Abstract
Purpose
Diffuse large B-cell lymphoma (DLBCL) is curable in 60% of patients treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP). MYC translocations, with or without BCL2 translocations, have been associated with inferior survival in DLBCL. We investigated whether expression of MYC protein, with or without BCL2 protein expression, could risk-stratify patients at diagnosis.
Patients and Methods
We determined the correlation between presence of MYC and BCL2 proteins by immunohistochemistry (IHC) with survival in two independent cohorts of patients with DLBCL treated with R-CHOP. We further determined if MYC protein expression correlated with high MYC mRNA and/or presence of MYC translocation.
Results
In the training cohort (n = 167), MYC and BCL2 proteins were detected in 29% and 44% of patients, respectively. Concurrent expression (MYC positive/BCL2 positive) was present in 21% of patients. MYC protein correlated with presence of high MYC mRNA and MYC translocation (both P < .001), but the latter was less frequent (both 11%). MYC protein expression was only associated with inferior overall and progression-free survival when BCL2 protein was coexpressed (P < .001). Importantly, the poor prognostic effect of MYC positive/BCL2 positive was validated in an independent cohort of 140 patients with DLBCL and remained significant (P < .05) after adjusting for presence of high-risk features in a multivariable model that included elevated international prognostic index score, activated B-cell molecular subtype, and presence of concurrent MYC and BCL2 translocations.
Conclusion
Assessment of MYC and BCL2 expression by IHC represents a robust, rapid, and inexpensive approach to risk-stratify patients with DLBCL at diagnosis.
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin's lymphoma and is curable in more than 60% of patients treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP).1 The best available clinical tool to risk-stratify patients with DLBCL at diagnosis is the International Prognostic Index (IPI); however, there remains marked heterogeneity in clinical outcomes within each risk group, and IPI variables do not provide insight into the underlying tumor biology. Gene expression profiling (GEP) can group DLBCL into prognostically different molecular subtypes based on cell-of-origin (COO) gene signatures, where the activated B-cell (ABC) type is associated with inferior overall survival (OS) compared with the germinal center B-cell (GCB) type.2,3 GEP is not available in most clinical laboratories; thus, immunohistochemical algorithms, such as the one proposed by Choi et al,4 have been developed assigning a COO subtype based on the expression of COO-related proteins.5,6 Unfortunately, the accuracy with which these algorithms correctly classify COO subtype or predict OS is variable among laboratories.4,6,7
Alterations in oncogenes and tumor suppressor genes can drive the pathogenesis of DLBCL.8,9 Two such oncogenes are MYC and BCL2, key regulators of cellular proliferation and apoptosis, respectively.10,11 Deregulation of MYC and BCL2 can result from chromosomal translocation or gene amplification, but it may also occur by other mechanisms, such as transcriptional upregulation downstream of NFκB pathway signaling.10,12,13
The presence of MYC translocation and high MYC mRNA expression have recently been associated with poor OS in patients with DLBCL treated with R-CHOP, raising questions about optimal management of these high-risk patients.14–16 However, many of these patients with MYC-positive DLBCL also coexpress high levels of BCL2 protein, which may be a confounding factor in this disease, given that the presence of concurrent MYC and BCL2 translocations—so-called double hits (DHITs)—are associated with a dismal outcome despite high-dose chemotherapy.14–19 Fluorescence in situ hybridization (FISH) has been useful at identifying MYC translocations but has failed to identify altered MYC expression by other mechanisms and is not available in all clinical laboratories. Recently, a novel monoclonal antibody that targets the N-terminus of the MYC protein was shown to provide sensitive and specific staining of nuclear MYC in paraffin embedded tissue, including DLBCL.20–22
Herein, we demonstrate that MYC protein expression by IHC represents a rapid and inexpensive marker to identify MYC overexpression in DLBCL, including patients harboring MYC translocations, and that the prognostic significance of MYC deregulation in R-CHOP–treated patients with DLBCL depends on its coexpression with BCL2 protein.
PATIENTS AND METHODS
Patient Population
We used pretreatment tumor biopsies taken from two independent cohorts of patients diagnosed with de novo DLBCL according to WHO classification (2008) criteria.1 Patients were initially selected because they were linked to clinical information, including baseline characteristics and outcome, were HIV negative, and were treated with curative intent with R-CHOP therapy (with or without radiation). Ethical approval was granted by the research ethics board of each institution, in accordance with the Declaration of Helsinki.
The training set consisted of 167 patients who were further selected based on the availability of both fresh frozen and formalin-fixed paraffin-embedded (FFPE) tissue, provided from 10 international institutions. A consensus diagnosis of DLBCL was confirmed by a panel of expert pathologists. A subset of these patients were previously reported by Lenz et al3 (n = 158), Savage et al14 (n = 49), Iqbal et al23 (n = 167), and Choi et al4 (n = 68). DLBCL molecular subtype (GCB, ABC, and unclassifiable) and molecular Burkitt's lymphoma, if present in DLBCL patient cases with high MYC expression, were assigned by GEP according to previously published protocols.3,24,25
The validation set consisted of 140 patients from the British Columbia Cancer Agency (BCCA) who were selected based on availability of FFPE tissue only. Nine patients were included in the study by Savage et al14; COO for these patients was assigned by IHC according to the Choi et al4 algorithm.
IHC Analysis
Cores of FFPE tissue were used to construct tissue microarrays at each participating institution. In both data sets, staining was performed on the Ventana platform (Roche, Basel, Switzerland) using routine staining protocols and the following antibodies: BCL2, clone 124 (Dako, Carpinteria, CA); MYC, Y69 (Epitomics, Burlingame, CA) and Ki67 (Dako; Appendix, online only). Protein expression was recorded in 10% increments as the percentage of positive tumor cells. Training set data were analyzed using X-Tile statistical software (http://www.tissuearray.org/rimmlab) to determine the optimum survival cut points for dichotomizing expression of MYC protein (≥ 40%), BCL2 protein (≥ 50%), and Ki67 index (≥ 90%).26 These cut points correspond to the maximum χ2 value of the Mantel-Cox test for OS between groups above and below the cut-point threshold.26 These same cut points were then carried through to the validation set.
The reproducibility of BCL2 and MYC protein expression by IHC was determined by comparing the results obtained by three different pathologists. In the training set, BCL2 expression in approximately half of the patients was scored by three pathologists at the University of Nebraska Medical Center (W.C.C., P.N.M., D.D.W.), with no disagreement; thus, the latter half were evaluated by a single pathologist (P.N.M.).23 MYC and Ki67 expression were determined by a single pathologist (G.W.S.) at the BCCA. In the validation set, three pathologists at the BCCA (G.W.S., K.L.T., R.D.G.) scored patient cases for BCL2, MYC, and Ki67 protein expression using the same antibodies and thresholds described for the training set. Concordance was achieved when all three pathologists assigned the same positive or negative expression value for a patient. Discordant patient cases were evaluated by all three pathologists at a multiheaded microscope to reach a consensus score.
FISH
Translocations involving BCL2 and MYC were identified by interphase FISH in FFPE tissue in tissue microarrays from both data sets using commercial dual-color break-apart probes from Abbott Molecular (Abbott Park, IL) according to previously described methods.14 Patient cases with break-apart signals in > 5% of nuclei were considered positive for the presence of a translocation. In the training and validation cohorts, FISH experiments were successful in 167 and 123 patient cases using the MYC probe and in 157 and 120 using the BCL2 probe, respectively.
GEP
GEP was performed on fresh frozen tissue from the training set using Affymetrix HG U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA).3 MYC mRNA expression was determined using log2 normalized expression values of probe set 202431_s_at and dichotomized into low versus high expression using a cutoff threshold determined by X-Tile (high, > 9.4).26
Statistical Analysis
Progression-free survival (PFS; PFS event, progression or death resulting from any cause) and OS (OS event, death resulting from any cause) were measured from the date of pathologic diagnosis and estimated using the Kaplan-Meier method. Differences were assessed using the log-rank test. The Cox proportional hazards model was used to determine hazard ratios (HRs) and CIs and whether variables were independent of the IPI by multivariate analysis. The χ2 test, Fisher's exact test, and Pearson correlation were used to determine association and correlation between variables. These statistical analyses were performed using SPSS (version 20.0; SPSS, Chicago, IL) and STATA software (version 8.2; STATA, College Station, TX).
RESULTS
MYC and BCL2 Expression in DLBCL
Within the training set, MYC protein was expressed in one third of patients with DLBCL (n = 48 of 167; 29%). In contrast, MYC translocations and high MYC mRNA expression were detected in only 18 (11%) of 167 and 19 (11%) of 167 patients, respectively, with nine overlapping both groups. MYC protein expression was exclusively nuclear in all cases (Figs 1A to 1C) and was reproducible among pathologists (concordance 94%). Incidence of MYC protein expression (52 of 140; 37%) and MYC translocation (16 of 123; 13%) in the validation set was similar to that in the training set (Table 1). When looking at MYC protein expression in the combined data sets, there was a wide distribution of MYC protein expression across samples (Appendix Fig A1, online only). The percentage of cells expressing MYC protein was significantly greater in patient cases with translocations than without translocations (mean percent positive cells, 61%; range, 20 to 100 v 29%; range, 0 to 100, respectively; P < .001). Although MYC protein expression correlated with a high proliferation rate (r = 0.41; P < .001), MYC protein expression, at our predetermined 40% threshold, was more sensitive than Ki67 at identifying patients with MYC translocations (25 of 34; 74% v 7 of 34; 21%). Alterations involving MYC were present in both the ABC and GCB subtypes of DLBCL, whether defined by GEP (training set) or by IHC (validation set; Table 2).
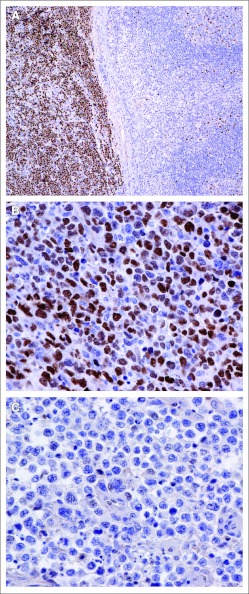
Fig 1.
Representative immunohistochemical analysis of MYC protein expression in diffuse large B-cell lymphoma (DLBCL). (A) Control: partial lymph node involvement by Burkitt's lymphoma (left) and follicular hyperplasia (right); there is bright nuclear staining of MYC protein in Burkitt's lymphoma cells, compared with < 5% of benign germinal center B cells stained for MYC (right). (B) MYC protein–positive DLBCL. (C) MYC protein–negative DLBCL.
Table 1.
Clinical Characteristics of Two Cohorts of Patients With DLBCL Treated With R-CHOP
| Characteristic | All Patients (N = 307) |
Training Cohort (n = 167) |
Validation Cohort (n = 140) |
P* | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||
| Age, years | |||||||
| Median | 63 | 62 | 65 | ||||
| Range | 17-92 | 17-92 | 19-90 | ||||
| > 60 | 179 of 307 | 58 | 85 of 167 | 51 | 94 of 140 | 67 | .004 |
| Ann Arbor stage > II | 162 of 296 | 55 | 81 of 161 | 50 | 81 of 135 | 60 | .107 |
| LDH > upper limit of normal | 126 of 297 | 42 | 67 of 167 | 40 | 59 of 130 | 45 | .364 |
| Extranodal sites ≥ two | 50 of 261 | 19 | 19 of 126 | 15 | 31 of 135 | 23 | .107 |
| ECOG PS > 1† | 78 of 282 | 28 | 36 of 148 | 24 | 42 of 134 | 31 | .190 |
| IPI score‡ | .021 | ||||||
| 0 or 1 | 114 of 288 | 40 | 72 of 154 | 47 | 42 of 134 | 31 | |
| 2 | 76 of 288 | 26 | 41 of 154 | 27 | 35 of 134 | 26 | |
| 3 | 54 of 288 | 19 | 22 of 154 | 14 | 32 of 134 | 24 | |
| 4 or 5 | 44 of 288 | 15 | 19 of 154 | 12 | 25 of 134 | 19 | |
| R-CHOP21 | 159 | 95 | 140 | 100 | |||
| R-CHOP14 | 8 | 5 | .759 | ||||
| Radiation | 37 | 22 | 29 | 21 | |||
| Cell-of-origin subtype | GEP | IHC§ | |||||
| ABC | 139 of 306 | 45 | 70 of 167 | 42 | 69 of 139 | 50 | .178 |
| GCB | 144 of 306 | 47 | 74 of 167 | 44 | 70 of 139 | 50 | |
| Unclassified‖ | 21 of 306 | 7 | 21 of 167 | 13 | |||
| Molecular Burkitt's lymphoma | 2 of 306 | 1 | 2 of 167 | 1 | |||
| MYC | |||||||
| MYC translocation | 34 of 290 | 12 | 18 of 167 | 11 | 16 of 123 | 13 | .561 |
| High mRNA expression | 19 of 167 | 11 | |||||
| High MYC protein expression | 100 of 307 | 33 | 48 of 167 | 29 | 52 of 140 | 37 | .094 |
| BCL2 | |||||||
| BCL2 translocation | 68 of 287 | 24 | 29 of 157 | 18 | 39 of 130 | 30 | .022 |
| High BCL2 protein expression | 160 of 304 | 53 | 73 of 164 | 44 | 87 of 140 | 62 | .002 |
| Median follow-up time, years¶ | 3.5 | 4.7 | .503 | ||||
| 5-year OS | 64 | 62 | |||||
NOTE. Bold font indicates significance.
Abbreviations: ABC, activated B-cell–like; DLBCL, diffuse large B-cell lymphoma; ECOG PS, Eastern Cooperative Oncology Group performance status; GCB, germinal center B-cell–like; GEP, gene expression profiling; IHC, immunohistochemistry; IPI, International Prognostic Index; LDH, lactate dehydrogenase; OS, overall survival; R-CHOP, rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone; R-CHOP14, R-CHOP 14-day cycle; R-CHOP21, R-CHOP 21-day cycle.
P values are for comparison between training and validation cohorts.
ECOG PS ranges from 0 to 4, where higher score indicates greater degree of impairment.
IPI score ranges from 0 to 5, with 0 indicating absence of prognostic factors and 5 indicating presence of all prognostic factors; IPI score was not calculated if more than one variable was unavailable.
IHC algorithm according to Choi et al.4
Unclassified indicates gene expression profiles are intermediate between ABC and GCB.
Median follow-up time for living patients was 3.5 years (range, 0.52 to 11.3 years) and 4.7 years (range, 1.0 to 8.0 years) for training and validation sets, respectively.
Table 2.
Differences in MYC and BCL2 Alterations According to Cell-of-Origin Subtype in Two Cohorts of Patients With DLBCL
| Cohort | ABC |
GCB |
Unclassified* |
mBL |
P† | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | ||
| MYC | |||||||||
| MYC translocation | |||||||||
| Training‡ | 6 of 70 | 9 | 10 of 74 | 14 | 1 of 21 | 5 | 1 of 2 | 50 | .346 |
| Validation§ | 6 of 62 | 10 | 10 of 61 | 16 | .268 | ||||
| High mRNA expression | |||||||||
| Training | 10 of 70 | 14 | 5 of 74 | 7 | 2 of 21 | 10 | 2 of 2 | 100 | .139 |
| MYC protein expression | |||||||||
| Training | 24 of 70 | 34 | 17 of 74 | 23 | 6 of 21 | 29 | 1 of 2 | 50 | .399 |
| Validation | 35 of 69 | 51 | 17 of 70 | 24 | .001 | ||||
| BCL2 | |||||||||
| BCL2 translocation | |||||||||
| Training | 4 of 70 | 6 | 23 of 74 | 31 | 2 of 11 | 10 | 0 | 0 | < .001 |
| Validation | 11 of 65 | 17 | 28 of 65 | 43 | .001 | ||||
| BCL2 protein expression | |||||||||
| Training | 44 of 70 | 63 | 22 of 74 | 30 | 7 of 21 | 35 | 0 | 0 | < .001 |
| Validation | 53 of 69 | 77 | 34 of 70 | 49 | .001 | ||||
| DHIT‖¶ | 5 of 14 | 36 | 9 of 14 | 64 | |||||
| MYC and BCL2 protein expression¶ | 42 of 55 | 76 | 13 of 55 | 24 | < .001 | ||||
| Absence of concurrent MYC and BCL2 protein expression or DHIT | 92 of 235 | 39 | 143 of 235 | 61 | |||||
NOTE. Bold font indicates significance.
Abbreviations: ABC, activated B-cell lymphoma; DHIT, double-hit lymphoma; DLBCL, diffuse large B-cell lymphoma; GCB, germinal center B-cell lymphoma; GEP, gene expression profiling; mBL, molecular Burkitt's lymphoma.
Unclassified indicates gene expression profiles are intermediate between ABC and GCB.
All P values refer to comparison between GCB and ABC subtypes.
In training set, cell-of-origin distinctions were assigned according to GEP and signatures previously described by Wright et al25 and Dave et al.24
In validation set, cell-of-origin distinctions were assigned according to immunohistochemical algorithm reported by Choi et al.4
DHIT refers to presence of concurrent MYC and BCL2 translocations.
Frequencies and P values refer to differences between molecular subtypes in combined data sets (training and validation), where patients were stratified according to presence of DHIT or presence of concurrent MYC and BCL2 proteins, which excluded DHITs. Cell-of-origin data were not available in one of 307 patient cases, and BCL2 protein was not available in three of 307 patient cases, one of which was DHIT.
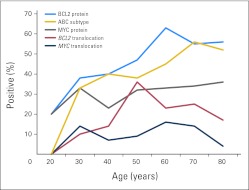
BCL2 protein expression was associated with the ABC subtype, and BCL2 translocation was associated with the GCB subtype (P = .001; Table 2). Both were more common in the validation cohort, which included older patients (P < .05; Table 1). Indeed, there was an age-related increase in the incidence of BCL2 protein expression, BCL2 translocation, and ABC subtype, but not MYC alterations (Appendix Fig A2, online only). Importantly, BCL2 protein expression by IHC was reproducible among pathologists, with a concordance of more than 91%.
Factors Associated With Clinical Outcome
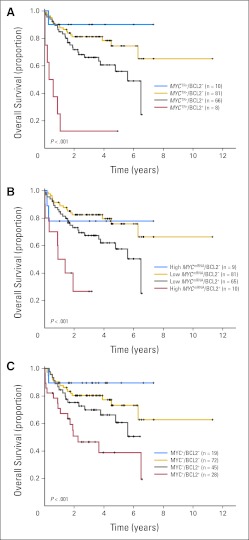
In univariate analysis, MYC protein expression and MYC translocation, alone, were not significantly associated with survival in the training set (P > .05). Factors that were associated with OS and PFS included elevated IPI score, ABC subtype, high MYC mRNA expression, and BCL2 protein expression (Appendix Table A1, online only). Next, we determined if BCL2 protein expression affected prognosis of MYC deregulation and whether there was an interaction between MYC and BCL2 variables. Presence of MYC translocation, high MYC mRNA expression, or high MYC protein expression, hereafter referred to as MYC positive, was only associated with inferior OS and PFS when BCL2 protein was coexpressed (BCL2 positive; Figs 2A to 2C; P < .001). Indeed, the interaction between these variables suggested that the negative prognostic impact of MYC and BCL2 was amplified when both variables were present, an effect that was consistent across platforms (protein, mRNA, or translocation; Appendix Table A1, online only). For example, the OS HR for coexpression of MYC-positive/BCL2-positive proteins was 3.2, compared with 0.47 for the MYC-positive/BCL2-negative group and 1.6 for the MYC-negative/BCL2-positive group (P = .001, P = .11, and P = .17, respectively). In the validation cohort, coexpression of MYC-positive/BCL2-positive proteins was also associated with inferior OS (HR, 1.6; P = .17), but there was no difference in outcome in the remaining patients (Appendix Fig A3, online only). Samples were then stratified into a high-risk group, defined by the presence of MYC-positive/BCL2-positive protein coexpression, and low-risk group, comprising the remaining patients. Presence of MYC-positive/BCL2-positive protein expression was associated with significantly inferior OS and PFS in both training and validation DLBCL cohorts (Figs 3A and 3B; P < .05). Importantly, the negative prognostic impact of MYC-positive/BCL2-positive status on OS persisted even after adjusting for IPI and COO in a Cox multivariant model (P ≤ .05; Appendix Table A2, online only).
Fig 2.
Overall survival (OS) of patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone based on alterations in MYC and BCL2 in the training set. Kaplan-Meier curves represent OS according to (A) presence of MYC translocation (TR) and BCL2 protein expression (BCL2+), (B) presence of high MYC mRNA expression and BCL2 protein expression, and (C) presence of MYC and BCL2 protein expression. Log-rank P < .001 for both OS and progression-free survival. Total evaluable patients for the analyses: (A) n = 165, (B) n = 165, and (C) n = 164.
Fig 3.
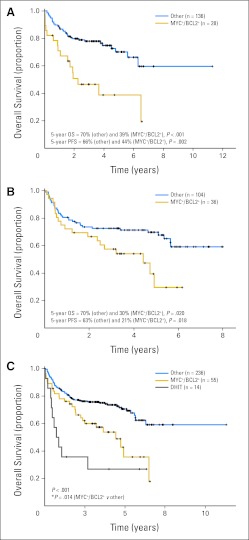
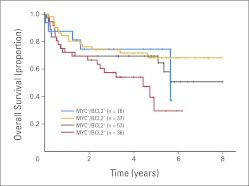
Overall (OS) and progression-free survival (PFS) of patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone according to presence of concurrent expression of MYC and BCL2 proteins (MYC+/BCL2+) and/or presence of concurrent MYC and BCL2 translocations. Kaplan-Meier curves represent OS according to presence of MYC+/BCL2+ in (A) the training cohort (n = 164) and (B) validation cohort (n = 140). (C) Both training and validation cohorts were combined because of the low frequency of double hits (DHITs). Patients were stratified according to presence of DHIT or MYC+/BCL2+ excluding DHITs. Four DHIT patient cases had no MYC protein expression; one had a missing value for BCL2 protein expression.
Concurrent Translocation of MYC and BCL2
There were 14 patient cases in the combined data set (14 of 290; 5%) fitting the diagnostic criteria of DLBCL but with concurrent translocation of MYC and BCL2 (ie, DHIT). Given this low incidence, both data sets were pooled for subsequent analyses. These patients had more adverse clinical risk factors, including higher levels of lactate dehydrogenase, worse performance status, and higher IPI scores than other patients with DLBCL (P < .05; Appendix Table A3, online only). DHIT samples expressed BCL2 protein (13 of 13); 10 (71%) of 14 samples expressed MYC protein, but only one (7%) of 14 had Ki67 > 90%. As expected, the clinical outcome of patients with DLBCL with DHIT status treated with R-CHOP in this study was extremely poor, with 5-year OS and PFS rates of 27% and 18%, respectively (Fig 3C). However, the outcome of the remaining MYC-positive/BCL2-positive patients (excluding those with DHIT status) was also poor compared with outcome among those who did not express MYC-positive/BCL2-positive proteins (Fig 3C; 5-year OS and PFS, 36% and 32% v 71% and 65%, respectively; P < .05). Interestingly, two of the three long-term survivors in the DHIT group had < 40% cells in their biopsy expressing MYC protein, suggesting that this may have prognostic relevance, analogous to what has been previously reported for BCL2 protein expression in DHITs,19 but too few patient cases preclude any meaningful conclusions.
DISCUSSION
Overexpression of MYC protein occurs in 33% of patient cases with DLBCL, of which only one third can be explained by the presence of MYC translocation. That MYC protein is expressed in both the GCB and ABC subtypes suggests an underlying biologic pathway independent of COO subtype. In addition to promoting cell-cycle progression, MYC plays an important role in metabolism, protein synthesis, stem-cell renewal, and mRNA regulation and can induce apoptosis by increasing p53 expression or amplifying apoptotic signaling pathways.27–30 Given this broad range of biologic activity, it is not surprising that deregulation of MYC is oncogenic.10 MYC protein expression has also been found to occur as a consequence of other genomic events (eg, inactivating p53), increased protein stability, activation of upstream signaling pathways (eg, NFκB), desensitization to inhibitory cytokines, and loss of host immunity.20,31,32 Given that many MYC-positive samples did not have a high proliferation rate in this study, these nonproliferative functions of MYC may contribute to the clinical and biologic attributes of DLBCL, possibly by inducing further DNA damage and genomic instability.33 Thus, MYC protein expression by IHC in DLBCL may represent a final integrator of all or most of the mechanisms that underlie MYC deregulation.
We found MYC and BCL2 IHC interpretation to be robust, reproducible, and largely unaffected by the vagaries introduced by differing fixation techniques used by the 10 institutions participating in this study. The cutoff scores for many biomarkers have often been arbitrary; in the case of BCL2, they have ranged from > 10% to > 75%, making comparison among studies difficult.34,35 As such, we defined our thresholds based on rational statistical methods in accordance with recommended guidelines.36 Our BCL2 threshold compares favorably with that defined by Tzankov et al,34 who used receiver operator curves to define optimal cutoff scores, but it is lower than the ≥ 75% threshold reported by Salles et al,35 who used a statistical approach similar to that of our study. The latter study investigated IHC markers in 2,451 DLBCL samples within the Lunenburg Lymphoma Biomarker Consortium, many of which were derived from young patients enrolled onto clinical trials, including studies evaluating R-CHOP administered every 14 days.35 These data suggest that age and treatment may be important factors to consider when defining the optimal thresholds of biomarkers in DLBCL. Indeed, we and others have shown that there are age-related changes in tumor biology and that treatment, such as rituximab, can influence the prognostic impact of several biomarkers in DLBCL, including BCL2.16,37–42
We also demonstrated that BCL2 protein expression is the main determinant of clinical outcome in MYC-positive DLBCL. This biologic effect was robust across different platforms (IHC, GEP, and FISH) and was validated in a second independent cohort, which was representative of a population-based registry, suggesting that this biomarker may be relevant even in older patients.16 Furthermore, coexpression of MYC and BCL2 proteins by IHC remained significant after adjusting for IPI, COO, and presence of DHIT. This is in keeping with the known biologic function of BCL2, which is a potent inhibitor of apoptosis and has been clearly shown to mediate chemotherapy resistance in MYC-positive lymphoma murine models.43 Although previous studies have demonstrated a negative prognostic effect of MYC translocation in DLBCL, a majority of patients' samples were BCL2 protein positive,14,16 and it is unknown whether those with MYC translocations, but lacking BCL2 expression, would share a similar fate. This is an important question if patients with MYC-positive DLBCL are to be considered for more aggressive therapy. Unfortunately, there are no data to support the use of any one regimen for the treatment of MYC-positive/BCL2-positive DLBCL at this time. The results from the MRC/NCRI (Medical Research Council/National Cancer Research Institute) LY10 trial and a recent report from Snurderl et al18 did not demonstrate a survival advantage in treating patients with DHIT lymphoma with high-dose regimens, most of which would not be well tolerated in older patients.44,18 The probable role that BCL2 has in this disease suggests that MYC-positive/BCL2-positive tumors may be amenable to chemotherapy regimens that include drugs targeting BCL2, such as BH3 mimetics, a strategy that has been shown to cure a subset of murine MYC-positive/BCL2-positive lymphomas.45–47
The primary aim of this study was to determine the role of MYC protein as a prognostic biomarker in DLBCL, not as a potential screening test to identify patients harboring MYC translocation. Our results confirm those of two recent studies demonstrating the excellent reproducibility of MYC protein by IHC among pathologists, the high correlation with presence of MYC translocation, and the weak correlation with Ki67. MYC protein by IHC in our study had a sensitivity of 74%, which was lower than the 100% reported by the other studies that evaluated it as a screening test for identifying patients with MYC translocations. Our study used a lower threshold (40% v 50%21 and 70%22), which was set based on the relationship between MYC protein and survival, not the presence of a translocation. Furthermore, the study by Kluk et al21 included too few patient cases to meaningfully address this issue (n = five translocations of 56 samples of de novo DLBCL), and that by Green et al22 included patient cases of Burkitt's lymphoma, which raised the baseline incidence of MYC translocations to 15%, a level higher than that observed in de novo DLBCL. Taken together, patients with MYC translocation are likely to express MYC protein. More importantly, our data suggest that MYC protein expression is more common than MYC translocation, and the MYC-positive/BCL2-positive immunophenotype is itself a powerful predictor of survival, independent of presence of MYC translocation.
In conclusion, this study has identified a group of clinically high-risk patients with DLBCL who coexpress MYC and BCL2 proteins, a clinical scenario that is more common than patients with lymphomas harboring the DHIT genotype (18% v 5%, respectively). Importantly, these biomarkers retained their prognostic significance in an independent cohort that consisted of patients from a population-based registry. Unlike with FISH, assessment of MYC and BCL2 protein expression by IHC represents a rapid, inexpensive, and reproducible technique that could be adopted by pathologists in most clinical centers. These promising results need to be validated prospectively in larger cohorts, using standardized staining and scoring methodologies, before implementation as prognostic biomarkers in clinical practice. Thus, MYC and BCL2 represent relevant biomarkers that should be tested in the context of clinical trials such that more effective therapies can be offered to these high-risk patients.
Appendix
Immunohistochemistry Protocol
Immunohistochemistry (IHC) for MYC was performed using the fully automated Ventana Discovery XT platform (Roche, Basel, Switzerland) at the British Columbia Cancer Agency. Slides containing formalin-fixed paraffin-embedded tissue sections 5-mm thick were heated to remove the paraffin from the tissue. Antigen retrieval was performed by heating the slides to 100°C in a Tris-based buffer solution (pH 8.0) called Ventana CC1 standard solution (Roche) for 28 minutes and cooled for 8 minutes. Slides were incubated with the primary antibody (cMYC clone Y69; Epitomics, Burlingame, CA) at a 1:50 dilution for 2 hours (no heat). After washing, slides were incubated for 16 minutes with the Ventana Ultramap rabbit secondary antibody (Roche). Unlike the protocol used by Ruzinova et al (Ruzinova MB, Caron T, Rodig SJ: Am J Surg Pathol 34:882-891, 2010), our method is fully automated, uses different slide preparation and antigen retrieval procedures, has different incubation times, and uses a less sensitive secondary antibody. IHC for BCL2 was performed using standard automated methods (Ventana; Roche) in two institutions—the University of Nebraska Medical Center (training) and British Columbia Cancer Agency—using two commercial antibodies (clones 124 and E17), as previously described (Savage KJ, Johnson NA, Ben-Neriah S, et al: Blood 114:3533-3537, 2009; Iqbal J, Meyer PN, Smith L, et al: Clin Cancer Res 17:7785-7795, 2011).
IHC Scoring
BCL2, MYC, and Ki67 protein expression were considered to be present if there was medium to bright protein staining within cells and considered absent if there was weak or no protein staining within cells. The percentage of positive cells was recorded in 10% increments.
IHC Thresholds
The optimal cutoff thresholds for deeming a patient case positive or negative were determined using X-Tile statistical software (http://www.tissuearray.org/rimmlab; Camp RL, Dolled-Filhart M, Rimm DL: Clin Cancer Res 10:7252-7259, 2004). X-Tile is a free software application that determines the optimal threshold based on the association of the biomarker with the optimal χ2 value for overall survival (OS). MYC protein expression alone was not significantly associated with OS or progression-free survival (P > .05) at any cut point. The distribution of MYC IHC data suggested a cut point at 40%, which also corresponded to the highest χ2 value by X-Tile. The distribution of MYC IHC in the validation cohort was similar to that in the training cohort, and using X-Tile, the optimal threshold was independently determined to be 40% (highest χ2 using OS as variable, 3.5; lowest P = .061). The threshold of 50% for BCL2 expression was determined to be most significantly associated with OS (X-Tile: χ2, 2; P = 1 at ≥ 30% cutoff v χ2, 9.7; P = .04 at ≥ 50% cutoff). This threshold was also confirmed to be the most significantly associated with survival using a different method described by Iqbal et al (Iqbal J, Meyer PN, Smith L, et al: Clin Cancer Res 17:7785-7795, 2011). Finally, no cutoff value could be obtained for Ki67 in X-Tile, because no threshold value was associated with OS or progression-free survival. Thus, both 80% and 90% threshold values were tested, because these have been used previously, but a Ki67 of 90% was most significantly associated with presence of MYC protein; thus, a 90% threshold was retained in the final analysis (Savage KJ, Johnson NA, Ben-Neriah S, et al: Blood 114:3533-3537, 2009; Miller TP, Grogan TM, Dahlberg S, et al: Blood 83:1460-1466, 1994).
Gene Expression Profiling
CEL files were normalized using robust multichip analysis, and samples were assigned a molecular subtype (molecular BL, germinal center B-cell type [GCB], activated B-cell type [ABC], or unclassifiable), according to the expression of classifier genes in the study by Dave et al (Dave SS, Fu K, Wright GW, et al: N Engl J Med 354:2431-2442, 2006; Irizarry RA, Bolstad BM, Collin F, et al: Nucleic Acids Res 31:e15, 2003). Unclassifiable refers to patient cases intermediating between ABC and GCB (ie, probability of being called ABC or GCB based on Bayesian classifier < 0.9; Wright G, Tan B, Rosenwald A, et al: Proc Natl Acad Sci U S A 100:9991-9996, 2003). The MYC probe set ID 202431_s_at was chosen because it was the only probe in the array situated in an exon of the MYC gene.
Table A1.
HRs for OS and PFS for Different Variables in Both Cohorts of Patients With DLBCL
| Variable | Training (n = 167) |
Validation (n = 140) |
GCB (n = 164) |
ABC (n = 139) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| IPI | ||||||||||||
| OS | 2.2 | 1.5 to 3.2 | < .001 | 1.7 | 1.1 to 2.4 | .005 | 2.3 | 1.6 to 3.4 | < .001 | 1.5 | 1.1 to 2.2 | .02 |
| PFS | 2.2 | 1.5 to 3.1 | < .001 | 1.7 | 1.2 to 2.5 | .005 | < .001 | .03 | ||||
| ABC subtype | ||||||||||||
| OS | 2.6 | 1.5 to 4.6 | < .001 | .18 | ||||||||
| PFS | 3.1 | 1.8 to 5.2 | < .001 | .06 | ||||||||
| MYC TR+ | ||||||||||||
| OS | .07 | .19 | .43 | 3.5 | 1.7 to 7.2 | < .001 | ||||||
| PFS | .06 | .19 | .40 | 3.8 | 1.9 to 7.6 | < .001 | ||||||
| High MYC mRNA | ||||||||||||
| OS | 2.2 | 1.1 to 4.6 | .03 | .08 | .69 | 2.8 | 1.2 to 6.5 | .02 | ||||
| PFS | .10 | .16 | .24 | 1.8 | 1.1 to 3.0 | .002 | ||||||
| MYC protein+ | ||||||||||||
| OS | 1.6 | 0.9 to 2.9 | .11 | 1.6 | 0.9 to 2.8 | .86 | .90 | .02* | ||||
| PFS | .33 | .37 | .65 | .10 | ||||||||
| Ki67 > 90%† | ||||||||||||
| OS | .16 | .73 | .23 | .28 | ||||||||
| PFS | .38 | .35 | .32 | .78 | ||||||||
| BCL2 protein+ | ||||||||||||
| OS | 2.4 | 1.3 to 4.2 | .002 | .57 | .72 | .11 | ||||||
| PFS | 2.4 | 1.4 to 4.2 | .001 | .30 | .36 | .08 | ||||||
| BCL2 TR+ | ||||||||||||
| OS | .89 | .17 | .61 | .10 | ||||||||
| PFS | .74 | .07 | .26 | .04* | ||||||||
| DHIT‡ | ||||||||||||
| OS | 12.5 | 4.6 to 33.4 | < .001 | .49 | 4.7 | 2.0 to 11.3 | < .001 | 1.9 | 1.0 to 3.8 | .05 | ||
| PFS | 10.4 | 3.9 to 27.5 | < .001 | .32 | 4.1 | 1.7 to 9.6 | .001 | .001 | ||||
| MYC TR and BCL2 protein+ | ||||||||||||
| OS | 1.3 | 1.1 to 1.6 | < .001 | .11 | .006* | 2.7 | 1.0 to 7.6 | .001* | ||||
| PFS | 1.3 | 1.1 to 1.5 | < .001 | .12 | .001* | < .001* | ||||||
| MYC mRNA and BCL2 protein+ | ||||||||||||
| OS | 1.3 | 1.1 to 1.6 | < .001 | .006* | .62 | |||||||
| PFS | 1.3 | 1.1 to 1.6 | < .001 | .01* | .29 | |||||||
| MYC protein and BCL2 protein+ | ||||||||||||
| OS | 1.3 | 1.0 to 1.6 | .001 | .12 | .06 | |||||||
| PFS | 1.3 | 1.1 to 1.6 | .001 | .19 | .13 | |||||||
| Cox model§ | ||||||||||||
| MYC translocation | ||||||||||||
| MYC−/BCL2+ | ||||||||||||
| OS | 1.9 | 1.1 to 3.5 | .03 | 1.0 | 0.5 to 1.9 | .99 | ||||||
| PFS | 1.9 | 1.1 to 3.4 | .02 | 1.1 | 0.6 to 2.1 | .72 | ||||||
| MYC+/BCL2− | ||||||||||||
| OS | 0.5 | 0.1 to 3.5 | .45 | 1.1 | 0.3 to 5.0 | .87 | ||||||
| PFS | 0.4 | 0.1 to 2.8 | .35 | 1.0 | 0.2 to 4.2 | .96 | ||||||
| MYC+/BCL2− | ||||||||||||
| OS | 11.0 | 4.5 to 26.8 | < .001 | 2.0 | 0.8 to 5.1 | .17 | ||||||
| PFS | 12.5 | 5.3 to 29.2 | < .001 | 2.2 | 0.9 to 5.4 | .08 | ||||||
| MYC mRNA | ||||||||||||
| MYC−/BCL2+ | ||||||||||||
| OS | 2.0 | 1.1 to 3.8 | .03 | |||||||||
| PFS | 2.1 | 1.2 to 3.7 | .01 | |||||||||
| MYC+/BCL2− | ||||||||||||
| OS | 1.2 | 0.3 to 5.4 | .77 | |||||||||
| PFS | 1.0 | 0.2 to 4.3 | .99 | |||||||||
| MYC+/BCL2+ | ||||||||||||
| OS | 6.6 | 2.7 to 16.2 | < .001 | |||||||||
| PFS | 5.3 | 2.2 to 12.7 | < .001 | |||||||||
| MYC protein | ||||||||||||
| MYC−/BCL2+ | ||||||||||||
| OS | 1.6 | 0.8 to 3.2 | .17 | 0.7 | 0.4 to 1.5 | .40 | ||||||
| PFS | 1.8 | 1.0 to 3.3 | .06 | 0.9 | 0.5 to 1.7 | .72 | ||||||
| MYC+/BCL2− | ||||||||||||
| OS | 0.47 | 0.1 to 2.1 | .11 | 0.8 | 0.3 to 2.3 | .73 | ||||||
| PFS | 0.38 | 0.1 to 1.6 | .19 | 0.7 | 0.2 to 2.0 | .51 | ||||||
| MYC+/BCL2+ | ||||||||||||
| OS | 3.2 | 1.6 to 6.4 | .001 | 1.6 | 0.8 to 3.2 | .17 | ||||||
| PFS | 2.8 | 1.4 to 5.4 | .002 | 1.7 | 0.9 to 3.2 | .12 | ||||||
| MYC+/BCL2+ proteins (dichotomized) | ||||||||||||
| OS | 2.9 | 1.6 to 5.3 | < .001 | 1.9 | 1.1 to 3.4 | .02 | ||||||
| PFS | 2.4 | 1.3 to 4.2 | .003 | 1.9 | 1.1 to 3.1 | .02 | ||||||
NOTE. HRs and 95% CIs were calculated using Cox regression model; P values are derived from log-rank test, except in Cox model evaluating interaction between variables. Cell of origin was defined by gene expression profiling and immunohistochemistry.
Abbreviations: ABC, activated B-cell lymphoma; DHIT, double-hit lymphoma; DLBCL, diffuse large B-cell lymphoma; GCB, germinal center B-cell lymphoma; HR, hazard ratio; IPI, International Prognostic Index; OS, overall survival; PFS, progression-free survival; TR, translocation.
Patient cases where P values for OS and PFS are > .05 by Cox regression but < .05 using log-rank test.
Ki67 protein > 80% was also tested (P > .05).
DHIT refers to presence of concurrent MYC and BCL2 translocations.
Controlling for interaction between variables (reference, MYC−/BCL2−).
Table A2.
HRs for OS and PFS of MYC and BCL2 Proteins in Two Multivariable Models
| Variable | Training (n = 167) |
Validation (n = 140) |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Model one | ||||||
| MYC−/BCL2+ | ||||||
| OS | 1.4 | 0.7 to 2.8 | .35 | 0.9 | 0.7 to 1.8 | .71 |
| PFS | 1.5 | 0.8 to 2.8 | .21 | 1.1 | 0.6 to 2.2 | .78 |
| MYC+/BCL2− | ||||||
| OS | 0.5 | 0.1 to 1.9 | .26 | 0.9 | 0.3 to 2.6 | .89 |
| PFS | 0.4 | 0.8 to 1.5 | .16 | 0.8 | 0.3 to 2.1 | .67 |
| MYC+/BCL2+ | ||||||
| OS | 2.1 | 1.0 to 4.3 | .04 | 1.7 | 0.8 to 3.3 | .16 |
| PFS | 1.8 | 0.9 to 3.5 | .11 | 1.7 | 0.9 to 3.2 | .13 |
| IPI | ||||||
| OS | 2.0 | 1.3 to 3.1 | .001 | 1.6 | 1.1 to 2.3 | .02 |
| PFS | 2.1 | 1.4 to 3.2 | < .001 | 1.7 | 1.1 to 2.4 | .004 |
| ABC subtype | ||||||
| OS | 1.9 | 1.0 to 3.5 | .04 | 1.0 | 0.9 to 1.1 | .66 |
| PFS | 2.2 | 1.3 to 4.0 | .005 | 1.0 | 0.9 to 1.1 | .62 |
| Model two | ||||||
| MYC+/BCL2+ proteins dichotomized | ||||||
| OS | 1.9 | 1.0 to 3.6 | .036 | 1.8 | 1.0 to 3.2 | .048 |
| PFS | 1.6 | 0.9 to 2.9 | .13 | 1.6 | 1.0 to 2.8 | .067 |
| IPI | ||||||
| OS | 2.0 | 1.3 to 3.0 | .001 | 1.6 | 1.1 to 2.3 | .013 |
| PFS | 2.0 | 1.3 to 2.9 | .001 | 1.7 | 1.2 to 2.3 | .003 |
| ABC subtype | ||||||
| OS | 2.1 | 1.1 to 3.8 | .017 | 1.0 | 0.9 to 1.1 | .67 |
| PFS | 2.6 | 1.5 to 4.5 | .001 | 1.0 | 0.9 to 1.1 | .62 |
NOTE. Reference group in first model includes patient cases lacking both MYC and BCL2 protein expression. In second model, patient cases are dichotomized into MYC+/BCL2+ coexpression versus all the other cases. Analysis excluded three patient cases where BCL2 protein expression was not available in training set.
Abbreviations: ABC, activated B-cell lymphoma; HR; hazard ratio; IPI, International Prognostic Index; OS, overall survival; PFS, progression-free survival.
Defined by gene expression profiling and immunohistochemistry.
Table A3.
Baseline Characteristics of Combined Cohorts of DLBCL Based on Concurrent Expression of MYC and BCL2 Proteins or Presence of Concurrent MYC and BCL2 Translocation
| Characteristic | Other (n = 236) |
MYC+/BCL2+ (n = 55) |
DHIT (n = 14)* |
P | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||
| Age > 60 years | 130 of 236 | 55 | 38 of 55 | 69 | 9 of 14 | 64 | .140 |
| Ann Arbor stage > II | 117 of 229 | 51 | 33 of 52 | 63 | 10 of 13 | 77 | .140 |
| LDH > upper limit of normal | 83 of 228 | 36 | 29 of 53 | 55 | 12 of 14 | 86 | < .001 |
| Extranodal sites ≥ two | 35 of 200 | 18 | 10 of 47 | 21 | 4 of 14 | 29 | .357 |
| ECOG PS > 1 | 52 of 217 | 24 | 17 of 50 | 34 | 8 of 13 | 62 | .007 |
| IPI score | .005 | ||||||
| 0 or 1 | 101 of 222 | 45 | 12 of 51 | 24 | 1 of 13 | 8 | |
| 2 | 57 of 222 | 26 | 16 of 51 | 31 | 2 of 13 | 15 | |
| 3 | 36 of 222 | 16 | 13 of 51 | 25 | 5 of 13 | 38 | |
| 4 or 5 | 28 of 222 | 13 | 10 of 51 | 20 | 5 of 13 | 38 | |
| Cell-of-origin subtype | |||||||
| ABC | 92 of 235 | 39 | 13 of 55 | 24 | 5 of 14 | 36 | < .001 |
| GCB | 143 of 235 | 61 | 42 of 55 | 76 | 9 of 14 | 64 | |
| MYC protein expression | 32 of 236 | 14 | 55 of 55 | 100 | 10 of 14 | 71 | < .001 |
| BCL2 protein expression | 92 of 236 | 39 | 55 of 55 | 100 | 13 of 13 | 1000 | < .001 |
| Ki67 > 90% | 27 of 229 | 12 | 19 of 54 | 35 | 1 of 14 | 7 | < .001 |
| Clinical Outcome | Other (n = 236) |
MYC+/BCL2+ (n = 55) |
DHIT (n = 14) |
P | |||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | ||
| 5-year survival, % | |||||||
| OS | 71 | 36 | 27 | < .001 | |||
| PFS | 65 | 32 | 18 | < .001 | |||
| Univariate model | |||||||
| OS | 2.1 | 1.3 to 3.1 | 3.95 | 2.0 to 7.7 | .004† | ||
| PFS | 1.9 | 1.2 to 2.8 | 4.1 | 2.1 to 7.8 | .004† | ||
| Multivariable Cox model‡ | |||||||
| OS | 1.8 | 1.1 to 2.8 | 2.7 | 1.3 to 5.3 | .01† | ||
| PFS | 1.7 | 1.1 to 2.5 | 2.8 | 1.4 to 5.3 | .02† | ||
| IPI | |||||||
| OS | 1.7 | 1.3 to 2.3 | < .001 | ||||
| PFS | 1.8 | 1.4 to 2.3 | < .001 | ||||
| ABC | |||||||
| OS | 1.0 | 0.9 to 1.0 | .74 | ||||
| PFS | 1.0 | 0.9 to 1.0 | .73 | ||||
NOTE. In this table, both cohorts were combined, and patients were stratified according to presence of concurrent MYC and BCL2 translocations (DHIT) or presence of MYC and BCL2 proteins (excluding DHIT). Five patient cases with DHIT either did not express MYC protein (n = 4 of 14) or could not be evaluated for BCL2 protein expression because of technical failure (n = 1 of 14). Other refers to DLBCL patient cases without concurrent MYC and BCL2 protein expression (MYC+/BCL2+) and without concurrent MYC and BCL2 translocations. Molecular subtypes were defined by gene expression profiling and the immunohistochemical algorithm described by Choi et al.4 Total number of evaluable patient cases was 305. Ki67 expression in Other and DHIT was not significantly different (P > .05). Bold font indicates significance.
Abbreviations: ABC, activated B-cell–like; DHIT, double-hit lymphoma; DLBCL, diffuse large B-cell lymphoma; ECOG PS, Eastern Cooperative Oncology Group performance status; GCB, germinal center B-cell–like; HR, hazard ratio; IPI, International Prognostic Index; LDH, lactate dehydrogenase; OS, overall survival; PFS, progression-free survival.
DHIT refers to presence of concurrent MYC and BCL2 translocations.
P values are for MYC+/BCL2+ protein; P values for DHIT were below listed values.
Four variables were included in the multivariable model: IPI, ABC subtype, MYC+/BCL2+, and DHIT. The hazard ratios and CIs derived from this model for the IPI and ABC are listed under Other.
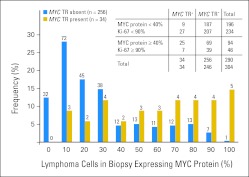
Fig A1.
Distribution of MYC protein expression in diffuse large B-cell lymphoma (DLBCL) according to presence of MYC translocation. Blue bars represent the proportion of patients with DLBCL expressing MYC protein when MYC translocation is absent; gold bars represent the proportion expressing MYC protein when MYC translocation is present.
Fig A2.
Age-related incidence of MYC and BCL2 deregulation in diffuse large B-cell lymphoma. ABC, activated B cell–like molecular subtype.
Fig A3.
Overall survival (OS) of patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in the validation cohort based on presence of MYC and BCL2 protein expression. Log-rank P > .05 for both OS and progression-free survival.
Footnotes
Processed as a Rapid Communication manuscript. See accompanying editorial on page 3433 and articles on pages 3439 and 3460; listen to the podcast by Dr Gopal at www.jco.org/podcasts
Supported by Awards No. 019005 from the NCIC and ST-PDF-01793 from the Michael Smith Foundation for Health Research for research fellow of the Terry Fox Foundation and by an award from the “Fonds de Recherche en Santé du Québec (N.A.J.); by the NCIC and Grant No. 019001 from the Terry Fox Foundation (J.M.C., R.D.G.); and by Grant No. TGT-53912 from the Terry Fox Foundation Strategic Health Research Training Program in Cancer Research at Canadian Institutes of Health Research (D.W.S., K.L.T.). Members of the Lymphoma/Leukemia Molecular Profiling Project are supported by Strategic Partnering to Evaluate Cancer Signatures Grant No. U01-CA-114778 from the National Cancer Institute.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Patrick Brunhoeber, Ventana Medical Systems (C) Consultant or Advisory Role: None Stock Ownership: None Honoraria: None Research Funding: Kerry J. Savage, Hoffmann-La Roche (Roche Canada); Joseph M. Connors, Hoffmann-La Roche (Roche Canada); Lisa M. Rimsza, Ventana Medical Systems; Randy D. Gascoyne, Hoffmann-La Roche (Roche Canada) Expert Testimony: None Other Remuneration: Lisa M. Rimsza, Ventana Medical Systems
AUTHOR CONTRIBUTIONS
Conception and design: Nathalie A. Johnson, Randy D. Gascoyne
Provision of study materials or patients: Graham W. Slack, Joseph M. Connors, Wing C. Chan, Lisa M. Rimsza, Rita M. Braziel, James R. Cook, Raymond R. Tubbs, Andreas Rosenwald, Jan Delabie,Randy D. Gascoyne
Collection and assembly of data: Nathalie A. Johnson, Graham W. Slack, Kerry J. Savage, Joseph M. Connors, David W. Scott, King L. Tan, Christian Steidl, Laurie H. Sehn, Wing C. Chan, Javeed Iqbal, Paul N. Meyer, Georg Lenz, George Wright, Lisa M. Rimsza, Carlo Valentino, Patrick Brunhoeber, Thomas M. Grogan, Rita M. Braziel, James R. Cook, Raymond R. Tubbs, Dennis D. Weisenburger, Elias Campo, Andreas Rosenwald, German Ott, Jan Delabie, Elaine S. Jaffe, Louis M. Staudt, Randy D. Gascoyne
Data analysis and interpretation: Nathalie A. Johnson, Susana Ben-Neriah, Sanja Rogic, Christian Steidl, Christina Holcroft
Manuscript writing: All authors
Final approval of manuscript: All authors
Affiliations
Nathalie A. Johnson and Christina Holcroft, Jewish General Hospital, Montreal, Quebec; Graham W. Slack, Kerry J. Savage, Joseph M. Connors, Susana Ben-Neriah, Sanja Rogic, David W. Scott, King L. Tan, Christian Steidl, Laurie H. Sehn, and Randy D. Gascoyne, British Columbia Cancer Agency, Vancouver, British Columbia, Canada; Wing C. Chan, Javeed Iqbal, Paul N. Meyer, and Dennis D. Weisenburger, University of Nebraska Medical Center, Omaha, NE; Georg Lenz, Oncology and Tumorimmunology, Charité–Universitätsmedizin, Berlin; Andreas Rosenwald and German Ott, University of Würzburg, Würzburg; German Ott, Robert-Bosch-Krankenhaus and Dr Margarete Fischer-Bosch Institute for Clinical Pharmacology, Stuttgart, Germany; George Wright, Elaine S. Jaffe, and Louis M. Staudt, National Cancer Institute, National Institutes of Health, Bethesda, MD; Lisa M. Rimsza, Carlo Valentino, Patrick Brunhoeber, and Thomas M. Grogan, University of Arizona, Tucson, AZ; Rita M. Braziel, Oregon Health and Science University, Portland, OR; James R. Cook and Raymond R. Tubbs, Cleveland Clinic, Cleveland, OH; Elias Campo, Hospital Clinic, Barcelona, Spain; and Jan Delabie, Radiumhospitalet, Oslo, Norway.
REFERENCES
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008. pp. 265–266. [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi WW, Weisenburger DD, Greiner TC, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–5502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 6.Meyer PN, Fu K, Greiner TC, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–207. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutiérrez-García G, Cardesa-Salzmann T, Climent F, et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood. 2011;117:4836–4843. doi: 10.1182/blood-2010-12-322362. [DOI] [PubMed] [Google Scholar]
- 8.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rui L, Schmitz R, Ceribelli M, et al. Malignant pirates of the immune system. Nat Immunol. 2011;12:933–940. doi: 10.1038/ni.2094. [DOI] [PubMed] [Google Scholar]
- 10.Adams JM, Harris AW, Pinkert CA, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 11.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 12.Iqbal J, Neppalli VT, Wright G, et al. BCL2 expression is a prognostic marker for the activated B-cell–like type of diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:961–968. doi: 10.1200/JCO.2005.03.4264. [DOI] [PubMed] [Google Scholar]
- 13.Iqbal J, Sanger WG, Horsman DE, et al. BCL2 translocation defines a unique tumor subset within the germinal center B-cell-like diffuse large B-cell lymphoma. Am J Pathol. 2004;165:159–166. doi: 10.1016/s0002-9440(10)63284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savage KJ, Johnson NA, Ben-Neriah S, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–3537. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 15.Rimsza LM, Leblanc ML, Unger JM, et al. Gene expression predicts overall survival in paraffin-embedded tissues of diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2008;112:3425–3433. doi: 10.1182/blood-2008-02-137372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrans S, Crouch S, Smith A, et al. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J Clin Oncol. 2010;28:3360–3365. doi: 10.1200/JCO.2009.26.3947. [DOI] [PubMed] [Google Scholar]
- 17.Aukema SM, Siebert R, Schuuring E, et al. Double-hit B-cell lymphomas. Blood. 2011;117:2319–2331. doi: 10.1182/blood-2010-09-297879. [DOI] [PubMed] [Google Scholar]
- 18.Snuderl M, Kolman OK, Chen YB, et al. B-cell lymphomas with concurrent IGH-BCL2 and MYC rearrangements are aggressive neoplasms with clinical and pathologic features distinct from Burkitt lymphoma and diffuse large B-cell lymphoma. Am J Surg Pathol. 2010;34:327–340. doi: 10.1097/PAS.0b013e3181cd3aeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson NA, Savage KJ, Ludkovski O, et al. Lymphomas with concurrent BCL2 and MYC translocations: The critical factors associated with survival. Blood. 2009;114:2273–2279. doi: 10.1182/blood-2009-03-212191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurel B, Iwata T, Koh CM, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluk MJ, Chapuy B, Sinha P, et al. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PloS one. 2012;7:e33813. doi: 10.1371/journal.pone.0033813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green TM, Nielsen O, de Stricker K, et al. High levels of nuclear MYC protein predict the presence of MYC rearrangement in diffuse large B-cell lymphoma. Am J Surg Pathol. 2012;36:612–619. doi: 10.1097/PAS.0b013e318244e2ba. [DOI] [PubMed] [Google Scholar]
- 23.Iqbal J, Meyer PN, Smith LM, et al. BCL2 predicts survival in germinal center B-cell-like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clin Cancer Res. 2011;17:7785–7795. doi: 10.1158/1078-0432.CCR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 25.Wright G, Tan B, Rosenwald A, et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camp RL, Dolled-Filhart M, Rimm DL. X-Tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 27.Kerosuo L, Piltti K, Fox H, et al. Myc increases self-renewal in neural progenitor cells through Miz-1. J Cell Sci. 2008;121:3941–3950. doi: 10.1242/jcs.024802. [DOI] [PubMed] [Google Scholar]
- 28.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 31.Wierstra I, Alves J. The c-myc promoter: Still MysterY and challenge. Adv Cancer Res. 2008;99:113–333. doi: 10.1016/S0065-230X(07)99004-1. [DOI] [PubMed] [Google Scholar]
- 32.Choi PS, van Riggelen J, Gentles AJ, et al. Lymphomas that recur after MYC suppression continue to exhibit oncogene addiction. Proc Natl Acad Sci U S A. 2011;108:17432–17437. doi: 10.1073/pnas.1107303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vafa O, Wade M, Kern S, et al. C-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 34.Tzankov A, Zlobec I, Went P, et al. Prognostic immunophenotypic biomarker studies in diffuse large B cell lymphoma with special emphasis on rational determination of cut-off scores. Leuk Lymphoma. 2010;51:199–212. doi: 10.3109/10428190903370338. [DOI] [PubMed] [Google Scholar]
- 35.Salles G, de Jong D, Xie W, et al. Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: A study from the Lunenburg Lymphoma Biomarker Consortium. Blood. 2011;117:7070–7078. doi: 10.1182/blood-2011-04-345256. [DOI] [PubMed] [Google Scholar]
- 36.McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat Clin Pract Urol. 2005;2:416–422. [PubMed] [Google Scholar]
- 37.Klapper W, Kreuz M, Kohler CW, et al. Patient age at diagnosis is associated with the molecular characteristics of diffuse large B-cell lymphoma. Blood. 2012;119:1882–1887. doi: 10.1182/blood-2011-10-388470. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Hernandez AM, Shibata D, et al. BCL2 translocation frequency rises with age in humans. Proc Natl Acad Sci U S A. 1994;91:8910–8914. doi: 10.1073/pnas.91.19.8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jardin F. Classification of diffuse large B-cell lymphoma by immunohistochemistry demonstrates that elderly patients are more common in the non-GC subgroup and younger patients in the GC subgroup (reply) Haematologica. 2012;97:e4. doi: 10.3324/haematol.2011.057240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thunberg U, Amini RM, Linderoth J, et al. BCL2 expression in de novo diffuse large B-cell lymphoma partly reflects normal differences in age distribution. Br J Haematol. 2009;146:683–684. doi: 10.1111/j.1365-2141.2009.07762.x. [DOI] [PubMed] [Google Scholar]
- 41.Mounier N, Briere J, Gisselbrecht C, et al. Rituximab plus CHOP (R-CHOP) overcomes bcl-2–associated resistance to chemotherapy in elderly patients with diffuse large B-cell lymphoma (DLBCL) Blood. 2003;101:4279–4284. doi: 10.1182/blood-2002-11-3442. [DOI] [PubMed] [Google Scholar]
- 42.Winter JN, Weller EA, Horning SJ, et al. Prognostic significance of Bcl-6 protein expression in DLBCL treated with CHOP or R-CHOP: A prospective correlative study. Blood. 2006;107:4207–4213. doi: 10.1182/blood-2005-10-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitt CA, Lowe SW. Bcl-2 mediates chemoresistance in matched pairs of primary E(mu)-myc lymphomas in vivo. Blood Cells Mol Dis. 2001;27:206–216. doi: 10.1006/bcmd.2000.0372. [DOI] [PubMed] [Google Scholar]
- 44.Mead GM, Barrans SL, Qian W, et al. A prospective clinicopathologic study of dose-modified CODOX-M/IVAC in patients with sporadic Burkitt lymphoma defined using cytogenetic and immunophenotypic criteria (MRC/NCRI LY10 trial) Blood. 2008;112:2248–2260. doi: 10.1182/blood-2008-03-145128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 46.Tse C, Shoemaker AR, Adickes J, et al. ABT-263: A potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 47.Mason KD, Vandenberg CJ, Scott CL, et al. In vivo efficacy of the Bcl-2 antagonist ABT-737 against aggressive Myc-driven lymphomas. Proc Natl Acad Sci U S A. 2008;105:17961–17966. doi: 10.1073/pnas.0809957105. [DOI] [PMC free article] [PubMed] [Google Scholar]