Abstract
The recently cloned NPR1 gene of Arabidopsis thaliana is a key regulator of acquired resistance responses. Upon induction, NPR1 expression is elevated and the NPR1 protein is activated, in turn inducing expression of a battery of downstream pathogenesis-related genes. In this study, we found that NPR1 confers resistance to the pathogens Pseudomonas syringae and Peronospora parasitica in a dosage-dependent fashion. Overexpression of NPR1 leads to enhanced resistance with no obvious detrimental effect on the plants. Thus, for the first time, a single gene is shown to be a workable target for genetic engineering of nonspecific resistance in plants.
Plants respond in a variety of ways to pathogenic microorganisms (1, 2). When the pathogen carries a specific avirulence (avr) gene and the plant host contains a cognate resistance (R) gene, a hypersensitive response occurs at the site of infection that results in inhibition of pathogen growth (1–6). Thus, a plant expressing a particular R gene is specifically resistant to pathogens expressing the corresponding avr gene. Several R genes conferring resistance to a variety of fungal, bacterial, and viral pathogens have recently been cloned from various plants, including Arabidopsis thaliana (4–6). The existence of conserved sequences among these genes suggests that they may function via common mechanisms. Indeed, the molecular events that occur after the specific avr-R interaction appear to be nonspecific. In addition to the hypersensitive response that blocks the local growth of an infecting pathogen, a secondary defense response can be triggered that renders uninfected parts of the plant resistant to a variety of normally virulent pathogens (7–9). This response is called systemic acquired resistance (SAR). The mechanisms of such induced resistance responses have been under intense study in recent years due to our basic interest in understanding immunity in plants and the possibility of identifying target genes for engineering long-lasting, broad-spectrum resistance in crops (10–15). Salicylic acid (SA) has been found to be an essential signal in the induction of SAR (16). In addition, exogenous application of SA or its analogs, such as 2,6-dichloroisonicotinic acid (INA) and benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester has been shown to induce SAR (17, 18). Often associated with the acquired resistance response is the induction of a group of pathogenesis-related (PR) genes; the roles of these genes in determining resistance have been inferred based on their expression patterns and sequence information (19–22), and demonstrated in some cases by observation of enhanced resistance in transgenic plants overexpressing a specific PR gene (23–27). However, the protection provided by a single PR gene is much narrower than that rendered by full-fledged SAR, and the degree of resistance is much less significant. Such experiments suggest that SAR is a result of the concerted expression of a battery of PR genes instead of the function of a single gene. Thus, genetic manipulation of the complete SAR response requires identification of genes involved in the SAR signal transduction pathway.
Using various genetic screens, the A. thaliana gene NPR1 (for nonexpresser of PR genes; also called NIM1 for nonimmunity or SAI1 for salicylic acid-insensitivity) was identified as a key regulator in transducing the SA signal leading to general acquired resistance responses, including SAR as well as local acquired resistance, the ability of plants to restrict the spread of virulent pathogen infections (10, 28–30). Mutations in the NPR1 gene result in a loss of resistance to virulent bacterial and fungal pathogens even when the plants are pretreated with SAR inducers (10). We recently cloned the NPR1 gene by a map-based strategy and found that it encodes a protein containing an ankyrin-repeat domain (31), which is found in many regulatory proteins, such as IκB and Cactus in animal immune responses (32). Expression studies demonstrated that although NPR1 is constitutively expressed in plants, its level can be further elevated by ≈2-fold after SA or INA treatment (31) or by pathogen infection (33). Upon SAR induction, activation of the NPR1 protein must also occur because constitutive expression of NPR1 in the absence of an inducer does not lead to constitutive expression of PR genes or resistance (31). These characteristics indicate that the SAR response may be enhanced through manipulation of NPR1 either at the level of expression or the level of protein activity or both. Here we report experiments investigating the possibility of generating disease resistance through overexpression of NPR1.
MATERIALS AND METHODS
Generation of NPR1 cDNA Transgenic Plants.
The NPR1 cDNA construct used in plant transformation was the same as described (31). Plant transformation and selection of transgenic plants also followed the procedure as described (31).
Analysis of PR1 Expression in NPR1 cDNA Transgenic Plants.
Total RNA was extracted from 30 2-week-old seedlings grown on Murashige–Skoog medium (34) containing 0.1 mM INA, and RNA blot analysis was performed by using A. thaliana PR1 and tobacco mitochondria β-ATPase as probes as described (10). The expression of PR1 was normalized against the level of β-ATPase.
ELISA Assay.
Forty 2-week-old seedlings grown on Murashige–Skoog medium were harvested and ground to fine powder in liquid nitrogen. Then 100 μl of 1× PBS containing 0.001% N-tosyl-l-phenylalanine chloromethyl ketone, 0.001% Nα-p-tosyl-l-lysine chloromethyl ketone, 0.133 μM phenylmethylsulfonyl fluoride, and 1 μM DTT was added to the powder. The extraction mixture was centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant was collected. The protein concentration of the supernatant was determined by using Bradford assay (35). For ELISA, 40 μg of total protein was mixed with an equal volume of 2× ELISA binding buffer (30 mM Na2CO3/91.5 mM NaHCO3/6.1 mM NaN3) and the total volume was brought up to 450 μl by addition of 1× ELISA binding buffer. Three serial 2-fold dilutions were made from this original mixture, and 100 μl duplicate aliquots of each dilution (including the original) were used for the assay. Polyclonal antibodies against NPR1, which were generated by using a synthesized 16-amino-acid oligopeptide from the C terminus of the protein as the antigen, were affinity-purified with the antigenic oligopeptide. Samples were incubated with the purified polyclonal antibodies against NPR1 for 1 hr, washed with 1× PBS containing 0.05% Tween 20 three times, then incubated with the secondary antibodies (alkaline phosphatase-conjugated anti-rabbit antiserum) for 1 hr and washed three times with 1×PBS containing 0.05% Tween 20. The substrate was added to the sample and the fluorescence was recorded after an overnight incubation at 4°C.
Immunoblot Analysis of NPR1 Protein.
Forty 2-week-old seedlings grown on Murashige–Skoog medium were ground to powder in liquid nitrogen to which an equal volume of universal lysis buffer (50 mM Tris/50 mM NaF/150 mM NaCl/0.5% Nonidet P-40/1 mM DTT/1 mM) was added. The samples were gently shaken at 4°C for 30 min and centrifuged at 4°C for 10 min. Protein concentrations in the supernatant were determined by using the Bradford assay (35). Two hundred micrograms of total protein were loaded onto an SDS/8% polyacrylamide gel and run in 250 mM Tris, 2 M glycine, and 2% SDS at room temperature at 200 V for 3 hr, and then electrotransfered onto a polyvinylidene difluoride membrane at 600 milliamp in a 4°C room for 3 hr. The membrane was blocked in 1×PBS containing 5% nonfat milk, and 0.05% Tween 20 overnight at 4°C. The membrane was then probed with the polyclonal antibodies against NPR1 at 1:800 for 1 hr and washed three times with the PBS-milk-Tween 20 solution. The membrane was then incubated with goat horseradish peroxidase conjugated anti-rabbit antibodies at 1:5000 for 1 hr and washed three times with PBS-milk-Tween 20. Chemofluorescence was detected with the enhanced chemiluminescence kit (Amersham). The x-ray film was exposed for 16 hr.
Analysis of NPR1 mRNA.
Poly(A)+ mRNA was extracted from 1 g of 2-week-old seedlings grown on Murashige–Skoog medium and RNA blot analysis was performed by using the A. thaliana NPR1 cDNA fragment as the probe (31). The expression of the NPR1 gene was normalized against the level of β-ATPase.
Trypan Blue Staining of A. thaliana Plants Infected with Peronospora parasitica.
Six days after the infection by P. parasitica strain Noco (P. parasitica Noco), the seedlings were boiled in one volume of lactophenol-trypan blue solution (23% phenol/25% glycerol/25% lactic acid/2.5 mg/ml trypan blue) and two volumes of 95% ethanol for 2 min then destained in chloral hydrate (2.5 mg/ml) overnight. The destained seedlings were then equilibrated in 80% glycerol and mounted for observation under a compound microscope.
Analysis of PR Gene Expression in Infected Plants.
Leaves of 4-week-old soil-grown wild-type and transgenic plants were infiltrated with Pseudomonas syringae pv. maculicola ES4326 (Psm ES4326) at OD600=0.001, and were collected at 0, 3, 6, 12, and 24 hr after inoculation. For each time point, leaves were collected from 10 individual plants. For the P. parasitica Noco infection, 2-week-old soil-grown wild-type and transgenic plants were sprayed with spores at 3 × 104 spores/ml and were collected at 0, 1, 3, and 6 days after infection. Total RNA was extracted from these infected plants and RNA blot analyses were performed by using PR1, PR2, PR5, and 18S rRNA as probes. PR1 and 18S rRNA were labeled using the random priming method (10). PR2 and PR5 were labeled using strand-biased PCR (36).
RESULTS
Isolation of A. thaliana Plants Expressing Different Levels of NPR1 Protein.
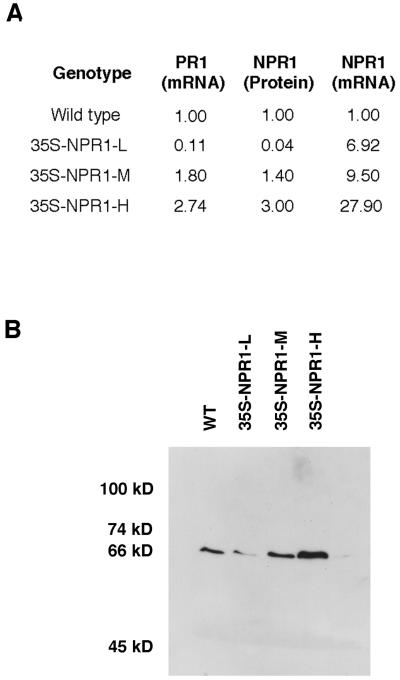
The NPR1 cDNA under the control of the constitutive 35S promoter of cauliflower mosaic virus was transformed into wild-type A. thaliana (Col-0). From 23 independent lines, progeny homozygous for the transgene were examined for the levels of the INA-induced PR1 gene expression. Because NPR1 regulates expression of PR genes in response to induction (10), plants expressing a higher level of PR1 are likely to contain a higher level of NPR1 protein. In these transgenic lines, we found a great deal of variation in the amount of PR1 gene expression after induction with INA. Based on the level of PR1 expression, we were able to place the plants into three groups (Fig. 1A). In 35S-NPR1-L, the PR1 expression is significantly lower (by ≈10-fold) than that of wild type, whereas in 35S-NPR1-M and 35S-NPR1-H, the PR1 expression is about 1.5- and 3-fold higher, respectively, than that of wild type. All the analyses described in this report were performed on two independent lines from each group that carry a single-site insertion of the transgene (as judged by the 3:1 segregation ratio of kanamycin resistance versus susceptibility in the F2 progeny). Because similar results were obtained from both lines, only one set of data are presented here. In these selected lines, the levels of NPR1 protein were measured by ELISA. It was found that the amount of NPR1 protein in 35S-NPR1-L is 25-fold lower than in the wild type, whereas both 35S-NPR1-M and 35S-NPR1-H have 1.5- to 3-fold higher levels than in the wild type (Fig. 1A). The NPR1 protein in transgenic plants was also visualized by using immunoblot analysis. A single band of 66 kDa, the expected molecular mass for NPR1, was detected (Fig. 1B). Quantification of the band intensity showed results agreeing with those obtained by using ELISA. Clearly, the levels of NPR1 protein in these plants correlate with the levels of the INA-induced PR1 gene expression; where there is more NPR1 protein, there is greater induction of the PR1 gene (Fig. 1 A and B). We also performed RNA blot analysis and detected high levels of NPR1 transcript in all transgenic lines. This is consistent with the fact that the NPR1 gene is driven by the constitutive 35S promoter but is inconsistent with the reduced levels of the NPR1 protein found in the 35S-NPR1-L lines. The discrepancy may be explained as a result of cosuppression, a phenomenon often observed in plants when a transgene is introduced that leads to suppression of both the endogenous gene and the incoming transgene (37). Even though different mechanisms have been proposed to explain the phenomenon (37), there are no published reports showing that cosuppression may occur without a significant reduction in the steady-state mRNA accumulation as seen in both the 35S-NPR1-L lines examined. We speculate that in 35S-NPR1-L, cosuppression takes place at the level of translation by an unknown mechanism. These 35S-NPR1-L lines are expected to mimic null npr1 mutants.
Figure 1.
Analysis of PR1 expression, NPR1 protein and mRNA in wild-type and NPR1 cDNA transgenic plants. (A) Comparison of INA-induced PR1 expression (as determined by Northern blot analysis), uninduced NPR1 protein (as measured by ELISA) and mRNA levels in NPR1 cDNA transgenic plants (as determined by Northern blot analysis) to those in wild type. All the analyses were performed twice with similar results. The transgenic plants were classified into three groups, designated as 35S-NPR1-L (low), 35S-NPR1-M (medium), and 35S-NPR1-H (high), respectively, based on the levels of induced PR1 expression. (B) Immunoblot showing the different levels of NPR1 protein in wild-type and NPR1 cDNA transgenic plants.
NPR1-Overexpressing Plants Display an Enhanced Resistance to a Bacterial Pathogen.
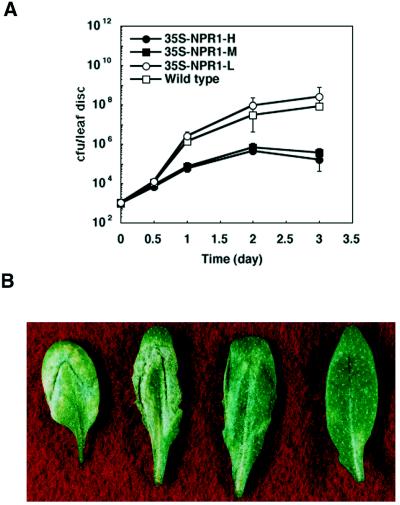
To determine if there is a correlation between the amount of NPR1 protein and disease resistance, we first tested the response of these transgenic plants to Psm ES4326, a virulent bacterial pathogen that causes leaf spots on wild-type A. thaliana plants (10). The 35S-NPR1-M and 35S-NPR1-H plants were found to exhibit a significantly elevated level of resistance to Psm ES4326, whereas 35S-NPR1-L plants are at least as susceptible as wild type to the pathogen. As shown in Fig. 2A, inhibition of pathogen growth in 35S-NPR1-M and 35S-NPR1-H plants was observed as early as 24 hr after the infection. Three days after inoculation, growth of the bacteria in the 35S-NPR1-M and 35S-NPR1-H lines was inhibited by 1,000-fold compared with that in the wild type and in 35S-NPR1-L (Fig. 2A). Furthermore, the difference in the resistance of these lines is also reflected by the varied degrees of disease symptoms. Three days after inoculation, fully developed water-soaked chlorosis appeared on the infected leaves of both wild-type and 35S-NPR1-L plants, whereas only small patches of yellowing were observed on the leaves of 35S-NPR1-M and 35S-NPR1-H plants (Fig. 2B). These observations indicate that the NPR1 protein controls resistance to this bacterial pathogen in a dosage-dependent manner.
Figure 2.
Analysis of disease resistance to the bacterial pathogen Pseudomonas syringae pv. maculicola ES4326 in wild-type and NPR1 cDNA transgenic plants. (A) Growth of Psm ES4326 in wild-type and NPR1 cDNA transgenic plants. The leaves of 4-week-old wild-type and the transgenic plants were inoculated with Psm ES4326 at OD600= 0.001. At 0, 0.5, 1, 2, and 3 days after the inoculation, infected leaves were collected and the bacterial growth was determined (10). Bars = 95% confidence limits of log-transformed data (38). Eight samples were taken for each time point. cfu, Colony forming unit. (B) Disease symptoms caused by Psm ES4326 in wild-type and NPR1 cDNA transgenic plants. From left to right, the order of the leaves is: wild type, 35S-NPR1-L, 35S-NPR1-M, and 35S-NPR1-H. The photograph was taken 3 days after infection.
NPR1-Overexpressing Plants Are More Resistant to an Oomycete Pathogen.
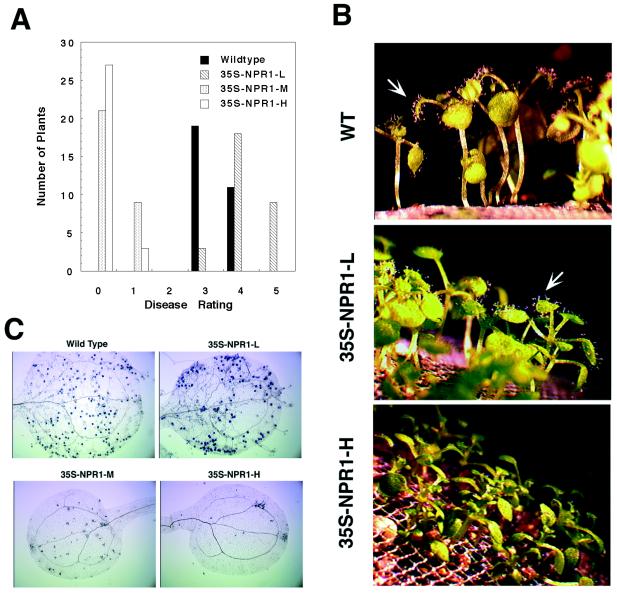
We then tested the response of these transgenic plants to P. parasitica Noco, an obligate oomycete pathogen that causes downy mildew in the A. thaliana Col-0 accession. Disease symptoms on each individual plant were scored based on the number of conidiophores that emerged after one week from each leaf, with “0” representing no disease symptoms and “5” representing the most severe symptoms (for details see Fig. 3A). Thirty infected plants of each line were sampled. All infected wild-type and 35S-NPR1-L plants had disease ratings of “3,” “4,” or “5;” whereas all 35S-NPR1-M and 35S-NPR1-H plants had ratings of “0” or “1.” Nonparametric statistical tests (38) of the data showed significant differences in disease symptoms between all sample pairs except between wild type and 35S-NPR1-L, or between 35S-NPR1-M and 35S-NPR1-H (Fig. 3 A and B). Although not statistically significant, more conidiophores were consistently observed in 35S-NPR1-L than in wild type and fewer conidiophores were consistently detected in 35S-NPR1-H than in 35S-NPR1-M, the same pattern as consistently observed for the growth of Psm ES4326. Larger sample sizes would be required to determine if these are significant differences. Trypan blue staining of the infected plants also showed differences in the growth of P. parasitica Noco in these plants (Fig. 3C). Fully developed disease symptoms were observed in both wild-type and 35S-NPR1-L leaves, as demonstrated by a heavy coverage of hyphae, conidiophores, and spores. In contrast, very few hyphae were detected on 35S-NPR1-M or 35S-NPR1-H leaves. It is obvious that the growth of P. parasitica Noco was strongly inhibited in both 35S-NPR1-M and 35S-NPR1-H plants. These data show that in addition to a bacterial pathogen, NPR1 also regulates resistance to an oomycete pathogen in a dosage-dependent fashion.
Figure 3.
Analysis of resistance to the oomycete pathogen Peronospora parasitica strain Noco in wild-type and NPR1 cDNA transgenic plants. (A) Disease ratings of wild-type and NPR1 cDNA transgenic plants after the infection with P. parasitica Noco. Two-week-old soil-grown seedlings of wild-type and transgenic plants were sprayed to imminent runoff with spores of P. parasitica Noco (∼3 × 104/ml). Six days after infection, 30 plants of each line were sampled to rate disease symptoms. Ratings were defined as follows: 0, no conidiophores on the plant; 1, no more than 5 conidiophores per infected plant; 2, 6–20 conidiophores on a few infected leaves; 3, 6–20 conidiophores on most of the infected leaves; 4, 5 or more conidiophores on all infected leaves; 5, 20 or more conidiophores on all infected leaves. The data were analyzed by using Mann–Whitney U tests (38). (B) Conidiophores observed in wild-type and NPR1 cDNA transgenic plants seven days after inoculation with P. parasitica Noco. Plants were examined under a dissecting microscope. (C) Trypan blue staining of P. parasitica-infected leaves of wild-type and NPR1 cDNA transgenic plants seven days after infection. Seedlings of wild-type and transgenic plants were stained with trypan blue and mounted for observation under a compound microscope (36).
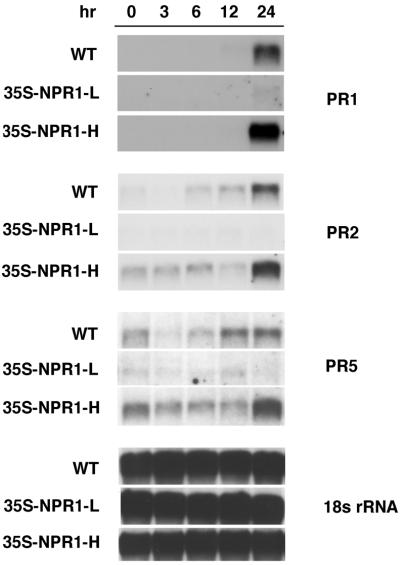
The Induction of PR Genes After a Pathogen Infection Is Stronger, but Not Quicker, in NPR1-Overexpressing Plants.
The enhanced disease resistance exhibited by 35S-NPR1-M and 35S-NPR1-H could be due to a higher expression level of PR genes and other unidentified defense-related genes in these plants. However, it is also possible that the enhanced disease resistance is a result of accelerated induction of these defense-related genes. To test this possibility, the PR gene induction pattern was examined in these transgenic plants after infection by Psm ES4326. As shown in Fig. 4, significant expression of PR1, BGL2 (PR2), and PR5 was observed 24 hr after infection in both the wild-type and the transgenic lines 35S-NPR1-M and 35S-NPR1-H. However, the levels of PR mRNA observed at the 24 hr time point varied in these plants, correlating with the levels of NPR1 protein; i.e., 2–3-fold higher in 35S-NPR1-M and 35S-NPR1-H, and 15-fold lower in 35S-NPR1-L compared with wild type (Fig. 4). In addition, it appears that the timing of the induction of PR gene expression correlates with the timing of the resistance response (Fig. 2). Inhibition of bacterial growth in NPR1-overexpressing plants was first evident 24 hr after the infection, and this corresponds to the earliest time point when strong PR gene expression was detected. The same analysis was also carried out 0, 1, 3, and 6 days after the infection by the oomycete pathogen P. parasitica Noco and a similar result was obtained as in those plants challenged with Psm ES4326. PR gene expression was not induced until 6 days after infection in both wild-type and transgenic plants, and the levels of PR mRNA correlated with the levels of NPR1 protein (data not shown). These findings suggest that overexpression of NPR1 results in stronger, rather than faster, induction of PR genes upon pathogen infection; this implies that a stronger induction of these PR genes and other unidentified defense-related genes is probably responsible for the enhanced disease resistance observed in NPR1-overexpressing plants.
Figure 4.
Analysis of PR gene expression in wild-type and NPR1 cDNA transgenic plants after infection by Psm ES4326. At 0, 3, 6, 12, and 24 hr after inoculation with Psm ES4326 at OD600 = 0.001, infected leaves were collected from ten individual plants and total RNA was extracted from the leaf tissue. RNA blot analyses were performed by using PR1, BGL2 (PR2), PR5, and 18S rRNA as probes (36).
DISCUSSION
Although 35S-NPR1-M and 35S-NPR1-H express ≈2–3-fold higher level of NPR1 protein than wild type, RNA blot analysis of these plants grown under noninducing conditions detected no increase in the basal level of expression of PR genes (Fig. 4). This indicates that NPR1 is normally inactive in the absence of induction. An inducing signal (SA, INA, or a pathogen) is necessary to activate NPR1, which then induces downstream PR gene expression. Therefore, NPR1-overexpressing plants will not express the disease resistance until they are challenged by a pathogen. This characteristic of activation only upon induction makes the NPR1 protein ideal for genetic engineering because constitutive expression of resistance is not only a waste of energy, likely to cause detrimental effects on plants, but also increases the selection pressure for more virulent pathogens. Indeed, in mutants such as cprs (which are constitutive expressers of PR genes) and SAR, reduced plant size and fertility are often observed (12, 36). Unlike these mutants, 35S-NPR1-H and 35S-NPR1-M display no significant difference in growth rate, morphology, and developmental timing compared with wild type and 35S-NPR1-L.
By overexpressing a single gene, NPR1, we were able to generate complete resistance to both the bacterial pathogen Psm ES4326 and the oomycete pathogen P. parasitica Noco, two very different pathogens virulent on wild-type A. thaliana. Because NPR1 is a “master” regulator of the downstream PR genes and probably other unidentified genes, elevated levels of NPR1 protein in 35S-NPR1-H and 35S-NPR1-M result in a more dramatic induction of all these downstream genes and simultaneous activation of multiple resistance mechanisms against very different pathogens. The spectrum of resistance established in 35S-NPR1-H and 35S-NPR1-M can be more fully explored as other SAR-affected pathogens of A. thaliana are identified and characterized.
It is interesting to note that a moderate (2–3-fold) increase in the NPR1 protein level and a corresponding increase in the expression of the downstream genes results in significantly enhanced resistance. This may be explained as a synergistic effect of the heightened expression of all, or a subset of, the downstream defense-related genes. Examples of such synergy have been reported previously (39, 40). In one study, simultaneous expression of a tobacco class I chitinase and a class I β-1,3-glucanase gene resulted in increased resistance to Fusarium oxysporum f.sp. lycopersici, whereas expression of either gene alone failed to confer resistance (39). In another study, coexpression in tobacco of barley antifungal proteins (a class II chitinase, a class II β-1, 3-glucanase, and a type I ribosome-inactivating protein) led to significantly higher resistance to a soilborn fungal pathogen Rhizoctonia solani compared with that found when each of these three genes was expressed alone (40). These findings indicate that the concerted actions of these defense-related genes are important in determining resistance and that manipulating regulatory genes such as NPR1 is a more effective approach of generating broad-spectrum resistance than overexpressing a particular PR gene, as has been attempted (23–27).
NPR1, which is likely a single-copy gene, determines resistance in a very different manner from that of a typical R gene, which often has a high number of homologs in the same genome (41). The presence or absence of an R gene in a plant determines whether it is heritably resistant to a specific pathogen carrying the cognate avr gene (1–6, 41). The plant and the pathogen may coevolve by regenerating new R and avr genes to create new specificity. Therefore, families of R genes exist in each plant genome to protect the plant from a wide range of pathogens (41). Because expression of one R gene confers resistance to only the particular pathogen carrying the corresponding avr gene, the feasibility of using R genes as targets for genetic engineering of durable, wide-spectrum resistance in plants is questionable. The nonspecific resistance conferred by NPR1, however, is determined by the dosage and the activity of NPR1, in contrast to the all-or-none hypersensitive response resistance determined by the R-avr interaction. As shown in this report, a threshold level of NPR1 protein is required to confer resistance. In uninduced wild-type plants, the level of NPR1 is not adequate to control infections by Psm ES4326 and P. parasitica Noco with the amounts of pathogen inoculant used. A further reduction in the NPR1 level as in 35S-NPR1-L only makes the plants slightly more susceptible to these pathogens compared with the wild type (Figs. 2 and 3). This is consistent with previous observations of the npr1 mutants (10, 31). Based on previous studies of npr1 mutant plants (29), when the amount of pathogen used in the experiments is reduced by 10-fold, a more dramatic difference in disease susceptibility between 35S-NPR1-L and the wild type is expected. Plants with increased levels of NPR1 (35S-NPR1-H and 35S-NPR1-M) become resistant to pathogens that are normally virulent on wild-type plants. Furthermore, agents that can increase the expression and activity of NPR1—such as SA, INA, or a necrotizing pathogen—are inducers of SAR. Once established, SAR should be difficult for a pathogen to overcome by evolutionary selection because it is conferred by multiple PR genes.
NPR1 homologs have been identified in many economically important plants, including canola, cabbage, broccoli, tobacco, tomato, potato, corn, and wheat (data not shown). Particularly, NPR1 homologous cDNA clones have been isolated and sequenced from tobacco and tomato, and the deduced proteins have been shown to share ≈70% amino acid sequence similarity to A. thaliana NPR1 (M. Kinkema and X.D., unpublished data). The ubiquitous existence of NPR1 in different plant species suggests that the findings with NPR1 in A. thaliana are likely to apply to other plants.
In this study, we explored a different approach for generating broad-spectrum disease resistance in plants. Instead of exogenous application of SAR-induced chemicals, we succeeded in enhancing the plants’ immunity by overexpressing a key regulator of the SAR signaling pathway, NPR1. It appears that this approach does not adversely affect the growth or development of the plants, making it an attractive method for controlling plant diseases.
Acknowledgments
We thank S. Bowling, J. Clarke, and M. Kinkema for helpful suggestions and comments on the manuscript. We also thank M. Kinkema for the unpublished data on NPR1 homologs in tobacco and tomato. This work was supported by U.S. Department of Agriculture Grant 95–37301-1917 to X. Dong.
ABBREVIATIONS
- INA
2,6-dichloroisonicotonic acid
- SA
salicyclic acid
- SAR
systemic acquired resistance
References
- 1.Lamb C J, Lawton M A, Dron M, Dixon R A. Cell. 1989;56:215–224. doi: 10.1016/0092-8674(89)90894-5. [DOI] [PubMed] [Google Scholar]
- 2.Lamb C J. Cell. 1994;76:419–422. doi: 10.1016/0092-8674(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 3.Keen N T. Annu Rev Genet. 1992;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 4.Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 5.Hammond-Kosack K, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bent A. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross A F. Virology. 1961;14:340–358. doi: 10.1016/0042-6822(61)90319-1. [DOI] [PubMed] [Google Scholar]
- 8.Kuc J. BioScience. 1982;32:854–860. [Google Scholar]
- 9.Ryals J A, Neuenschwander U H, Willits M G, Molina A, Steiner H-Y, Hunt M D. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao H, Bowling S A, Gordon A S, Dong X. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawton K, Uknes S, Friedrich L, Gaffney T, Alexander D, Goodman R, Métraux J-P, Kessmann H, Ahl-Goy P, Gut-Rella M, Ward E, Ryals J. In: Mechanisms of Defense Responses in Plants. Fritig B, Legrand M, editors. Dordrecht, The Netherlands: Kluwer Academic; 1993. pp. 422–432. [Google Scholar]
- 12.Bowling S A, Guo A, Cao H, Gordon A S, Klessig D F, Dong X. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietrich R A, Delany T P, Uknes S J, Ward E R, Ryals J A, Dangl J L. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg J T, Guo A, Klessig D F, Ausubel F M. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 15.Weymann K, Hunt M, Uknes S, Neuenschwander U, Lawton K, Steiner H-Y, Ryals J. Plant Cell. 1995;7:2013–2022. doi: 10.1105/tpc.7.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 17.Métraux J-P, Ahl-Goy P, Staub T, Speich J, Steinemann A, Ryals J, Ward E. In: Advances in Molecular Genetics of Plant-Microbe Interactions. Hennecke H, Verma D P S, editors. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic; 1991. pp. 432–439. [Google Scholar]
- 18.Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel K-H, Ostendorp M, Staub T, Ward E, Kessmann H, Ryals J. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Loon L C, Van Kammen A. Virology. 1970;40:199–211. doi: 10.1016/0042-6822(70)90395-8. [DOI] [PubMed] [Google Scholar]
- 20.Ward E R, Uknes S J, Williams S C, Dincher S S, Wiederhold D L, Alexander D C, Ahl-Goy P, Metraux J-P, Ryals J A. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yalpani N, Silverman P, Wilson T M A, Kleier D A, Raskin I. Plant Cell. 1991;3:809–818. doi: 10.1105/tpc.3.8.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broglie K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Mauvais C J, Broglie R. Science. 1991;254:1194–1197. doi: 10.1126/science.254.5035.1194. [DOI] [PubMed] [Google Scholar]
- 24.Alexander D, Glascock C, Pear J, Stinson J, Ahl-Goy P, Gut-Rella M, Goodman R M, Ryals J. In: Advances in Molecular Genetics of Plant-Microbe Interactions. Nester E W, Verma D P S, editors. Vol. 3. Dordrecht, The Netherlands: Kluwer Academic; 1993. pp. 527–533. [Google Scholar]
- 25.Alexander D, Goodman R M, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E, Ryals J. Proc Natl Acad Sci USA. 1993;90:7327–7331. doi: 10.1073/pnas.90.15.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D, Raghothama K G, Hasegawa P M, Bressan R A. Proc Natl Acad Sci USA. 1994;91:1888–1892. doi: 10.1073/pnas.91.5.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Q, Maher E A, Masoud S, Dixon R A, Lamb C J. Bio/Technology. 1994;12:807–812. [Google Scholar]
- 28.Delaney T, Friedrich L, Ryals J. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glazebrook J, Rogers E E, Ausubel F M. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah J, Tsui F, Klessig D F. Mol Plant--Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 31.Cao H, Glazebrook J, Clarke J D, Volko S, Dong X. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 32.Peters L L, John K M, Lu F M, Eicher E M, Higgins A, Yialamas M, Turtzo L C, Otsuka A J, Lux S E. J Cell Biol. 1995;130:314–330. doi: 10.1083/jcb.130.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner H-Y, Johnson J, Delaney T, Jesse T, Vos P, Uknes S. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 35.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Bowling S A, Clarke J D, Liu Y, Klessig D F, Dong X. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matzke M A, Matzke A J M. Plant Physiol. 1995;107:679–685. doi: 10.1104/pp.107.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokal R R, Rohlf F J. Biometry. 2nd Ed. New York: Freeman; 1981. [Google Scholar]
- 39.Jach G, Gornhardt B, Mundy J, Logemann J, Pinsdorf P, Leah R, Schell J, Maas C. Plant J. 1995;8:97–109. doi: 10.1046/j.1365-313x.1995.08010097.x. [DOI] [PubMed] [Google Scholar]
- 40.Jongedijk E, Tigelaar H, van Roekel J S C, Bres-Vloemans S A, Dekker I, van den Elzen P J M, Cornelissen B J C, Melchers L S. Euphytica. 1995;85:173–180. [Google Scholar]
- 41.Crute I R, Pink A C. Plant Cell. 1996;8:1747–1755. doi: 10.1105/tpc.8.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]