugen’s protocol on Affymetrix Genome U133 Plus 2.0 GeneChips (Affymetrix Santa Clara, CA). The arrays were washed and processed on a Fluidics Station 450 and then scanned on the Genechip Scanner 3000.
Raw expression values were normalized using Robust Multichip Averaging (RMA), as well as our internal-gene normalization method [20]. There were 123 Y chromosome probesets on Affymetrix Human U133 plus 2.0 arrays, of which, 42 probesets (representing 24 genes) were located in the 2 PARs, and 81 probesets (representing 56 genes) were located in the NRY region. To reduce false positive results, the probesets with intensity lower than the background level in all samples were excluded (log2 ≤ 5.5), and the probesets identified as “absent” using the MAS 5.0 expression summary algorithm were also excluded. Probe sets for the X-linked homologues to selected NRY genes were included in the analyses to determine if the homologues had different responses to stroke.
Differences in demographic data between groups were analyzed using Fisher’s exact test and a two-tailed t-test where appropriate. To demonstrate that transcripts were differentially expressed between men with strokes compared to controls, an analysis of covariance (ANCOVA) was performed, adjusting for age and batch. Probesets with a p value < 0. 05 and a FC > ∣1.2∣ were considered significant.
Results
Demographics and probesets
There were 40 male stroke subjects and 41 control males in the study (Table 1). There were no significant differences in race (p = 0.74) between the groups (Table 1). There were significant differences in the ages of the control and (50.2, SEM 2.7) and ischemic stroke (66.9, SEM 2.0) subjects (p < 0.05). There were significant differences in hypertension but no significant differences in hyperlipidemia or diabetes (Table 1).
Table 1.
Demographics of male ischemic stroke patients and male control subjects.
| Control | Stroke | |
|---|---|---|
| Number of Subjects | 41 | 40* |
| Age** (mean ± SEM, years) | 50.2±2.7 | 66.9±2.0 |
| Race (Caucasian, %) | 85.4% | 90.0% |
| Hypertension** (%) | 13 (31.7) | 23 (57.5) |
| Hyperlipidemia (%) | 12 (29.3) | 9 (22.5) |
| Diabetes (%) | 4 (9.8) | 7 (17.5) |
| NIHSS 3h (Q1, median, Q3) | N/A | 6.75,11.5,16 |
| NIHSS 24h (Q1, median, Q3) | N/A | 3, 9,14.25 |
| NIHSS 5d (Q1, median, Q3) | N/A | 2,6,9.5 |
N = 40 subjects at ≤ 3h, 36 subjects at 5h, 37 subjects at 24h. Q=quartile, NIHSS=National Institutes of Health Stroke Scale.
p<0.05
Of the 123 Y chromosome probe sets, 58 were identified as “present” using MAS5.0 and had expression values above background. These included 25 probesets (representing 15 genes) that targeted the NRY region, and 33 probe sets (representing 20 genes) that targeted the PAR region (Supplementary Table 1). Nine additional probesets were included in the analyses to determine if the X chromosome homologues of the regulated NRY genes were similarly regulated following ischemic stroke. Thus, a total of 67 probesets were included in the analyses (Supplementary Table 1).
Y-linked homologues in the NRY region
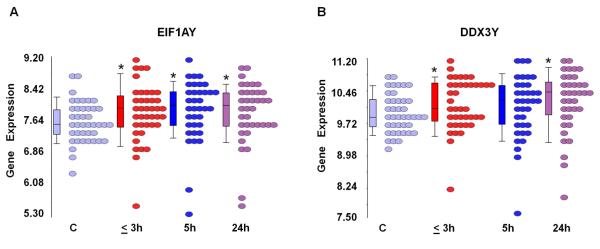
Two NRY genes (represented by 4 probe sets) changed expression in males with stroke compared to the control males (Figure 1, Supplementary Table 2). Of these, EIF1AY (eukaryotic translation initiation factor 1A, Y-linked, represented by 2 probe sets) was up-regulated at all three time points after stroke (Figure 1A). DDX3Y (DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked, represented by 2 probe sets) was up regulated at 3h and 24h after stroke (Figure 1B, Supplementary Table 2). These findings did not address whether there were similar changes in expression of the X chromosome homologues of these NRY genes. Indeed, we found that the male X chromosome homologue EIFAX was not regulated following ischemic stroke; and, the male X chromosome homologue DDX3X was up regulated only at 24h following stroke (Supplementary Table 2).
Figure 1.
Expression of genes in the NRY region of the Y chromosome differentially expressed following ischemic stroke. RNA expression (log2-transformed intensity values) is shown on the Y axis for EIF1AY (A) and DDX3Y (B). Control male subjects and male patients at different time points following ischemic stroke (≤ 3h, 5h, 24h) are shown on the X axis, each circle represents a single patient. Note: * = significantly changed following stroke compared to control (C) (p<0.05, FC>∣1.2∣).
Testis-Specific genes in the NRY region
There were no testis-specific genes that significantly changed expression after acute ischemic stroke.
X-Y identical genes in PAR regions
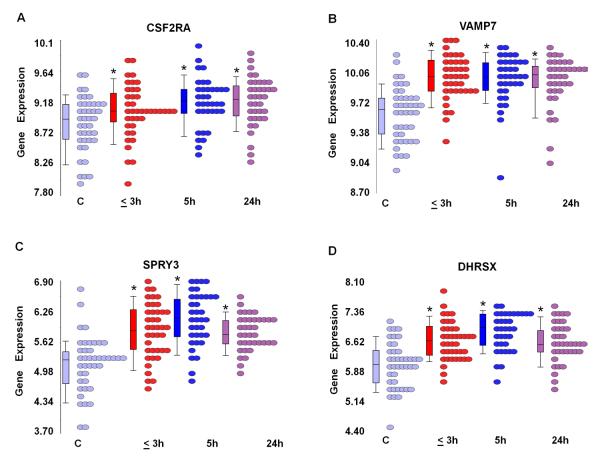
Four PAR genes (represented by 7 probe sets) were significantly differentially expressed in male stroke patients compared to male controls at 3h after stroke. Five genes (represented by 8 probe sets) were significantly differentially expressed at 5h after stroke. Four genes (represented by 6 probe sets) were significantly differentially expressed at 24h after stroke (p<0.05, FC > ∣1.2∣) (Supplementary Table 2). Of these, four PAR genes (represented by 7 probe sets) were up-regulated in male stroke patients compared to male controls at all three time points including colony stimulating factor 2 receptor, alpha, low-affinity (CSF2RA) (Figure 2A ), vesicle-associated membrane protein 7 (VAMP7) (Figure 2B), sprouty homolog 3 (SPRY3) ( Figure 2C), and dehydrogenase / reductase (SDR family) X-linked (DHRSX) (Figure 2D) (Supplementary Table 2).
Figure 2.
Expression of identical X and Y chromosome genes differentially expressed in males following ischemic stroke. RNA expression (log2-transformed intensity values) is shown on the Y axis for CSF2RA (A), VAMP7 (B), SPR3(C) and DHRSX (D). Control males and male patients at different time points following ischemic stroke (≤ 3h, 5h, 24h) are shown on the X axis, each circle represents a single patient. Note: * = significantly changed following stroke compared to control (C) (p <0.05, FC>∣1.2∣).
Discussion
This is the first study to demonstrate changes of Y chromosome gene expression in whole blood following ischemic stroke in males. Some of the changes of gene expression following stroke occurred for Y chromosome specific genes located in the NRY region of the chromosome. This suggests that the Y chromosome might contribute to sex differences in ischemic stroke.
Genes in the NRY region
Two coding genes, DDX3Y and EIF1AY, were up regulated in ischemic stroke and both were on the NRY regions that do not recombine with the X chromosome. This may suggest sex-specific involvement in stroke for these genes. DDX3Y is an ATP-dependent RNA helicase, involved in RNA metabolism including secondary structure alteration, splicing, spliceosome assembly and translation initiation [17, 21]. EIF1AY (Y-linked member of the EIF-1A family of genes) is involved in translation initiation [21]. DDX3Y and EIF1AY encode for human male-specific minor histocompatibility antigens which contribute to immune-mediated disease [22]. EIF1AY is also associated with autism [23]. Since these genes regulate translation initiation, alternative splicing, splicesome assembly, and are expressed in blood cells, they appear to have a role in the immune system following stroke.
The specificity of the DDX3Y and EIF1AY changes of Y-linked gene expression are emphasized by the fact that the X-linked homologues had different patterns of expression. The X-linked homologue EIF1AX did not change expression following stroke in either males (current study) or females [24] ) unlike EIF1AY. The X-linked homologue DDX3X only changed expression at 24h after stroke in males in current study, and female-specific up-regulation was observed on DDX3X at all time point after stroke[24] These data show NRY Y-linked gene expression that is specific to males, and suggest that at least some X and Y chromosome homologues may not be functionally equivalent as suggested by others [16, 17].
X-Y identical genes in PAR regions
Even though PAR genes are identical on X and Y chromosomes, and most of the PAR genes escape X-inactivation (excluding VAMP7), the process is not “all or none”. Thus, gene expression levels could differ for the same genes expressed on the X and Y chromosomes, with different functional consequences in males and females [12, 13]. Though we could not discriminate whether PAR gene expression could be attributed to the X or Y chromosome or to both based upon the experimental design of this study, a few PAR genes may have different expression patterns in males with stroke compared to the literatures and female-specific PARs genes in stroke in our another study [24]PAR genes up regulated following stroke in males included CSFR2A, SPRY3, VAMP7 and DHRSX. The CSFR2A gene encodes the alpha subunit of colony stimulating factor 2 (GM-CSF) receptor for GM-CSF. CSFR2A controls production, differentiation and function of granulocytes and macrophages. However, even though CSFR2A is higher in males with stroke compared to control males in this study, CSF2RA mRNA is reported to be higher in normal females compared to normal males [13]. Since GM-CSF induces angiogenesis, improves cerebral blood flow and protects against ischemic injury [25, 26], this may suggest that GM-CSF modulation of the vasculature via CSFR2A may be more important in females than males. SPRY3 was also up regulated in males. However, it antagonizes endothelial cell activation by fibroblast growth factor (FGF) and vascular endothelial growth factor (VEGF) [27]. The finding that CSFR2 and SPRY3 are both up-regulated in male stroke patients suggests that leukocytes can synthesize factors that both positively and negatively regulate the vasculature following stroke.
VAMP7 encodes a transmembrane protein that is a member of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family. Inactivation of both X and Y homologues of this gene has been demonstrated [14], which might result in dosage difference between males and females. The encoded protein is involved in the fusion of transport vesicles to their target membranes. VAMP7 mediates glucose transporter GLUT4 translocation in cultured cardiomyocytes. This could modulate glucose metabolism in immune and brain cells following ischemic stroke [28] since VAMP7 is widely distributed in the adult rat brain and transports exocytotic vesicles [29]. Importantly, it plays a key role in the secretion of stored mediators in vesicles for eosinophils (e.g. eosinophil toxin) and neutrophils (e.g. myeloperoxidase, lactoferrin and MMP9) [30] which might modulate leukocyte responses to injured endothelium and other brain cells following ischemia.
DHRSX and SPRY3 were significantly up regulated in males with stroke, but they were not identified as differentially expressed in females with stroke[24]. This could suggest Y specific regulation of DHRSX and SPRY3 following stroke and/or male specific up regulation of these genes on the single male X chromosome. Regardless, it demonstrates male specific changes of gene expression related to the sex chromosomes.
Limitations
Though the male to male comparisons limited the scope of the study, it ensured that the changes of gene expression related only to males and may help explain why the genes reported here were not detected in previous studies that included both males and females in the analyses [31]. Second, the sample sizes were small and several demographic variables were not well matched. Though age and hypertension were not well matched in current study, age was considered in the analysis, and when hypertention was co-variated in the ANCOVA models it did not affect the results reported here (data not shown). a Even so, larger and better matched cohorts will be needed in future studies. It will be important to match age, blood pressure, lipid profiles, blood glucose and the many other factors that play important roles in the pathophysiology of stroke. Thus, the current results should be considered preliminary since this discovery type study will need to be confirmed in a future independent study. Finally, since all the stroke patients were treated within 3h after stroke onset, some of the changes of expression observed at 5 and 24 h may be related to treatment.
Supplementary Material
Acknowledgments, Sources of Funding
This study was supported by NS056302 (FRS) and PO21040N635110 (JPB) from NIH/NINDS and the American Heart Association Bugher Foundation (FRS). Dr. Glen Jickling is a fellow of the Canadian Institutes of Health Research (CIHR). Dr. Yingfang Tian, Huichun Xu and Brad Ander were Bugher Fellows. This publication was also made possible by Grant Number UL1 RR024146 from the National Center for Medical Research to the CTSC at UC Davis. Dr. Bushnell and the SAVVY study were supported by NIH/NINDS K02 NS058760.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict(s) of Interest/Disclosure(s) All the authors declared no conflicts of interest.
References
- 1.Forster A, Gass A, Kern R, et al. Gender differences in acute ischemic stroke: etiology, stroke patterns and response to thrombolysis. Stroke. 2009;40:2428–2432. doi: 10.1161/STROKEAHA.109.548750. [DOI] [PubMed] [Google Scholar]
- 2.Reid JM, Dai D, Gubitz GJ, et al. Gender differences in stroke examined in a 10-year cohort of patients admitted to a Canadian teaching hospital. Stroke. 2008;39:1090–1095. doi: 10.1161/STROKEAHA.107.495143. [DOI] [PubMed] [Google Scholar]
- 3.Caso V, Paciaroni M, Agnelli G, et al. Gender differences in patients with acute ischemic stroke. Womens Health (Lond Engl) 6:51–57. doi: 10.2217/whe.09.82. [DOI] [PubMed] [Google Scholar]
- 4.Allen CL, Bayraktutan U. Risk factors for ischaemic stroke. Int J Stroke. 2008;3:105–116. doi: 10.1111/j.1747-4949.2008.00187.x. [DOI] [PubMed] [Google Scholar]
- 5.Silva GS, Lima FO, Camargo EC, et al. Gender differences in outcomes after ischemic stroke: role of ischemic lesion volume and intracranial large-artery occlusion. Cerebrovasc Dis. 30:470–475. doi: 10.1159/000317088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seshadri S, Beiser A, Kelly-Hayes M, et al. The lifetime risk of stroke: estimates from the Framingham Study. Stroke. 2006;37:345–350. doi: 10.1161/01.STR.0000199613.38911.b2. [DOI] [PubMed] [Google Scholar]
- 7.Kent DM, Buchan AM, Hill MD. The gender effect in stroke thrombolysis: of CASES, controls, and treatment-effect modification. Neurology. 2008;71:1080–1083. doi: 10.1212/01.wnl.0000316191.84334.bd. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Dziennis S, Hurn PD, Alkayed NJ. Mechanisms of gender-linked ischemic brain injury. Restor Neurol Neurosci. 2009;27:163–179. doi: 10.3233/RNN-2009-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol. 2010;10:594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 10.Siegel C, Turtzo C, McCullough LD. Sex differences in cerebral ischemia: possible molecular mechanisms. J Neurosci Res. 2010;88:2765–2774. doi: 10.1002/jnr.22406. [DOI] [PubMed] [Google Scholar]
- 11.Dewing P, Chiang CW, Sinchak K, et al. Direct regulation of adult brain function by the male-specific factor SRY. Curr Biol. 2006;16:415–420. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 13.Talebizadeh Z, Simon SD, Butler MG. X chromosome gene expression in human tissues: male and female comparisons. Genomics. 2006;88:675–681. doi: 10.1016/j.ygeno.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Esposito M, Ciccodicola A, Gianfrancesco F, et al. A synaptobrevin-like gene in the Xq28 pseudoautosomal region undergoes X inactivation. Nat Genet. 1996;13:227–229. doi: 10.1038/ng0696-227. [DOI] [PubMed] [Google Scholar]
- 15.Lau YF, Zhang J. Expression analysis of thirty one Y chromosome genes in human prostate cancer. Mol Carcinog. 2000;27:308–321. doi: 10.1002/(sici)1098-2744(200004)27:4<308::aid-mc9>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 16.Ditton HJ, Zimmer J, Kamp C, et al. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet. 2004;13:2333–2341. doi: 10.1093/hmg/ddh240. [DOI] [PubMed] [Google Scholar]
- 17.Rosner A, Rinkevich B. The DDX3 subfamily of the DEAD box helicases: divergent roles as unveiled by studying different organisms and in vitro assays. Curr Med Chem. 2007;14:2517–2525. doi: 10.2174/092986707782023677. [DOI] [PubMed] [Google Scholar]
- 18.Pancioli AM, Broderick J, Brott T, et al. The combined approach to lysis utilizing eptifibatide and rt-PA in acute ischemic stroke: the CLEAR stroke trial. Stroke. 2008;39:3268–3276. doi: 10.1161/STROKEAHA.108.517656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamova B, Xu H, Jickling G, et al. Gene expression profiling of blood for the prediction of ischemic stroke. Stroke. 41:2171–2177. doi: 10.1161/STROKEAHA.110.588335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamova BS, Apperson M, Walker WL, et al. Identification and validation of suitable endogenous reference genes for gene expression studies in human peripheral blood. BMC Med Genomics. 2009;2:49. doi: 10.1186/1755-8794-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro-Costa P, Plancha CE, Goncalves J. Genetic dissection of the AZF regions of the human Y chromosome: thriller or filler for male (in)fertility? J Biomed Biotechnol. 2010:936569. doi: 10.1155/2010/936569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miklos DB, Kim HT, Miller KH, et al. Antibody responses to H-Y minor histocompatibility antigens correlate with chronic graft-versus-host disease and disease remission. Blood. 2005;105:2973–2978. doi: 10.1182/blood-2004-09-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serajee FJ, Mahbubul Huq AH. Association of Y chromosome haplotypes with autism. J Child Neurol. 2009;24:1258–1261. doi: 10.1177/0883073809333530. [DOI] [PubMed] [Google Scholar]
- 24.Stamova B, Tian Y, Jickling G, et al. The X-Chromosome Has a Different Pattern of Gene Expression in Women Compared With Men With Ischemic Stroke. Stroke. 2011 doi: 10.1161/STROKEAHA.111.629337. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buschmann IR, Busch HJ, Mies G, Hossmann KA. Therapeutic induction of arteriogenesis in hypoperfused rat brain via granulocyte-macrophage colony-stimulating factor. Circulation. 2003;108:610–615. doi: 10.1161/01.CIR.0000074209.17561.99. [DOI] [PubMed] [Google Scholar]
- 26.Bath PM, Sprigg N. Colony stimulating factors (including erythropoietin, granulocyte colony stimulating factor and analogues) for stroke. Cochrane Database Syst Rev. 2007 doi: 10.1002/14651858.CD005207.pub3. CD005207. [DOI] [PubMed] [Google Scholar]
- 27.Cabrita MA, Christofori G. Sprouty proteins: antagonists of endothelial cell signaling and more. Thromb Haemost. 2003;90:586–590. doi: 10.1160/TH03-04-0217. [DOI] [PubMed] [Google Scholar]
- 28.Schwenk RW, Dirkx E, Coumans WA, et al. Requirement for distinct vesicle-associated membrane proteins in insulin- and AMP-activated protein kinase (AMPK)-induced translocation of GLUT4 and CD36 in cultured cardiomyocytes. Diabetologia. 53:2209–2219. doi: 10.1007/s00125-010-1832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coco S, Raposo G, Martinez S, et al. Subcellular localization of tetanus neurotoxin-insensitive vesicle-associated membrane protein (VAMP)/VAMP7 in neuronal cells: evidence for a novel membrane compartment. J Neurosci. 1999;19:9803–9812. doi: 10.1523/JNEUROSCI.19-22-09803.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Logan MR, Lacy P, Odemuyiwa SO, et al. A critical role for vesicle-associated membrane protein-7 in exocytosis from human eosinophils and neutrophils. Allergy. 2006;61:777–784. doi: 10.1111/j.1398-9995.2006.01089.x. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y, Xu H, Du X, et al. Gene expression in blood changes rapidly in neutrophils and monocytes after ischemic stroke in humans: a microarray study. J Cereb Blood Flow Metab. 2006;26:1089–1102. doi: 10.1038/sj.jcbfm.9600264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.